Abstract
Objective
De-identified clinical data in standardized form (eg, diagnosis codes), derived from electronic medical records, are increasingly combined with research data (eg, DNA sequences) and disseminated to enable scientific investigations. This study examines whether released data can be linked with identified clinical records that are accessible via various resources to jeopardize patients' anonymity, and the ability of popular privacy protection methodologies to prevent such an attack.
Design
The study experimentally evaluates the re-identification risk of a de-identified sample of Vanderbilt's patient records involved in a genome-wide association study. It also measures the level of protection from re-identification, and data utility, provided by suppression and generalization.
Measurement
Privacy protection is quantified using the probability of re-identifying a patient in a larger population through diagnosis codes. Data utility is measured at a dataset level, using the percentage of retained information, as well as its description, and at a patient level, using two metrics based on the difference between the distribution of Internal Classification of Disease (ICD) version 9 codes before and after applying privacy protection.
Results
More than 96% of 2800 patients' records are shown to be uniquely identified by their diagnosis codes with respect to a population of 1.2 million patients. Generalization is shown to reduce further the percentage of de-identified records by less than 2%, and over 99% of the three-digit ICD-9 codes need to be suppressed to prevent re-identification.
Conclusions
Popular privacy protection methods are inadequate to deliver a sufficiently protected and useful result when sharing data derived from complex clinical systems. The development of alternative privacy protection models is thus required.
The discovery of genetic and clinical associations is an integral facet of personalized medicine. In this context, researchers need access to large quantities of patient-level data,1 such that various organizations have established data warehouses tied to biorepositories.2 3 At the same time, organizations are increasingly required to share patient data beyond their borders. In the USA, for example, the National Institutes of Health requires data collected or analyzed under National Institutes of Health-sponsored genome-wide association studies (GWAS) to be made publicly available in resources such as the DataBase of Genotypes and Phenotypes (dbGaP) for future researchers.4
To address privacy concerns5 and ensure compliance with regulations, policies that limit the sharing of patient-specific genomic data in a personally identifiable form are emerging. For instance, the NIH recently specified that data submitted to dbGaP should adhere to data-sharing regulations,3 akin to the Health Insurance Portability and Accountability Act of 1997 (HIPAA) privacy rule.6 From a technical stance, protection is often achieved by removing explicit identifiers, such as Social Security Numbers, from sensitive biomedical data and modifying the values of quasi-identifiers; ie, attributes that in combination can link to external resources to uncover patient identity, such as demographics7 and health-provider visits.8
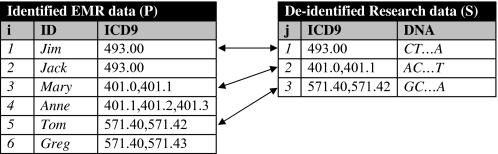
To the best of our knowledge, this work is the first to illustrate that clinical features released in the form of standardized codes can also facilitate privacy violations. This is due, in part, to the fact that such information resides in sources external to the released data, such as the identified electronic medical record (EMR) systems from where they were derived. Consider, for example, figure 1 which illustrates identified EMR data for a number of patients and a research sample. This sample contains the International Classification of Disease (ICD) version 9 codes and DNA sequences of three of these patients, namely Jim, Mary and Tom, and needs to be released in a de-identified form to support a GWAS. However, releasing this sample may result in breaching the privacy of Tom and Mary, because the identities of these patients can be uniquely associated with their DNA sequences when the research sample is linked to the EMR data using ICD-9 codes.
Figure 1.
Data linkage attack based on ICD-9 codes. EMR, electronic medical record.
We emphasize that this data linkage scenario is realistic, because data collected from EMR systems, such as diagnosis codes, is increasingly disseminated in sources including biorepositories and publicly available hospital discharge datasets,7 and poses a serious privacy threat because identified genomic information may be misused or abused.9 At the same time, existing privacy regulations and research methodologies are not designed to deal with diagnosis codes residing in EMR systems, and focus on data with much simpler semantics.
We investigate the feasibility of the above data linkage scenario using patient diagnosis codes derived from the EMR system of the Vanderbilt University Medical Center. Our analysis is realistic in that we focus on a sample of patient records that will be deposited into dbGaP and is currently involved in a GWAS funded by the National Human Genome Research Institute as part of the electronic MEdical Records and GEnomics (eMERGE) network at the NIH. Our results indicate that: (1) the majority of patients' diagnosis codes are unique not only within the sample, but also with respect to the larger population of the 1.2 million patients from which they were derived; and (2) popular techniques, such as suppression and generalization, fail to prevent re-identifying patients, or excessively distort the released information to the point that it loses its clinical utility for GWAS. Given the success of the attack, we conclude this work with a discussion on potential policy and technology approaches to mitigate this attack.
Background
Medical data privacy
The anonymization of health and genetic information through perturbation (ie, the changing of patient-specific quasi-identifier values) is typically achieved using one of the following approaches:
Suppression:10 11 the removal of patient records or particular quasi-identifier values;
Generalization:12 the replacement of quasi-identifier values with more general, but semantically consistent values; and
Randomization:13 the addition of noise to quasi-identifier values.
Methods that utilize the above approaches can be grouped into categories according to the type of data they perturb. The first category, motivated by the initial investigation of Sweeney,7 which showed that 87% of US citizens are expected to be unique with the quasi-identifier of {zip-code, gender, date of birth}, addresses the protection of demographic data. Work in this area tends to assume that data are stored in a relational table in which each patient is represented by a single record and associated with a small set of quasi-identifier attributes, typically less than 10.12 Our research differs along two dimensions. First, we treat person-specific disease information, in the form of ICD-9 codes, as quasi-identifiers. Second, to handle the complex data stored in EMR systems, we assume that each patient can be associated with a large number of records, such as one record per hospital visit, as well as distinct ICD-9 codes, often over 50 per patient.
A second category of previous research attempts to prevent linkage through DNA sequences using generalization14 15 or randomization16 of features, such as single nucleotide polymorphisms. In contrast, we consider the perturbation of diagnosis codes and leave DNA sequences intact. We assume that the only type of data contained in both EMR systems and biorepositories is diagnosis codes. We acknowledge this as a limitation of this study and leave alternative linkage scenarios, such as linking data based on DNA and demographics, to future studies.
Finally, a third category of previous research has shown that patients' hospital-visit patterns, or ‘trails’, may suffice as a quasi-identifier.17 It was recently demonstrated that parts of trails can be suppressed to thwart this attack.8 This approach is orthogonal to our work because we disclose a set of ICD-9 codes for each patient devoid of information related to visit trails.
We note that privacy can also be achieved using cryptography-based methods,18 19 which are typically applied to a setting in which a number of users query encrypted data contained in a centralized or federated repository. The privacy goal of these methods is to answer queries without decrypting the data, so that users learn nothing beyond query results. Using such methods, a researcher may learn, for example, the number of people diagnosed with 571.43, without accessing the original data shown in the left part of figure 1 but only an encrypted form of it. We emphasize that cryptography-based methods adopt a fundamentally different notion of privacy from the one considered in this work, and offer no protection against re-identification in their own right. For instance, although answering the above query may lead to re-identifying Greg, who is the only patient associated with 571.43 in the left part of figure 1, cryptography-based methods would allow an answer to this query to be retrieved. They thus need to be combined with perturbation-based methods to prevent re-identification.20
Re-identification through ICD-9 codes
To formally represent the privacy problem studied in this paper, let U be the set of ICD-9 codes stored in an EMR system. The set P={p1, …, pn} denotes a database of transactions on a patient population. Each transaction pi in P is a tuple 〈IDi, Qi〉, where IDi is a set of explicit identifiers for a patient i∈{1,…,n} whose information is contained in the EMR, and Qi a set of items from U. For example, P, shown in the left table of figure 1 contains five transactions, the first of which has ID1={Jim} and Q1={493.00}. Also, let S={s1, …, sm} denote a second database that represents the information released to a centralized research repository. S contains transactions of the form 〈Qj, DNAj〉, where Qj is a set of items from U corresponding to a patient j∈{1,…, m} whose transaction also appears in P, and DNAj is this patients' DNA record (eg, SNP or sequence data). For instance, three of the transactions contained in P, those corresponding to Jim, Mary and Tom, are released in S, which is depicted on the right table of figure 1.
We represent the number of transactions pi that have exactly the same set Qj with a transaction sj as dis(Qj), which we refer to as the distinguishability of Qj. Using this notation, a patient j is said to be uniquely identifiable when the dis(Qj)=1. Large distinguishability values imply higher privacy. For example, the distinguishability of Q1={493.00}, which corresponds to the ICD-9 codes associated with the first transaction in S, is 2. This is because two transactions in P, those corresponding to Jim and Jack, have a set of items equal to {493.00}. Similarly, the distinguishability of the set Q2 = {401.0, 401.1}, which corresponds to the ICD-9 codes of the second tuple in S, equals 1. Thus, Mary is considered to be uniquely identifiable.
Methods
Materials
For this study, we selected patient records from the de-identified version21 of StarChart,22 the EMR system of the Vanderbilt University Medical Center. This dataset contains diagnosis codes on 1 174 793 patients and each patient is associated with a unique random number that serves as an ID. We consider the entire set of patient records as the population table P. The de-identified sample table S we study in this paper was selected from P and was created for the purposes of a GWAS on native electrical conduction within the ventricles of the heart. S contains 2762 patient records and represents a ‘heart healthy’ sample that contains no previous heart disease, no heart conduction abnormalities, no electrolyte abnormalities, and no concurrent use of medications that can interfere with conduction. Table 1 provides summary statistics for the number of diagnosis codes per patient, as well as certain demographics, in P and S.
Table 1.
Statistics for the population table P, the sample S, and the BioVU subpopulation
| Patients' attributes | Statistics/values | Population P | Sample S | BioVU |
| Distinct ICD-9 codes per patient | Maximum | 370 | 301 | 323 |
| Median | 3 | 23 | 14 | |
| IQR | 7 | 39 | 25 | |
| Year of birth | Median | 1944 | 1955 | 1947 |
| IQR | 63.5 | 21 | 43.5 | |
| Gender | Male | 46% | 38% | 41% |
| Female | 54% | 62% | 59% | |
| Ethnicity | White | 58% | 82% | 76% |
| Hispanic | 2% | 1% | 1% | |
| Black | 11% | 12% | 11% | |
| Asian | 1% | 1% | 1% | |
| Unknown | 28% | 4% | 11% |
IQR, interquartile range.
Although the patients in S are different than those in P with respect to the number of diagnosis codes (a median of 23 as opposed to 3), the data linkage attack we describe is feasible when different samples used in GWAS are shared. This is because the patients that are most suitable for clinical research will, on average, contribute a greater quantity of data to the EMR. However, P is composed of every patient record in the Vanderbilt EMR system dating back to the 1980s. As the tertiary provider and only level 1 trauma center for its region, many Vanderbilt patients have only one admission or outpatient clinic visit. As such, P includes STAT records for which the patient has a very small number of diagnosis codes.
To conduct de-identified clinical research with DNA samples, Vanderbilt recently established a biobank linked to the de-identified EMR.21 The samples collected for this biobank are derived from outpatient visits in which a patient's blood was drawn for routine laboratory study. The subpopulation of P that have de-identified DNA samples available for study is called BioVU and, given the nature of sample collection, consists of recent ‘complete-capture’ EMR records. DNA sequences that take part in GWAS and will be disseminated to repositories such as dbGaP are derived from the BioVU subpopulation, including the sample S we use in this work.
To examine the extent to which S is representative of clinical research cohorts, we investigated how it relates to BioVU with respect to the distribution of ICD-9 codes per patient. We hypothesize that if the heart healthy sample contains similar records to BioVU, then it provides a good indication for the re-identification risk of the latter. BioVU contains over 70 000 patient records (as of November 2009) and, for comparison to S and P, table 1 also provides aggregate statistics for the BioVU subpopulation.
As can be seen, the median and interquartile range of ICD-9 codes are similar for the records of the BioVU dataset and S, but very different between either of these groups and P. To further support the hypothesis that records beyond S are susceptible to the data linkage attack, we examined more than 10 cohorts derived from BioVU that correspond to different clinical phenotypes, finding similar results (beyond the scope of this paper). The median number of distinct ICD-9 codes (by five-digit, category, section, or chapter) and length of follow-up in each of these cohorts are similar to S. In fact, the counts of distinct ICD-9 sections and chapters in S is actually less than most of the other clinical phenotype cohorts. This indicates that the S is in fact a representative sample for studying the privacy risk and that the risk level would be comparable when other samples derived from the BioVU dataset or other EMR-derived populations are released.
We recognize that an alternative hypothesis is that demographic bias in S led to a large number of ICD-9 codes (and thus a smaller distinguishability score), but we find this to be unsubstantiated. More specifically, table 1 illustrates the median and interquartile range for year of birth, as well as the distribution of patients' gender and ethnicity values. Note that all datasets contain patients having similar values with respect to these demographics.
We thus believe that the results obtained using S will generalize to other cohorts selected for GWAS studies.
Risk measure and protection techniques
The primary objective of this study is to measure the likelihood of achieving a linkage based on diagnosis codes when: (1) no protection measures are applied and (2) perturbation techniques are applied to the de-identified data S shared for research. In the first scenario, we compute the privacy risk as the probability of re-identifying a patient through diagnosis codes. We consider the distinguishability of each Qj for j∈{1, …, 2762}, such that the risk is expressed as 1/dis(Qj). Again, consider Q1={493.00} from the right table (sample) of figure 1. As this set appears twice in the left table (population) of figure 1 the probability of associating DNA1 to Jim is 1/2.
In the second scenario, we examine the perturbation techniques of suppression and generalization. More specifically, we investigate the policy of suppressing diagnosis codes that fail to appear frequently in S; that is, they appear in less than 5%, 15%, and 25% of the patient records in S. This strategy is similar in principle to the one proposed by Vinterbo et al10 and Eguale et al,23 and effectively assumes that each patient is represented by a single record with one diagnosis code. In other words, by suppressing a diagnosis code that appears in less than 5% of transactions, for example, each remaining code appears in a ‘group’ of size at least 0.05×2762=138. The intuition behind this strategy is that when codes appear frequently in the de-identified table S released to the centralized research repository, their combination will appear in many transactions in the populations' EMR data contained in P. We then suppress the same diagnosis codes from P and measure the distinguishability of each Qj.
Beyond suppression, we consider uniformly generalizing all ICD-9 codes in the de-identified sample S according to the ICD-9's internal hierarchical organization. The ICD-9 taxonomy of diseases and diagnoses are all three-digit disease codes (called categories, eg, 250: ‘diabetes mellitus’) followed by two possible digits of further specification (eg, 250.02: diabetes mellitus, type II, uncontrolled, without complication). The three-digit codes are organized into 17 chapters (eg, endocrine, nutritional, metabolic, immunity), which are further organized into 120 sections (eg, 250–259.99: diseases of other endocrine glands). In this study, we ‘rolled-up’ the fully specified five-digit ICD-9 codes to their three-digit representations. For example, each of the ICD-9 codes 571.40 and 571.41 would be replaced by the common code 571. In doing so, the set Q3 shown in the right table of figure 1 is generalized to {571, 571}. Note, many ICD-9 codes will appear more than once, especially for chronic diagnoses or common symptoms, such as chest pain. Recording the multiple instances of a code is important for clinical research because it helps establish the phenotype and severity of disease. For instance, Q3={401, 401} and Q4={401, 401, 401} in the left part of figure 1 remain distinguishable after generalization. The intuition behind generalization is that an attacker is incapable of distinguishing between ICD-9 codes that differ in their rightmost digits. We apply this form of generalization to the de-identified sample S and the larger patient population P using the same method. Doing so to the tables shown in figure 1, for example, result in increasing the distinguishability of Q3={571, 571} from 1 to 2 because the sets Q5 and Q6 in the left part of figure 1 are both generalized to {571, 571}. To measure the effect of applying generalization in terms of achieving privacy, we also define privacy gain type 1 as the difference between the percent of patients that are re-identifiable when the sample contains original and generalized data.
Finally, we consider a combination of perturbation methods to protect data by applying generalization on the suppression result. The rationale behind this strategy is that generalization will further improve the level of privacy protection achieved by suppression. We note that the applying suppression on the result of generalization is also possible, but it does not affect the results significantly, because suppression is much more powerful than generalization in terms of protecting privacy in our setting. To quantify the effect of applying generalization on the top of suppression in terms of achieving privacy, we define privacy gain type 2 as the difference between the percentage of patients that are re-identifiable when only suppression or both suppression and generalization have been applied to the data sample. A privacy gain type 2 of 0% implies that applying generalization on the top of suppression has no effect in improving privacy.
Data utility measures
The protection of patient privacy is a regulatory and ethical requirement, but at the same time, it may limit the scientific usefulness of the resulting records. It is thus important to measure the loss in ‘utility’ when applying protection strategies. There are many data utility measures that have been proposed,7 12–14 but are not applicable to our setting because they either deal with generalized7 12 14 or nominal data only.13
In this work, we examine the utility at a dataset level by using the percentage of retained diagnoses at increasingly generalized levels, namely fully-specified five-digit ICD-9 codes, three-digit ICD-9 codes, and ICD-9 sections. Retaining a small percentage of diagnosis information after perturbation may indicate a significant loss of utility. In addition, we examine whether the retained data are useful for analysis at a more fine-grained level. In this respect, we report the descriptions for the retained diagnosis information at the various levels of aggregation.
We also quantify data utility at a patient level by measuring how suppression affects the distribution of ICD-9 codes. This is achieved by measuring the difference between the size of sets Qi and Q'i in S for i=1,…,m, where Q'i is the result of applying suppression to Qi. We call this measure size loss (SL) and note that it quantifies the number of suppressed ICD-9 codes. In addition, to measure the fraction of ICD-9 codes that are lost due to suppression, we consider relative size loss (RSL), which is computed by normalizing the SL measure by the size of Qi. As an example of computing SL and RSL measures, consider suppressing the ICD-9 code 401.1 from Q2={401.0,401.1} shown in the left part of figure 1 to obtain Q'2={401.1} as a result. In this case, the SL and RSL scores can be computed as 2–1=1 and (2–1)/2=0.5, respectively.
Results
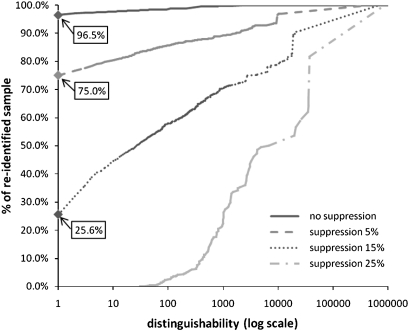
We report the risk of associating a patient's record from the de-identified sample S with the corresponding identity using all five-digit ICD-9 codes of the 1.2 million patient records contained in the population table P in figure 2. This summarizes the percentage of patients in the sample (y-axis) below a certain distinguishability score (x-axis) with respect to the larger population. Notice that, over 96% of patients are vulnerable to re-identification when no perturbation is invoked. The attack may thus be successful in practice.
Figure 2.
Risk of re-identification using five-digit ICD-9 codes. A distinguishability of 1 means that a patient is uniquely identifiable.
Next, we repeated the experiment after suppressing all ICD-9 codes that appeared in at most 5%, 15% and 25% of transactions in the sample from both tables. The result, summarized in figure 2, suggests that approximately 75% and 26% of patients can be re-identified when the 5% and 15% suppression threshold is applied, respectively. The point at which no patient can be re-identified is the 25% threshold.
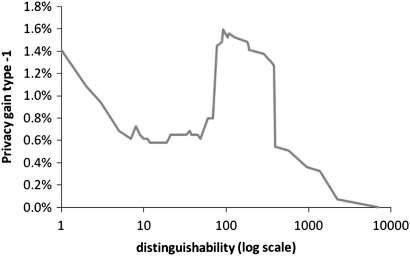
We then generalized ICD-9 codes in the sample to their three-digit representation. As can be seen in figure 3 the level of privacy gain type 1 is less than 2%, which indicates that the effect of generalization on reducing the risk of re-identifiability is insignificant.
Figure 3.
Privacy gain type 1 computed between original and generalized ICD-9 codes to their three-digit representation.
As a third protection strategy, we applied suppression followed by generalization. We report the percentage of patients vulnerable to re-identification and the privacy gain type 2 for various suppression thresholds in table 2. Notice, generalization had marginal influence on re-identification risk and the uniqueness of all patient records was prevented only when the suppression threshold was at 25%.
Table 2.
Impact of generalization after suppression on identifiability of the population
| Suppression threshold | Identified patients | Privacy gain type 2 |
| 5% | 70.4% | 4.6% |
| 10% | 48.2% | 7.4% |
| 15% | 16.3% | 9.3% |
| 20% | 0.25% | 0.6% |
| 25% | 0.0% | 0.0% |
Beyond privacy protection, we measured the utility of the resulting records. Table 3 reports the percentage of retained disease information when applying suppression using thresholds ranging from 5% to 25%. The percentage of five-digit ICD-9 codes, three-digit ICD-9 codes and ICD-9 sections retained is at most 2%, 7% and 25.4%, respectively. These results indicate that suppression may lead to a substantial loss of clinical information, even when a low threshold is invoked. Moreover, applying suppression with a threshold of 25%, the point at which no patient is unique, results in retaining very little information, as only 0.1%, 0.7% and 3.2% of five-digit ICD-9 codes, three-digit ICD-9 codes and ICD-9 sections, respectively, are retained. In addition, we report the disease information retained in table 4. This information may be too general to be of assistance for GWAS, because large categories of diagnoses have been suppressed.
Table 3.
Percentage of retained diagnosis coding information after suppression
| Suppression threshold | ICD-9 codes retained | ||
| Five-digit codes | Three-digit codes | ICD-9 sections | |
| 5% | 1.8% | 6.7% | 25.4% |
| 10% | 0.7% | 2.6% | 11.9% |
| 15% | 0.3% | 1.5% | 7.9% |
| 20% | 0.2% | 0.8% | 4.0% |
| 25% | 0.1% | 0.7% | 3.2% |
Table 4.
The complete set of ICD-9 codes that show up in at least 25% of the research sample
| Five-digit codes | Three-digit codes | ICD-9 sections | ||
| 401.1: Benign essential hypertension | → | 401: Essential hypertension | → | Hypertensive disease |
| 780.79: Other malaise and fatigue | → | 780: Other soft tissue | → | Rheumatism excluding the back |
| 729.5: Pain in limb | → | 729: Other disorders of soft tissues | → | Rheumatism excluding the back |
| 789.0: Abdominal pain | → | 789: Other abdomen/pelvis symptoms | → | Symptoms |
| 786.5: Chest pain | → | 786: Respiratory system | → | Symptoms |
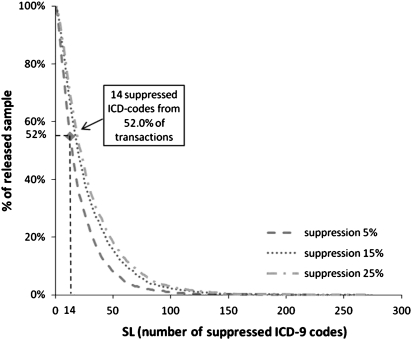
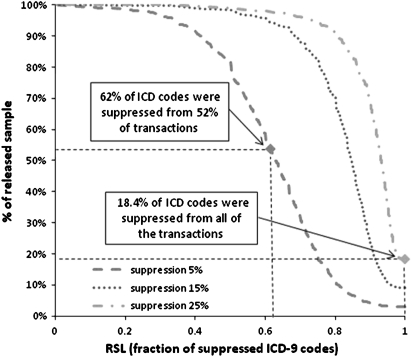
Finally, we evaluated data utility per patient by reporting SL scores for three suppression levels, namely 5%, 15% and 25%, in figure 4. Increasing the level of suppression applied results in less data utility, due to the trade-off between utility and privacy.12 Nevertheless, 14 ICD-9 codes were suppressed from more than 52% of the transactions in all tested suppression levels. Figure 5 reports the RSL scores for the same suppression levels. As can be seen, the percentage of suppressed ICD-9 codes of the transactions was more than 60% in all suppression levels, while all of the ICD-9 codes of more than 18% of the transactions had to be suppressed at a suppression level of 25%, which prevents re-identification. These results verify that suppression may heavily distort data, reducing data utility significantly.
Figure 4.
Number of suppressed ICD-9 codes versus percentage of released sample for various levels of suppression. SL, size loss.
Figure 5.
Fraction of suppressed codes versus percentage of released sample for various levels of suppression. RSL, relative size loss.
Discussion
One of the primary reasons the perturbation techniques considered in this study fail to achieve protection is because they assume that a patient is represented by a single record harboring one diagnosis code. This assumption is far from reality, because a patient is often associated with a set of diagnosis codes. The suppression and generalization of infrequent ICD-9 codes thus does not limit the distinguishability of sets of codes. As our sample contains sets of ICD-9 codes that appear very infrequently, substantial data suppression is required to reduce distinguishability with respect to the larger population, which excessively distorts data in the process.
At the same time, it is important to recognize that the use of diagnosis codes for re-identification in the real world is partly mitigated through existing data access policies. For instance, users of the research-use de-identified version of the EMR at Vanderbilt University Medical Center21 are required to sign a confidentiality agreement that explicitly prohibits re-identification attempts, and record-level data in dbGaP are available only to institutional review board-approved researchers.3 Nonetheless, such policies provide no formal privacy protection guarantees.
To provide such guarantees, anonymization methods for sparse and high-dimensional data that are proposed by the data mining community can be employed.24 However, these methods do not exploit the semantics of EMR data and are therefore unlikely to preserve data utility in the context of biomedical research. Designing methods that preserve both data privacy and the utility of EMR data is an interesting topic of future research.
Towards this goal, we point out several directions for future work. First, we intend to investigate the considered attack after relaxing the strong assumptions on data availability and the data user's external knowledge made in this work. For instance, instead of assuming that a data recipient knows all of the diagnosis codes for a patient, we plan to evaluate re-identification risks when only ICD-9 codes contained in publicly available hospital discharge databases are used. In this context, an attacker would be limited to less than 10 diagnosis codes per patient visit. Intuitively, as there will be less data available for an attacker to use, the re-identification risk is expected to drop.24
Second, it is necessary to consider a more robust data utility model. In particular, the goal of sharing the Vanderbilt patient records studied in this project is to facilitate GWAS. To have a more precise portrayal of the impact on GWAS accuracy, it will be necessary to measure the strength of associations that can be derived from anonymized data.
Finally, we note that this work considers only diagnosis codes and none of the patient demographics that are common to medical information shared for secondary purposes, such as gender, race, or age. Although it has been shown that such combinations would only increase the re-identification risk,7 10 quantifying the risk posed by disclosing certain combinations of these attributes together with diagnosis codes is required to develop practical policies that mitigate the risk of patient re-identification.
Conclusions
Our work illustrated that the re-identification of patient-specific data through standardized clinical data is a practical privacy threat. We demonstrated the feasibility of such an attack with real patient clinical information. Furthermore, we showed that neither suppression of rare codes (a requirement of the HIPAA privacy rule), nor generalization, sufficiently protects records while retaining clinically meaningful information.
Acknowledgments
The authors would like to thank Dr Daniel Masys, Dr Dan Roden, the members of the Health Information Privacy Laboratory, and the anonymous referees for comments and discussions. They also thank Xiaoming (Sunny) Wang for technical support in accessing the de-identified data used in this study.
Footnotes
Funding: This research was funded by grant U01HG004603 of the National Human Genome Research Institute and 1R01LM009989 of the National Library of Medicine.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Gurwitz D, Lunshof J, Altman R. A call for the creation of personalized medicine databases. Nat Rev Drug Discov 2006;5:23–6 [DOI] [PubMed] [Google Scholar]
- 2.Barbour V. UK Biobank: a project in search of a protocol? Lancet 2003;361:1734–8 [DOI] [PubMed] [Google Scholar]
- 3.National Institutes of Health Policy for sharing of data obtained in NIH supported or conducted genome-wide association studies. NOT-OD-07-088. Bethesda, MD, USA: NIH, 2007 [Google Scholar]
- 4.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet 2007;39:1181–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGuire A, Fisher R, Cusenza P, et al. Confidentiality, privacy, and security of genetic and genomic text information in electronic health records: points to consider. Genet Med 2008;10:495–9 [DOI] [PubMed] [Google Scholar]
- 6.US Department of Health and Human Services, Office for Civil Rights Standards for protection of electronic health information; final rule. Washington, DC, USA: Federal Register, 20 February 2003; 45 CFR:Pt. 164. [Google Scholar]
- 7.Sweeney L. k-anonymity: a model for protecting privacy. Int J Uncertain Fuzz Knowledge-Based Systems 2002;10:557–70 [Google Scholar]
- 8.Malin B. A computational model to protect patient data from location-based re-identification. Artif Intell Med 2007;40:223–39 [DOI] [PubMed] [Google Scholar]
- 9.Rothstein M, Epps P. Ethical and legal implications of pharmacogenomics. Nat Rev Genet 2001;2:228–31 [DOI] [PubMed] [Google Scholar]
- 10.Vinterbo S, Ohno-Machado L, Dreiseitl S. Hiding information by cell suppression. Proc AMIA Symp 2001:726–30 [PMC free article] [PubMed] [Google Scholar]
- 11.Meyerson A, Williams R. On the complexity of optimal k-anonymity. Proceedings of ACM symposium on principles of database systems 2004:223–8 [Google Scholar]
- 12.Samarati P. Protecting respondents identities in microdata release. IEEE Trans Knowl Data Eng 2001;13:1010–27 [Google Scholar]
- 13.Agrawal R, Srikant R. Privacy-preserving data mining. Proc ACM SIGMOD Int Conf on Management of Data 2000:439–50 [Google Scholar]
- 14.Lin Z, Hewett M, Altman RB. Using binning to maintain confidentiality of medical data. Proc AMIA Symp 2002:454–8 [PMC free article] [PubMed] [Google Scholar]
- 15.Malin B. Protecting genomic sequence anonymity with generalization lattices. Methods Inf Med 2005;44:687–92 [PubMed] [Google Scholar]
- 16.Lin Z, Owen AB, Altman RB. Genomic research and human subject privacy. Science 2004;305:183. [DOI] [PubMed] [Google Scholar]
- 17.Malin B, Sweeney L. How (not) to protect genomic data privacy in a distributed network: using trail re-identification to evaluate and design anonymity protection systems. J Biomed Inform 2004;37:179–92 [DOI] [PubMed] [Google Scholar]
- 18.Kantarcioglu M, Jiang Y, Liu Y, et al. A cryptographic approach to securely share and query genomic sequences. IEEE Trans Inf Technol Biomed 2008;12:606–17 [DOI] [PubMed] [Google Scholar]
- 19.Hacigümüş H, Iyer B, Li C, et al. Executing SQL over encrypted data in the database-service-provider model. Proc ACM SIGMOD Int Conf on Management of Data 2002:216–27 [Google Scholar]
- 20.Kantarcioglu M, Jiang W, Malin B. A privacy-preserving framework for integrating person-specific databases. Proc privacy in statistical databases 2008:298–314 [Google Scholar]
- 21.Roden D, Pulley J, Basford M, et al. Development of a large scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stead W, Bates R, Byrd J, et al. Case study: the Vanderbilt University Medical Center information management architecture. In: Van De Velde Rudi, Degoulet Patrice, eds. Clinical information systems: a component-based approach. New York, USA: Springer-Verlag, 2003 [Google Scholar]
- 23.Eguale T, Bartlett G, Tamblyn R. Rare visible disorders/diseases as individually identifiable health information. AMIA Annu Symp Proc 2005:947. [PMC free article] [PubMed] [Google Scholar]
- 24.Narayanan A, Shmatikov V. Robust de-anonymization of large sparse datasets. Proc IEEE symposium on security and privacy 2008:111–25 [Google Scholar]