Abstract
The authors report on the development of the Cancer Tissue Information Extraction System (caTIES)—an application that supports collaborative tissue banking and text mining by leveraging existing natural language processing methods and algorithms, grid communication and security frameworks, and query visualization methods. The system fills an important need for text-derived clinical data in translational research such as tissue-banking and clinical trials. The design of caTIES addresses three critical issues for informatics support of translational research: (1) federation of research data sources derived from clinical systems; (2) expressive graphical interfaces for concept-based text mining; and (3) regulatory and security model for supporting multi-center collaborative research. Implementation of the system at several Cancer Centers across the country is creating a potential network of caTIES repositories that could provide millions of de-identified clinical reports to users. The system provides an end-to-end application of medical natural language processing to support multi-institutional translational research programs.
Introduction
Translational research encompasses the dynamic cycle of laboratory studies, clinical studies and epidemiology in service of advancing clinical medicine. The development of informatics tools and infrastructure to support translational research has been the subject of several large-scale national projects.1–4 Translational sciences often require detailed clinical information, for example, to link molecular information to disease phenotype. Clinical expression of disease such as disease stage, disease severity, and response to treatment also provide crucial information for case identification and correlative studies. Unfortunately, almost all clinical outcome information of this kind is stored as unstructured or semi-structured free-text rather than coded, structured data. Natural language processing (NLP) has been used by numerous investigators to code and extract information from clinical documents.5–7 Informatics tools that build on NLP methods are needed to support clinical and translational research within a multi-institutional environment. However, few systems of this kind are currently in existence.
Background
Value of pathology information and tissue
Tissue specimens provide an extremely important resource for researchers and may be collected prospectively or retrospectively. Prospectively collected research specimens in tissue banks are usually only available in small numbers but may be highly annotated with manually extracted clinical information. In contrast, clinical remainders of tissues and fluids provide a much greater pool of possible translational research specimens, but are typically associated with few or no clinical annotations. Almost all information about these specimens must be derived from free-text clinical reports such as the surgical pathology report (SPR). Large volume archives of clinically derived tissues associated with information in the accompanying SPR could provide a rich resource for translational research, if the archive could be made searchable in a manner compliant with the Health Insurance Portability and Accountability Act (HIPAA).8
System history
The caTIES system evolved from a text processing system that we originally developed for the Shared Pathology Informatics Network (SPIN), which proposed to develop a network of institutions sharing de-identified data and tissue through coded SPR.9 Although the vision of the SPIN network was not realized beyond a prototype linking the four contributing institutions, the goal of the project to enable translational research across institutions fostered foundational research in this area. The caTIES project continues this goal but extends the previous system by (1) integrating with the Cancer Biomedical Informatics Grid (caBIG)1 2 architecture, common object representation and controlled vocabulary, (2) providing graphical interfaces and methods for query based retrieval and selection of cases, and (3) implementing a regulatory policy for federated data and tissue sharing through an ‘honest broker’ mechanism.
Design objectives
Establish a federation of de-identified, concept-coded clinical text archives built on a grid architecture
Data sharing should use the federated model, enabling local authority over management of data. Data must be stripped of all 18 required patient identifiers, to ensure compliance with HIPAA ‘safe-harbor’ practices. To improve sensitivity and specificity of retrieval, documents must be preprocessed to create concept codes for present and absent diseases, pathologic findings, anatomic locations, surgical procedures and other important medical concepts.
Support exploration of large document datasets using concept-based query and result visualization methods
The interface must balance the need for a simple and obvious concept-based search capability in most cases with greater expressivity in some cases. For some use cases, researchers must be able to find tissue and documents based on complex Boolean logic in addition to temporal relationships between documents.
Enforce a regulatory model of clinical data use for translational research based on Institutional Review Board protocols and honest brokers
Previous efforts towards development of an inter-institutional network of document archives for research purposes have used an open ‘airport’ model, requiring that institutions agree to provide data to all interested users across all institutions.9 In contrast, we considered it essential to (1) bind all requests for data to a local Institutional Review Board (IRB) protocol, and (2) enable institutions to decide to supply data to outside researchers on a study-by-study basis, Additionally, we sought to (3) provide sufficient policies and procedures regarding identity provisioning and auditing to mitigate the risk of sharing de-identified data, and (4) create a rigorous security infrastructure to promote trust among organizations.
Facilitate collaborative translational research across institutions
The potential benefits of sharing data across organizations are even greater when researchers across organizations can work together to manage datasets of documents and tissues. The system should enable such virtual datasets unencumbered by organizational boundaries.
Promote easy adoption and customization by using open source frameworks, tools, vocabularies, and algorithms
Informatics tools for translational research are typically deployed in resource limited environments. To ease adoption, customization, and long term maintenance, the system should be built on open-source frameworks, tools and algorithms wherever possible, and use freely available vocabularies for concept-coding.
Enable interoperability within a larger community of research systems
The system should function within the context of caBIG to promote interoperability between caTIES and other cancer research systems.
System description
caTIES is a suite of clients, services, and datastores connected by and implemented on caBIG architectural blueprints. The system establishes a set of caBIG services that sufficiently govern caTIES behavior. A caTIES service network may function autonomously or may connect to outside service subscribers, such as caBIG.
Datastores
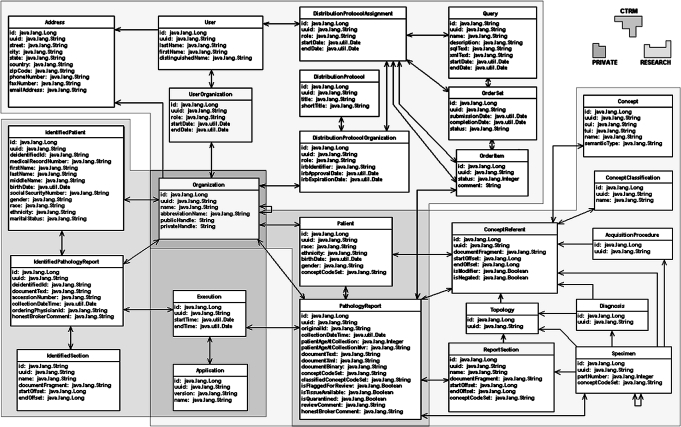
caTIES establishes a single logical data model sufficient to house all caTIES data (figure 1). At each datastore, some parts of the schema may remain unpopulated but the schema is deployed as a whole. caTIES uses three primary datastores: (1) the private datastore, (2) the research datastore, and (3) the Collaborative Tissue Resource Manager (CTRM) datastore (figure 2). Each organization hosts one private datastore and one research datastore. In the typical configuration, the private and research datastores reside on different machines. The caTIES network hosts a single publicly accessible CTRM for use by all organizations.
Figure 1.
Object model for private, de-identified and CTRM datastores.
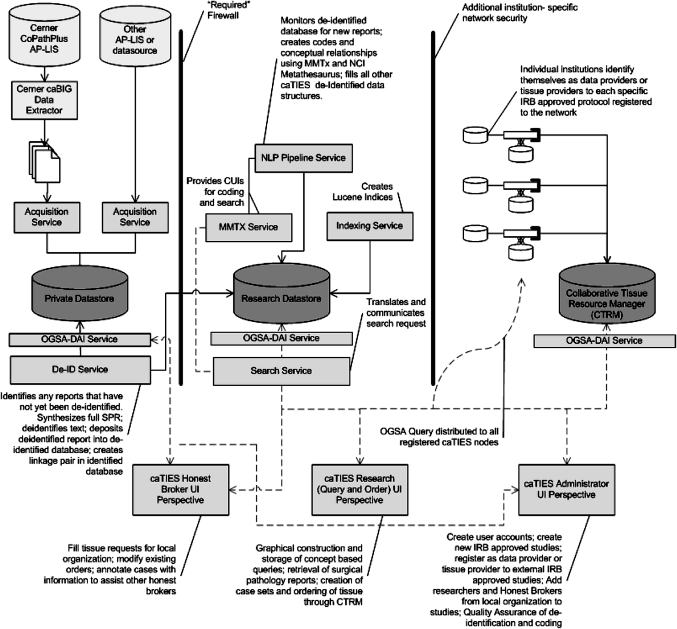
Figure 2.
Information architecture showing suite of services, datastores and clients.
The private datastore is the recipient of data derived from clinical systems such as the Anatomic Pathology Laboratory Information System (AP-LIS). It contains identified free text as well as dates, patient medical record numbers and specimen accession numbers. It is only available for access by honest brokers within the organization hosting the specific private datastore.
The research datastore contains de-identified free text reports, along with other unrestricted information such as gender, and age if less than 90. The research datastore is also the target of the NLP Pipeline Service, which creates and stores conceptual annotations with each free-text report. The schema of this database includes the Consented High Performance Index and Retrieval of Pathology Specimens (CHIRPS) SPIN submission schema9 permitting interoperability between caTIES and SPIN.
The CTRM datastore manages the collaborative construction and manipulation of tissue studies. Researchers build tissue order sets and electronically interact with honest brokers at external organizations. Honest brokers are disinterested third-parties, who are responsible for determining availability of biospecimens, filling orders for biospecimens, and providing additional de-identified outcomes data.
caTIES uses hibernate object relational mapping technology, providing a flexible façade for multi-platform relational database management systems (RDBMS) access.
Data preparation services
The data preparation phase runs as a series of operating system-based services that transform data from free-text documents stored in clinical systems to concept-annotated de-identified documents stored in the relational database. caTIES services run continually, release machine resources when not in use, and revive on machine restart.
Data preparation encompasses four tasks, performed by four corresponding services, in the following order: (1) acquisition, (2) de-identification, (3) concept-coding, and (4) indexing.
Acquisition services
Data may be transferred from AP-LIS or document repositories using a variety of acquisition services. Because of the heterogeneity of clinical systems, caTIES adopters are tasked with populating the private datastore before starting the caTIES services. Adopters may use existing tools provided by vendors, or may write their own data transfer mechanisms, targeting the caTIES logical schema.
To assist adopters, we currently support a data transfer mechanism based on a Cerner data warehouse product that extracts data from any of the three Cerner AP-LIS systems. Two of the four institutions collaborating in the caBIG caTIES pilot implemented this method of data transfer. The third institution wrote its own Health Level 7 (HL7) interface, which directly utilizes the institutional HL7 router feed. The fourth institution created database specific queries to upload identified data. Additional AP-LIS specific acquisition services are being considered for future development.
De-identification
The caTIES de-identification service removes the 18 identifiers required by HIPAA, and creates and stores randomly generated Universally Unique Identifiers linked to the original identifiers, to support a method for re-identification that is, permissible under HIPAA. At our institution this functionality is achieved using DeID, a commercially available de-identification system. However, caTIES is designed to permit easy uncoupling of the default de-identifier. Adopters can use any system providing similar functionality by implementing a simple Java interface. We have benchmarked this capability using the Harvard scrubber.10 The choice of a default commercial system was motivated by the need for a well-established, formally evaluated method for de-identification.11 12 As newer systems for de-identification mature, we expect that open-source de-identification will replace the default commercial system.
Concept coding
The caTIES coding pipeline service (table 1) produces conceptual annotations on free-text documents. Coding is performed by a sequence of modular processing resources generally applied in the following order:
Resetter: clears document, deletes existing annotations.
Tokeniser: tokenizes words, numbers, punctuation and spaces.
Chunker: parses reports into sections, parts, sentences, and phrases.
Spell-checker (excluded by default): identifies erroneous spelling and suggests frequency based correction.13
RegEx: annotates a pre-defined set of attribute and value pairs such as tumor grade and stage.
Vocabulary concept tagger: annotates fragment of free text to associated concept from a controlled terminology using MetaMap Transfer (MMTx).14
Semantic-type filter: removes concepts associated with unwanted semantic types.
NegEx: implements the NegEx negation detection algorithm to tag explicitly negated concepts.15
Semantic-type categorization: extracts body parts, procedures, diseases and findings based on vocabulary semantic types.
Physical model deducer: uses rudimentary nearest neighbor discourse level reasoning to arrange named entities into a decomposition and topologic hierarchy.
Extractor: converts the hierarchy to valid Extensible Markup Language (XML) as defined by the CHIRPS XML schema definition.9
Table 1.
caTIES coding pipeline service components
| Order | Procesing resource | Function | Resource type | Authors | GATE mechanism | Imports |
| 1 | Resetter | Clears document of existing annotations | Off-the-shelf GATE component | Sheffield | Java | None |
| 2 | Tokeniser | Finds words, numbers, punctuation and spaces | Off-the-shelf GATE component | Sheffield | Java, JAPE | None |
| 3 | Spell Checker | Makes unsupervised spelling correction based on best guess. | Custom component | U Pittsburgh, Harvard | Java | Gspell, Harvard Frequency Filter |
| 4 | Case Insensitive Gazatteer | Finds words from lists, stopwords, pre, post and pseudo negation tags | Custom component | U Pittsburgh | Gazetteer | NegEx Terms |
| 5 | Case Sensitive Gazetteer | Site specific section headers | Custom component | U Pittsburgh | Gazetteer | None |
| 6 | Chunker | Parses reports into sections, parts, sentences, and phrases, | Custom component | U Pittsburgh | JAPE | None |
| 7 | RegEx | Uses regular expressions to find attribute value pairs | Custom component | Harvard | Java | None |
| 8 | MMTx Concept Coder | Annotates fragment of free text to associated concept from controlled terminology using MMTx with the NCI Metathesaurus as a custom datasource, and based on vocabulary sources defined by the user | Custom component | U Pittsburgh, Regenstrief | Java | MMTx, NCI Metathesaurus |
| 9 | Concept Filter | Filters unwanted semantic types | Custom component | U Pittsburgh, Regenstrief | JAPE | None |
| 10 | NegEx | Implements NegEx negation detection algorithm to tag explicitly negated concepts | Custom component | U Pittsburgh | JAPE | None |
| 11 | Concept Categorizer | Extracts organs, procedures, diseases based on semantic type | Custom component | U Pittsburgh, Regenstrief | JAPE | None |
| 12 | Physical Model Deducer | Employs rudimentary discourse level reasoning to infers compositional topology of concepts | Custom component | U Pittsburgh | Java | None |
| 13 | CHIRPS Extractor | Populates the CHIRPS schema sufficient to populate a SPIN or caTIES node | Custom component | U Pittsburgh | Java | None |
GATE, General Architecture for Text Engineering; JAPE, Java Annotations Pattern Engine; MMTx, MetaMap Transfer; NCI, National Cancer Institute.
The core language-processing functionality of the system is achieved using the open-source General Architecture for Text Engineering platform.16 Implementation details of the coding pipeline service are provided in table 1.
For concept coding, caTIES uses MMTx pre-configured with the National Cancer Institute (NCI) Metathesaurus.17 Use of the NCI Metathesaurus is a condition of participation in the caBIG. However, users outside of caBIG may choose any other vocabulary or vocabulary subset that can be used with MMTx, by configuring MMTx differently prior to installation of caTIES.
caTIES coding services have been designed to run in parallel to take advantage of multiple processors available at an organization, greatly reducing the total time for coding massive document sets.
Indexing
The caTIES indexing service creates a text search engine index for fast access to documents based on the characteristics of the document text and conceptual codes. This index must fulfill the requirement of fast substring searching independent of an underlying RDBMS. CaTIES uses Lucene 2.3 for its information retrieval engine.18
The caTIES SPR index is streamlined for temporally constrained, patient level query by mapping the composite primary key of patient unique identifier and SPR collection date and time to the range of long numbers. This mechanism requires additional bookkeeping time and space in the accompanying RDBMS but it is otherwise transparent to the user.
In addition to the conceptual document index, caTIES maintains an ancestor index that associates NCI Thesaurus concepts with their ancestry. Here ancestry is defined to be all concepts in the transitive closure along the reverse isa-relationship of the NCI Thesaurus. The ancestor index provides ancestors both at SPR index time and later during client query formulation.
Information retrieval services
For information retrieval across organizations, caTIES uses a grid service architecture based on the Open Grid Service Architecture (OGSA).19 Grid services are stateful webservices that provide more functionality than the basic webservices they are built upon. The caTIES client communicates with three services to search for and retrieve documents. All caTIES services are implemented using the Globus Toolkit Webservices Resource Framework (GT4)—a reference implementation of the OGSA specification.
MMTx service
The caTIES MMTx service derives conceptual search criteria on the client side, based on a user query string. Users may modify concepts interactively.
Search service
The caTIES search service communicates the search criteria (including Boolean logic, temporal relationships, and concepts) to the server. On the server side this request is converted from SPIN query XML to Lucene query language. Hits from the search are organized into a response payload that consists of report unique identifiers and some report header information. Subsequent drill down into report specifics occurs on future server requests.
OGSA-data access and integration (DAI) service
The caTIES OGSA-DAI service provides a Web services conduit for basic Structured Query Language (SQL) Data Manipulation Language and Description Definition Language (DDL) interactions with a data source. OGSA-DAI is an extension to the core functionality of the Globus Toolkit, that provides access to a wide range of databases including MySQL, DB2, Oracle, Postgres, SQL Server, and XIndice, as well as indexed text files. Thus, caTIES may be implemented with any of these database management systems.
User interface, query and results visualization
The caTIES user interface (UI) is composed of four role-based perspectives: researcher, preliminary user, administrator, and honest broker. At login, the caTIES client loads the appropriate perspective for the user. The user can switch between perspectives if she is registered with more than one role in the system.
The caTIES client is a Java application deployed using Java WebStart. Open source libraries used in the construction of the client application include (1) JGraph library20 for displaying the Diagram query view, (2) GlazedLists library21 for displaying the results table and (3) JFreeChart library22 for constructing pie/bar charts for the results.
Researcher perspective
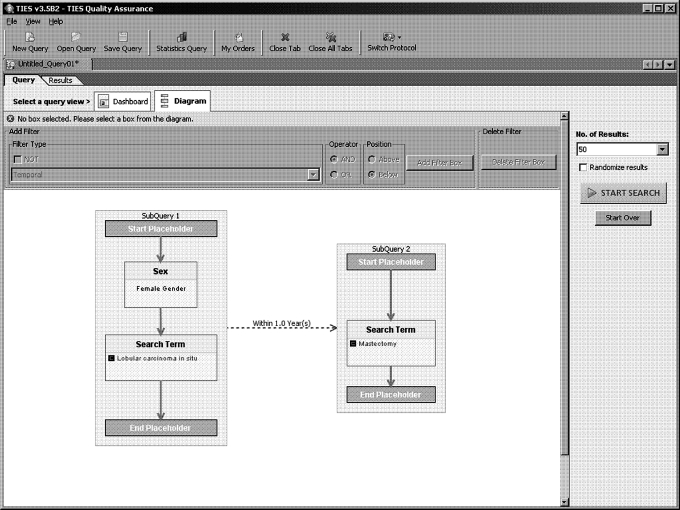
The researcher perspective supports query construction and execution, and order management for the distribution protocol. caTIES supports both query by text and query by concept. Users can constrain queries by demographic variables such as age and gender. Standard Boolean constructs including AND, OR and NOT can be used to combine all of the above constraints. Additionally, users can formulate temporal queries based on the timing of diagnostic reports. An example of a temporal query is: “Find all females who had Lobular Carcinoma in Situ, followed by mastectomy within 1 year” (figure 3).
Figure 3.
User interface—diagram method for query construction.
Queries can be modeled using two views: dashboard and diagram. The dashboard view allows for simple text-box driven query construction. The diagram view permits more expressive nested Boolean query construction using a filter-flow metaphor (figure 3). Views are synchronized so that a query in the diagram view always matches the query in the dashboard view. However, since the diagram view is more expressive, not all queries modeled in the diagram view can be viewed in the dashboard view.
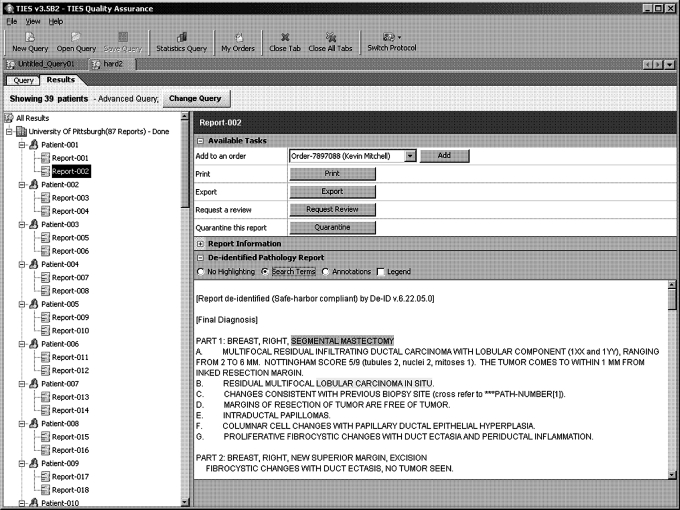
Results are visualized in tabular and tree format. In the tree format, they are hierarchically organized by owning organization, and then by patient. Selecting a report in this tree provides detailed document information and annotations (figure 4). The tabular view lists all reports by key criteria (eg, age, gender, concepts) and can be reorganized by sorting.
Figure 4.
User interface—results visualization.
Preliminary user
The preliminary user perspective is identical to the researcher perspective except that it returns only aggregate level data (histograms and pie charts). No record level data can be obtained. Preliminary users typically obtain access without IRB approval, to collect data preparatory to research.
Administrator perspective
The Administrator UI perspective is used by system administrators and honest brokers to accomplish administrative functions. It supports user account creation, registration of new IRB approved studies, registration of the institution as data provider or tissue provider to external IRB approved studies, and addition of researchers and honest brokers to studies from the administrator's local organization. In addition, it supports quality assurance of de-identification and concept coding. Reports flagged by users for potential errors in de-identification or coding may be reviewed by honest brokers using the Quality Assurance tab. Documents flagged for de-identification errors are quarantined and unavailable for subsequent use until the error is corrected or released.
Honest broker perspective
The honest broker UI perspective enables impartial individuals such as tissue bankers and cancer registrars to assist researchers in filling requests for tissue or further clinical data. On login, the honest broker perspective provides a queue of unfilled requests. Honest brokers can view data from the private (identified) database of their own institution only, in order to fill orders or provide further data in a de-identified manner.
Collaborative study management
caTIES uses a protocol-based model for collaborative research across a network of organizations. The paradigm is based on a fundamental assumption that exchange of de-identified data and/or tissue between any repository and any researcher requires two IRB protocols—(1) by the organization establishing a de-identified repository for providing data or tissue to one or more researchers, and (2) by the researcher for searching a de-identified repository established at one or more organizations. Differences among IRBs in regulation of data-sharing and materials transfer create the requirement for maximal local control over participation. Thus, organizations who host caTIES nodes may agree to provide data or tissue on external protocols on a study-by-study basis. In previous work, we have validated these assumptions in interviews with IRB and regulatory officials at six US cancer centers.23 The model of privacy, security, and collaboration needs for a research grid derived from this interview study differs dramatically from the open (‘airport’) model of collaboration that has been previously used.9
Access to caTIES must occur within the context of a valid (time-sensitive) approved IRB protocol. All users are bound to one or more IRB approved protocols at the time of user registration. When a protocol is registered by an administrator for a researcher seeking to obtain data or tissue, the administrator registers the home institution as a Data Consumer or Tissue Consumer respectively. The home institution becomes a Data Provider to this local IRB protocol automatically. And if the administrator registers the organization as a Tissue Consumer, then the home organization automatically becomes a Tissue Provider to this local IRB protocol.
Once the protocol is registered, other caTIES nodes may agree to participate on this study protocol. Honest brokers must determine whether a given protocol registered at an external organization meets the constraints of the repository IRB protocol for sharing data that has been approved at the providing organization. In previous work, we determined that many organizations may require only assurance that an external researcher has appropriate credentials and IRB protocol (which can be established at the time of provisioning), but that requirements for data sharing may be more stringent at some organizations.23 The approach we developed enables participation within the bounds of local regulatory requirements.
Within the constraints of this model, caTIES has many features that support collaborative research between organizations hosting caTIES nodes. For example, researchers from different institutions can be a part of the same study protocol, and thus they may create queries and order sets that can be viewed and edited by other researchers on the protocol who may reside at different institutions.
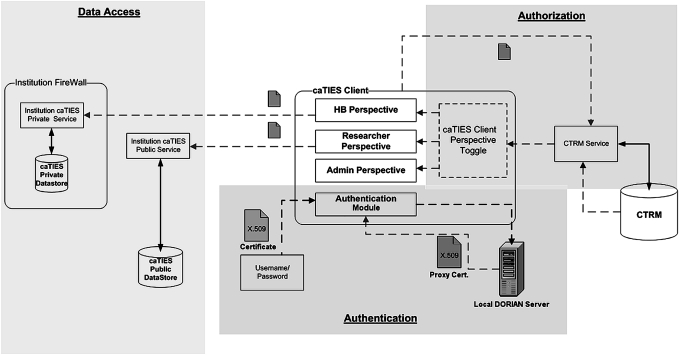
Security architecture
caTIES uses a series of security enforcement layers (figure 5) to lock out unauthorized resource access. Security enforcement layers include:
Figure 5.
Security architecture showing authentication, authorization and access layer.
Physical layer
Physical security of data is supported by the complete separation of de-identified and identified data (which reside on different machines in the typical configuration).
Network layer
At the network layer, caTIES uses the security model of the Globus Toolkit and OGSA-DAI (figure 5). The Globus Toolkit uses Grid Security Infrastructure (GSI)24 for enabling secure authentication and communication over an open network. GSI provides a number of useful services for Grids, including mutual authentication and single sign on. GSI is based on public key encryption, X.509 certificates, and the Secure Sockets Layer communication protocol. Extensions to these standards have been added for single sign on and delegation. The Globus Toolkit's implementation of the GSI adheres to the Generic Security Service application programming interface,25 which is a standard interface for security systems promoted by the Internet Engineering Task Force.
All caTIES Grid Services are configured as secure grid services. CaTIES secure grid services authenticate and authorize based on a local Dorian installation. Dorian26 provides the caGRID Identity Provider and Identify Federation Service interfaces. Authentication is dependent on a valid Security Assertion Markup Language assertion, and generates a proxy certificate or grid identity. Entries for each authorized user are also stored in the grid maps at each caTIES node, providing an additional level of security.
Application layer
At the application layer, caTIES maintains security mechanisms for restricting access based on the user's authorization attributes (figure 5). The caTIES CTRM client application dictates authorization using information in its CTRM datastore and embedded business logic. Users are granted restricted access based on their authorized resource set.
Database layer
At the database layer, caTIES restricts access to data using RDBMS standard mechanisms (figure 5). RDBMS roles and their table access privileges mirror the high level authorization roles of caTIES: administrator, honest broker, researcher, and preliminary user.
Security policy
Sharing of data between organizations requires agreements, policies and standard operating procedures among participants with regard to the adequacy of de-identification, provisioning of credentials, requirements for IRB review, and auditing of data and protocols. The caTIES project has developed a set of human processes and policies to support the functioning of a caTIES network, which are publicly available.27 The security policy was derived from an in depth interview-based study which used problem scenarios to elicit security and privacy requirements.23
Deployment and installation
The caTIES installer provides a common front end for installing and configuring all caTIES services and datastores. The caTIES website provides access to currently supported releases of caTIES, installation, administration and user manuals, demonstration videos, and other information to assist new users. The software is available on SourceForge28 which also hosts the caTIES user forums.29 caTIES is released under the caBIG open source license.
Status report
Evaluation
Previous evaluations of the early components of the caTIES pipeline have already been reported.30 31 The current evaluation focused on determining the deployed performance of the system using (1) studies of query response timing, and (2) metrics of basic information retrieval, using a set of 30 standard queries of clinical significance (tables 2a and 2b). Queries were invented for three general categories of complexity. Simple queries had no more than two concepts and no temporal relationships or negations. Moderate complexity queries had more than two concepts or negated concepts but no temporal relationships. Complex queries had more than two concepts or negated concepts with temporal relationships. We first tested the query set to determine the length of time to query completion in the deployed system at University of Pittsburgh. At the time of query response testing, the Pittsburgh repository contained more than 1.4 million documents, and was deployed on an IBM HS22 Blade Server with the following specifications: 2×Intel Xeon Processor X5550 (Quad Core), 24 GB Memory, 2×73 GB 15K SAS Drives (mirrored) internal disks, IBM DS3400 300 GB disk storage, 15K Serial Attached SCSI (SAS) drives, running VMWare vSphere 4.0 Standard Edition.
Table 2a.
Response time and performance metrics, low and medium complexity queries
| Response time over three retrievals | Performance metrics | |||||||||||
| # | Complexity | Query | Number reports retrieved | Mean time to first results (sec) | SD | Mean time to all results (sec) | SD | Number of Reports or Report Sets (complex) classified | Agreement | TP | FP | Precision |
| 1 | Low | Men, 60–80 with prostatic adenocarcinoma on prostatectomy | 1792 | 1.08 | 0.62 | 4.63 | 1.92 | 50 | 0.98 | 49 | 1 | 0.98 |
| 2 | Low | Women, 30–50 with atypical endometrial hyperplasia | 792 | 0.70 | 0.19 | 0.70 | 0.19 | 33 | 1.00 | 33 | 0 | 1.00 |
| 3 | Low | Patients, 20–50 with phaeochromocytoma | 54 | 0.95 | 0.31 | 0.96 | 0.31 | 50 | 0.98 | 49 | 1 | 0.98 |
| 4 | Low | Patients with hemangiosarcoma of scalp | 17 | 0.49 | 0.13 | 0.49 | 0.13 | 17 | 1.00 | 17 | 0 | 1.00 |
| 5 | Low | Patients 10–30, with cystosarcoma phylloides | 18 | 0.59 | 0.07 | 0.59 | 0.07 | 18 | 0.94 | 16 | 2 | 0.89 |
| 6 | Low | Patients with superficial spreading melanoma, metastatic | 5 | 0.46 | 0.08 | 0.46 | 0.08 | 5 | 1.00 | 5 | 0 | 1.00 |
| 7 | Low | Patients with medullary carcinoma in thyroid gland | 27 | 0.59 | 0.26 | 0.60 | 0.26 | 27 | 0.96 | 26 | 1 | 0.96 |
| 8 | Low | Patients with adenocarcinoma in brain | 156 | 0.65 | 0.33 | 0.89 | 0.44 | 50 | 1.00 | 50 | 0 | 1.00 |
| 9 | Low | Men with invasive ductal carcinoma of breast | 29 | 0.53 | 0.15 | 0.53 | 0.15 | 29 | 1.00 | 29 | 0 | 1.00 |
| 10 | Low | Patients, >60 with Hodgkins disease | 549 | 0.64 | 0.17 | 0.84 | 0.22 | 50 | 0.94 | 34 | 16 | 0.68 |
| All Low complexity queries | 3439 | 0.67 | 0.20 | 1.07 | 1.26 | 329 | 0.98 | 308 | 21 | 0.94 | ||
| 11 | Moderate | Patients with prostatic hypertrophy and PIN on prostate biopsy | 17 | 0.67 | 0.23 | 0.67 | 0.23 | 17 | 1.00 | 17 | 0 | 1.00 |
| 12 | Moderate | Patients with either scar or radial scar, and intraductal papilloma on mastectomy or excisional biopsy of breast | 680 | 0.74 | 0.28 | 2.36 | 0.46 | 50 | 1.00 | 50 | 0 | 1.00 |
| 13 | Moderate | Patients, 40–60 with tubulovillous adenoma and adenocarcinoma in colon or rectum | 220 | 0.68 | 0.18 | 1.34 | 0.17 | 50 | 0.94 | 49 | 1 | 0.98 |
| 14 | Moderate | Patients with lung fibrosis secondary to systemic lupus | 4 | 0.47 | 0.09 | 0.47 | 0.09 | 4 | 1.00 | 4 | 0 | 1.00 |
| 15 | Moderate | Patients with adenomyosis on endocervical biopsy or hysterectomy | 1069 | 0.59 | 0.24 | 4.15 | 1.72 | 50 | 1.00 | 50 | 0 | 1.00 |
| 16 | Moderate | Patients with prostatic adenocarcinoma, and PIN but no perineural invasion | 39 | 0.53 | 0.12 | 0.54 | 0.12 | 39 | 0.97 | 38 | 1 | 0.97 |
| 17 | Moderate | Patients with papillary carcinoma of thyroid in the setting of multi-nodular goiter | 60 | 0.50 | 0.06 | 0.51 | 0.06 | 50 | 1.00 | 50 | 0 | 1.00 |
| 18 | Moderate | Patients with osteosarcoma of femur or tibula showing tumor necrosis | 9 | 0.52 | 0.16 | 0.52 | 0.16 | 9 | 0.78 | 7 | 2 | 0.78 |
| 19 | Moderate | Patients with lobular carcinoma in situ and microcalcifications undergoing a procedure in which a sentinel lymph node was biopsied | 87 | 0.66 | 0.17 | 0.66 | 0.17 | 50 | 0.96 | 48 | 2 | 0.96 |
| 20 | Moderate | Patients 40–60 with cirrhosis or fibrosis and hepatocellular carcinoma on liver biopsy | 85 | 0.74 | 0.21 | 0.75 | 0.21 | 50 | 0.88 | 43 | 7 | 0.86 |
| All Moderate Complexity Queries | 2270 | 0.61 | 0.10 | 1.20 | 1.19 | 369 | 0.96 | 356 | 13 | 0.96 | ||
FP, False Positive; PIN, prostatic intraepithelial neoplasia; TP, True Positive.
Table 2b.
Response time and performance metrics, high complexity queries
| Response time over three retrievals | Performance metrics | |||||||||||
| # | Complexity | Query | Number reports retrieved | Mean time to first results (sec) | SD | Mean time to all results (sec) | SD | Number of Reports or Report Sets (complex) classified | Agreement | TP | FP | Precision |
| 21 | High | Patients with sclerosing cholangitis on liver biopsy who have also had ulcerative colitis on another procedure | 4 | 15.74 | 1.15 | 15.75 | 1.16 | 2 | 0.50 | 0 | 2 | 0.00 |
| 22 | High | Women diagnosed with LCIS who had a subsequent mastectomy within 1 year | 1034 | 18.87 | 6.64 | 36.24 | 31.59 | 39 | 0.97 | 36 | 3 | 0.92 |
| 23 | High | Patients with dysplastic nevi who were diagnosed with melanoma after an interval of at least 1 year | 148 | 29.13 | 23.98 | 29.14 | 23.97 | 33 | 0.88 | 25 | 8 | 0.76 |
| 24 | High | Patients with diagnosis GERD or Barrett's esophagus who later had esophagectomy showing adenocarcinoma | 136 | 30.91 | 25.73 | 33.35 | 29.92 | 28 | 1.00 | 28 | 0 | 1.00 |
| 25 | High | Men with anaplastic astrocytoma who later developed glioblastoma multiforme | 15 | 16.00 | 2.18 | 16.00 | 2.18 | 7 | 1.00 | 7 | 0 | 1.00 |
| 26 | High | Patients with both schwannomas and meningiomas | 10 | 16.55 | 2.79 | 16.55 | 2.79 | 4 | 1.00 | 4 | 0 | 1.00 |
| 27 | High | Patients with tissue documented Berger's disease who later underwent kidney transplantation | 6 | 16.21 | 2.04 | 16.21 | 2.04 | 3 | 0.33 | 1 | 2 | 0.33 |
| 28 | High | Patients with DFSP and a second procedure for local extension or recurrence within 3 months. | 19 | 20.19 | 8.90 | 20.19 | 8.90 | 9 | 1.00 | 8 | 1 | 0.89 |
| 29 | High | Patients with colonic adenocarcinoma who also have had Invasive ductal carcinoma of breast | 24 | 24.85 | 16.17 | 24.85 | 16.17 | 11 | 0.91 | 10 | 1 | 0.91 |
| 30 | High | Patients with renal carcinoma in kidney tissue who also have lung tissue with metastatic renal cell carcinoma | 59 | 16.61 | 2.95 | 16.62 | 2.94 | 26 | 0.88 | 23 | 3 | 0.88 |
| All high complexity queries | 1455 | 20.50 | 5.74 | 22.49 | 7.88 | 162 | 0.93 | 142 | 20 | 0.88 | ||
| All queries | 7164 | 7.26 | 10.15 | 8.25 | 11.29 | 860 | 0.96 | 806 | 54 | 0.94 | ||
DFSP, dermatofibroma sarcoma protuberans; FP, False Positive; GERD, Gastroesophageal Reflux Disease; LCIS, Lobular Carcinoma in Situ; TP, True Positive.
Results show mean and SD for three attempts to account for network traffic fluctuation (table 2). For simple and moderate queries, caTIES responds in sub-second time. Temporal queries do take substantially longer but still respond within 20 s on average and within 1 min in almost all cases.
Next, we tested the information retrieval aspects of the system. In this study, we determined only the precision of the system (table 2). Two authors of this manuscript, a pathologist (RC) and a knowledge engineer with expertise in pathology (MC) separately coded results of all 30 queries as true positive or false positive. All reports (or report sets for complex queries) were coded unless more than 50 reports or report sets were returned, in which case the judges coded only the first 50 reports or report sets returned by the system. Judges achieved an overall inter-rater reliability of 96% agreement. Results show high precision for simple and moderate queries (average 0.94–0.96), which drops slightly for the more complex temporal queries (average 0.88). Performance is expected to degrade for these queries since coding a true positive for temporal queries requires that both reports returned are true positive for each of the two clauses in the query.
Error analysis (table 3) was performed on all reports marked as false positive by either judge. A total of 73 cases were analyzed. The most common errors related to retrieval of documents in which the search concept was erroneously coded by the system because a substring of the more complex concept was recognized by MMTx. In many cases, these errors occur because the more complex concepts are post-coordinated concepts (eg, “post-mastectomy scar”) and are not represented in the vocabulary. Another common source of errors related to erroneous clinical diagnoses—specimens are sometimes labeled with a clinical diagnosis, which is subsequently corrected by pathological examination. Diagnostic uncertainty was a third cause of error, and is a common problem in retrieving clinical documents. Other error categories observed (in decreasing frequency) include: initials incorrectly coded as abbreviations, concepts identified in the report that are in fact historical, conceptual relationships not properly scoped, and errors in negation detection. Of note, the majority of observed errors could be eliminated by (1) limiting search to specific report sections of the report and by (2) extending the negation detection to include newer algorithms which account for uncertainty. Future versions of the system will include these modifications.
Table 3.
Analysis of errors
| Error type | No of errors | % Total errors |
| Substring of more complex concept incorrectly coded (eg, report for “post-mastectomy scar” retrieved for query “mastectomy”) | 17 | 23.29% |
| Information provided in clinical diagnosis is incorrect or incongruent with pathological diagnosis (eg, report describing specimen labeled by clinicians as “DFSP” is retrieved for query “DFSP” even though pathologic diagnosis was not DFSP) | 15 | 20.55% |
| Finding or diagnosis is expressed as uncertain (eg, “cannot exclude”) | 14 | 19.18% |
| Initials in report are misinterpreted as abbreviation and thus miscoded (eg, report with initials “HL” retrieved for query “Hodgkin's Lymphoma”) | 11 | 15.07% |
| Concepts present in report are historical (eg, report describing “previous history of renal cell carcinoma” retrieved for query “renal cell carcinoma”) | 8 | 10.96% |
| Correct concepts but incorrect conceptual relationships (eg, report containing “prostate cancer without perineural invasion, and urothelial cancer with perinerual invasion” is returned for query “prostate cancer with perineural invasion” | 6 | 8.22% |
| Negated concepts incorrectly identified as present (eg, report containing “neither prostatic intraepithelial carcinoma (PIN) nor carcinoma is seen” is returned for query containing “neither prostatic intraepithelial carcinoma (PIN)” | 2 | 2.74% |
| Total no of errors | 73 | 100% |
Deployment
At University of Pittsburgh, caTIES is deployed as a production system, supported by the information systems help-desk and applications trainer. Deployment of caTIES at our institution is governed by the Health Sciences Tissue Bank which oversees the policy aspects of the system, using existing human honest broker systems approved by our Institutional Review Board. The system has met the security and privacy requirements of the University of Pittsburgh Medical Center (UPMC) to operate as a ‘UMPC approved clinical system’.
CaTIES has also been deployed at three other caBIG funded institutions including University of Pennsylvania, Thomas Jefferson University, and Washington University St Louis as part the caBIG caTIES pilot. Additionally, caTIES has been deployed by a Midwestern stand-alone cancer center, a Midwestern university affiliated cancer center, and by members of a Western US health consortium, with minimal assistance from the developers. A growing number of other institutions are evaluating and deploying the system without our assistance.
Discussion
The caTIES system provides an example of an end-to-end medical NLP application that could be used to support multi-institutional collaboration and translational science. The system has a strong policy foundation, expressive user interface, and builds on existing open-source tools and vocabularies. Results of our studies show that it retrieves documents and document-sets quickly, and operates with high precision.
Lessons learned
The successful deployment of this translational research system required that we to adopt the security and privacy practices of the more highly regulated health information environment. Acceptance of this repository at our institution took over 1 year and required substantial interaction with IRB, hospital privacy and security officers, honest broker services, and the health sciences tissue bank. The use of a data stewardship model was an important step in reaching consensus among stakeholders. Despite the fact that the data was de-identified, we determined that the system must achieve the same security status as any clinical system in our environment. Automated de-identification is not risk-free and the potential for unregulated use of data must be minimized.
Despite the successful deployment of the caTIES system across multiple individual institutions, including our own, one key functionality of the system has not been used beyond demonstration purposes—no institutions are currently using the grid communications system to support ongoing, multi-institutional data sharing. To reach this goal, we must have a trust fabric with suitable policies and processes for sharing data and tissue. The policy groundwork for such a federation has already been established for our system,23 and more general frameworks and national policies are emerging.32 33 But the practical implementation of such a data sharing network will likely require a great deal more work even after such frameworks become mature, available and widely accepted.
Future work
Future versions of the open-source caTIES software will include support for other relational database management systems and operating systems, and will enable individuals deploying the system to more easily specify vocabularies within the Unified Medical Language System. Enhanced methods for data transfer from clinical systems are planned for future releases. Additionally, we expect to provide similar capabilities for coding a select set of other document types, including radiology reports.
An important aspect of ongoing work is to establish a community of institutions committed to achieving a true data sharing network of caTIES nodes using existing national frameworks. The use of the system to support a nationwide virtual paraffin tissue bank is considered the key long term project goal.
Acknowledgments
We thank Lucy Cafeo at the University of Pittsburgh Department of Biomedical Informatics for expert preparation and review of the manuscript. We also thank the many collaborators, developers, and adopters who contributed to the ideas implemented in the current system, including: Jules Berman, Frank Manion, David Carell, Linda Schmandt, Aditya Nemlekar, Michael Becich, Mark Watson, Rakesh Najaragan, Michelle Bisceglia, Rajiv Dhir, Anil Parwani, Jack London, Ian Fore, George Komatsoulis, Lawrence Wright, John Quigley, Dave Fenstermacher, Qing Zeng, Gunther Schadow, David Berkowitz and Henry Chueh.
Footnotes
Funding: Work on the caTIES system has been funded by multiple sources including the National Cancer Institute R01 CA132672, caBIG program under the Tissue Bank and Pathology Tools Workspace task order to University of Pittsburgh (caBIG contract #79207CBS10), and also by a Clinical and Translational Sciences Award to the University of Pittsburgh (U54 RR023506-01). Earlier work was funded by the National Cancer Institute Shared Pathology Informatics Network (U01 CA 091343). Other Funders: National Cancer Institute; Tissue Bank and Pathology Tools Workspace; Clinical and Translational Sciences Award; National Cancer Institute Shared Pathology Informatics Network.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.caBIG Strategic Planning Workspace The Cancer Biomedical Informatics Grid (caBIG): infrastructure and applications for a worldwide research community. Stud Health Technol Inform 2007;129:330–4 [PubMed] [Google Scholar]
- 2.Buetow KH. An infrastructure for interconnecting research institutions. Drug Discov Today 2009;14:605–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heller C, de Melo-Martin I. Clinical and Translational Science Awards: can they increase the efficiency and speed of clinical and translational research? Acad Med 2009;84:424–32 [DOI] [PubMed] [Google Scholar]
- 4.McGowan J. Is the CTSA initiative mandating a role for knowledge informatics? AMIA Annu Symp Proc 2008:1207–8 [PubMed] [Google Scholar]
- 5.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haug PJ, Ranum DL, Frederick PR. Computerized extraction of coded findings from free-text radiologic reports. Work in progress. Radiology 1990;174:543–8 [DOI] [PubMed] [Google Scholar]
- 7.Hripcsak G, Friedman C, Alderson PO, et al. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med 1995;122:681–8 [DOI] [PubMed] [Google Scholar]
- 8.Health Insurance Portability and Accountability Act of 1996; Public-Law 104-191 Available at: http://aspe.hhs.gov/admnsimp/pl104191.htm (accessed 16 March 2010). [PubMed] [Google Scholar]
- 9.Drake TA, Braun J, Marchevsky A, et al. A system for sharing routine surgical pathology specimens across institutions: the Shared Pathology Informatics Network. Hum Pathol 2007;38:1212–25 [DOI] [PubMed] [Google Scholar]
- 10.Beckwith BA, Mahaadevan R, Balis UJ, et al. Development and evaluation of an open source software tool for deidentification of pathology reports. BMC Med Inform Decis Mak 2006;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol 2004;121:176–86 [DOI] [PubMed] [Google Scholar]
- 12.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crowell J, Zeng Q, Ngo L, et al. A frequency-based technique to improve the spelling suggestion rank in medical queries. J Am Med Inform Assoc 2004;11:179–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. AMIA Annu Symp Proc 2001:17–21 [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 16.Cunningham H, Maynard D, Bontcheva K, et al. GATE: an Architecture for Development of Robust HLT. In Proceedings of the 40th Anniversary Meeting of the Association for Computational Linguistics (ACL-02). Philadelphia, PA: ACL Press, 2002:168–175 [Google Scholar]
- 17.NCI Metathesaurus Available at: http://ncim.nci.nih.gov/ (accessed 16 March 2010).
- 18.Lucene Available at: http://lucene.apache.org/java/docs/ (accessed 16 March 2010).
- 19.Globus Toolkit Available at: http://www.globus.org/ogsa/ (accessed 16 March 2010).
- 20.J Graph Available at: http://www.jgraph.com/ (accessed 16 March 2010).
- 21.Glazed Lists Available at: http://www.publicobject.com/glazedlists/ (accessed 16 March 2010).
- 22.JFreeChart Available at: http://www.jfree.org/jfreechart/ (accessed 16 March 2010).
- 23.Manion FJ, Robbins RJ, Weems WA, et al. Security and privacy requirements for a multi-institutional cancer research data grid: an interview-based study. BMC Med Inform Decis Mak 2009;9:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Globus Security Infrastructure Available at: http://www.globus.org/security/overview.html (accessed 16 March 2010).
- 25.Generic Security Service API Available at: http://www.ietf.org/rfc/rfc2743.txt (accessed 16 March 2010).
- 26.Langella S, Hastings S, Oster S, et al. Sharing data and analytical resources securely in a biomedical research Grid environment. J Am Med Inform Assoc 2008;15:363–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.caBIG™ Policies and Procedures for Operation of a Public caTIES Node. Available at: http://gforge.nci.nih.gov/frs/download.php/1867/DSIC_Deliveragle_Security_Deliverable_11pdf (accessed 16 March 2010).
- 28.caTIES Available at: http://sourceforge.net/project/showfiles.php?group_id=180605&package_id=241990 (accessed 16 March 2010).
- 29.caTIES User Forums Available at: http://sourceforge.net/projects/caties/forums/forum/626701 (accessed 16 March 2010).
- 30.Liu K, Mitchell KJ, Chapman WW, eds. Automating tissue bank annotation from pathology reports—comparison to a gold standard expert annotation set. AMIA Annu Symp Proc 2005:460–4 [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell KJ, Becich MJ, Berman JJ, et al., eds. Implementation and evaluation of a negation tagger in a pipeline-based system for information extraction from pathology reports. Proc MEDINFO 2004;11(Pt 1):663–7 [PubMed] [Google Scholar]
- 32.caBIG Working Groups Available at: https://cabig.nci.nih.gov/working_groups/DSIC_SLWG/DSIC_Products (accessed 16 March 2010).
- 33.Safran C, Bloomrosen M, Hammond WE, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. J Am Med Inform Assoc 2007;14:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]