Abstract
MetaMap is a widely available program providing access to the concepts in the unified medical language system (UMLS) Metathesaurus from biomedical text. This study reports on MetaMap's evolution over more than a decade, concentrating on those features arising out of the research needs of the biomedical informatics community both within and outside of the National Library of Medicine. Such features include the detection of author-defined acronyms/abbreviations, the ability to browse the Metathesaurus for concepts even tenuously related to input text, the detection of negation in situations in which the polarity of predications is important, word sense disambiguation (WSD), and various technical and algorithmic features. Near-term plans for MetaMap development include the incorporation of chemical name recognition and enhanced WSD.
Keywords: Indexing, information retrieval, Medline, MetaMap, Metathesaurus, natural language processing, UMLS
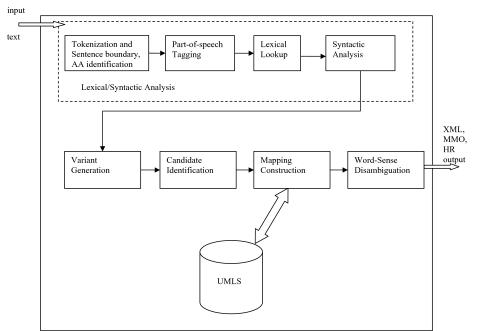
MetaMap1 is a widely available program providing access from biomedical text to the concepts in the unified medical language system (UMLS) Metathesaurus. MetaMap arose in the context of an effort to improve biomedical text retrieval, specifically the retrieval of MEDLINE/PubMed citations.2 3 It provided a link between the text of biomedical literature and the knowledge, including synonymy relationships, embedded in the Metathesaurus. Early MetaMap development was guided by linguistic principles which provided both a rigorous foundation and a flexible architecture in which to explore mapping strategies and their applications.4–6 A system diagram showing MetaMap processing is shown in figure 1. Input text undergoes a lexical/syntactic analysis consisting of:
Figure 1.
MetaMap system diagram. HR, human readable; MMO, MetaMap machine output; UMLS, unified medical language system.
tokenization, sentence boundary determination and acronym/abbreviation identification;
part-of-speech tagging;
lexical lookup of input words in the SPECIALIST lexicon7; and
a final syntactic analysis consisting of a shallow parse in which phrases and their lexical heads are identified by the SPECIALIST minimal commitment parser.7
Each phrase found by this analysis is further analyzed by the following processes:
variant generation, in which variants of all phrase words are determined (normally by table lookup);
candidate identification, in which intermediate results consisting of Metathesaurus strings, called candidates, matching some phrase text are computed and evaluated as to how well they match the input text;
mapping construction, in which candidates found in the previous step are combined and evaluated to produce a final result that best matches the phrase text; and, optionally,
word sense disambiguation (WSD), in which mappings involving concepts that are semantically consistent with surrounding text are favored.8
The evaluation performed on both the candidates and the final mappings is a linear combination of four linguistically inspired measures: centrality; variation; coverage; and cohesiveness. The evaluation process begins by focusing on the association, or mapping, of input text words to words of the candidates. Centrality, the simplest of the measures, is a Boolean value which is one if the linguistic head of the input text is associated with any of the candidate words. The variation measure is the average of the variation between all text words and their matching candidate words, if any. Coverage and cohesiveness measure how much of the input text is involved in the mapping (the coverage) and in how many chunks of contiguous text (the cohesiveness). The four measures are combined linearly giving coverage and cohesiveness twice the weight of centrality and variation, and the result is normalized to a value between 0 and 1000. Complete details of the evaluation process can be found in the technical document, MetaMap evaluation.9
MetaMap is highly configurable across multiple dimensions, including:
data options, which choose the vocabularies and data model to use;
output options, which determine the nature and format of the output generated by MetaMap; and
processing options, which control the algorithmic computations to be performed by MetaMap.
The data options allow the user to choose the UMLS data (eg, 2009 for the 2009AA release) for use by MetaMap, and the desired level of filtering to employ.
MetaMap's relaxed data model uses:
lexical filtering, which excludes most Metathesaurus strings mapped to a concept which are essentially identical to another string for the same concept; and
manual filtering, which excludes unnecessarily ambiguous terms, as determined by a detailed annual study.
MetaMap's strict model supplements the above filtering regimen with:
syntactic filtering, which excludes complex expressions with underlying grammatical structure (eg, ‘accident caused by caustic and corrosive substances’), which MetaMap would normally be unable to find anyway because they span multiple phrases.
Typical output options include:
hiding or displaying the semantic types or concept unique identifiers (CUI) of all displayed concepts;
hiding or displaying candidates or mappings, where MetaMap will not even compute these elements unless some other option requires them;
generating XML output rather than the default human-readable output;
excluding or restricting output to concepts of specified semantic types; and
excluding or restricting output to concepts drawn from specified vocabularies.
Some of MetaMap's most useful processing options include:
controlling the types of derivational variants used in lexical variant generation (no variants at all, adjective/noun variants only, or all variants);
turning on and off MetaMap's WSD module;
term processing, which causes MetaMap to process each input record, no matter how long, as a single phrase, in order to identify more complex Metathesaurus terms;
allowing overmatches so that, for example, the input text medicine will map to any concept containing the word medicine, medical or any other variant of medicine; and
allowing concept gaps so that, for example, the text obstructive apnea will map to concepts ‘obstructive sleep apnoea’ and ‘obstructive neonatal apnea’, which are considered too specific for normal processing.
Note that the combination of the last three options (together with hiding the mappings) is known as MetaMap's browse mode. It is generally used to explore the Metathesaurus both broadly and deeply as opposed to the more normal mode in which the ‘best match’ to the input text is sought. See the Genre and task issues section below for more information about browse mode. Details of all aspects of MetaMap processing can be found in the technical documents at the MetaMap portal.10
As mentioned earlier, the final phase of MetaMap's lexical/syntactic processing involves computing a shallow parse, dividing the input text into phrases, which form the basis of MetaMap's subsequent processing. Each phrase's human-readable output by default consists of three parts:
the phrase itself;
the candidates, a list of intermediate results consisting of Metathesaurus strings matching some or all of the input text. In addition, the preferred name of each candidate is displayed in parentheses if it differs from the candidate, and the semantic type of the candidate is also shown; and finally,
the mappings, combinations of candidates matching as much of the phrase as possible.
Most elements of human-readable output, whether default or optional, can be shown or hidden based on MetaMap options. In addition, by default MetaMap displays only those mappings that receive the highest score. MetaMap's default human-readable output generated from the input text obstructive sleep apnea is shown in figure 2. In this example MetaMap identified 11 Metathesaurus candidates, the best of which received a perfect score of 1000, and by itself formed the top-scoring mapping. If all mappings had been requested, then several more mappings including one consisting of the combination of ‘apnea, sleep’ and ‘obstructive’ would have been displayed.
Figure 2.
An example of MetaMap's human-readable output.
MetaMap possesses a number of strengths and weaknesses. Among its strengths are its thoroughness, characterized by its aggressive generation of word variants, and its linguistically principled approach to its lexical and syntactic analyses as well as its evaluation metric for scoring and ranking concepts. It is also adept at constructing partial, compound mappings when a single concept is insufficient to characterize input text phrases. As evidenced by the above description of some of its options, it is highly configurable; its behavior can be easily customized depending on the task being addressed. Finally, because its lexicon and target vocabulary can be replaced with others from another domain, it has the property of domain independence.
One of MetaMap's weaknesses is that it can be applied only to English text. MetaMap's English-centric nature is evident throughout its implementation, not just in its lexical and syntactic algorithms. Also, a negative consequence of its thoroughness is that it is relatively slow. In its current implementation, it is not appropriate for real-time use; and it would require a major fine-grained parallel re-implementation in order to overcome this weakness. The efficiency enhancements described below in the Algorithm tuning section, with rare exceptions, allow MetaMap to process a given MEDLINE citation in well under a minute, although complex phrases, for example:
‘from filamentous bacteriophage f1 PCR polymerase-chain reaction PDB Protein Data Bank PSTI human pancreatic secretory trypsin inhibitor RBP retinol-binding protein SPR surface plasmon resonance TrxA E. coli thioredoxin’
can still require hours of computation because they generate many hundreds of thousands of potential mappings. It is examples such as this that make it clear that re-implementing the MetaMap algorithm to process phrases in parallel would not in general be sufficient to sanction the use of MetaMap for real-time or high-volume applications. That would require sub-phrasal parallelization, which for mapping construction represents a non-trivial challenge. Although MetaMap was originally designed for tasks that can easily be accomplished using our scheduler facility, which uses multiple servers to provide document-level parallelization, it is likely that we will undertake a fine-grained parallelization effort in the future.
Perhaps MetaMap's greatest weakness is its reduced accuracy in the presence of ambiguity. MetaMap employs a WSD algorithm8 to reduce ambiguity, but it is clear that further disambiguation efforts will be needed to solve the problem satisfactorily, especially as the Metathesaurus is becoming ever more ambiguous. We return to this issue in the Conclusion.
After some experience with MetaMap, it became clear that it could be applied to tasks other than retrieval, namely text mining,11–13 classification, question answering,14 knowledge discovery,15 and concept-based indexing,16–20 among others. In addition, research efforts involving MetaMap have extended to groups outside the National Library of Medicine (NLM).21–32
Requests for access to MetaMap from the biomedical informatics community grew over the years, often coinciding with particular research questions. Besides describing MetaMap in general, a major purpose of this paper is to highlight some of these research topics. We emphasize the challenges throughout MetaMap's development history and conclude with a discussion of plans for future MetaMap extensions, including chemical name recognition and enhanced WSD.
In order to provide more context for the paper, we note that MetaMap has been used by NLM researchers and outside users since 1994 and is currently available via web access, a downloadable Java implementation (MMTx), an application programming interface, and most recently, a downloadable version of the complete Quintus Prolog implementation of MetaMap itself.10 MetaMap was originally developed using Quintus Prolog, which is available from and maintained by the Swedish Institute of Computer Science (http://www.sics.se). Quintus Prolog provides an interactive, Emacs-based, source-linked debugger, a C interface, incremental compilation, and the ability to create runtime binaries which can be used without a Quintus Prolog license. We are currently porting MetaMap to SICStus Prolog, which offers better support for application deployment under Windows. Finally, we note that MetaMap and MMTx are two versions of the same program. MetaMap, the original program, was developed using Prolog because the language lent itself well to prototyping natural language processing (NLP) applications. We created the Java-based MMTx as a way to distribute MetaMap while separating development from production efforts and because of its platform independence and zero cost. We have since discovered that, due to MMTx's tokenization/lexicalization routines, the two programs produce slightly different results despite concerted efforts to reconcile them. We have also learned that almost no MetaMap users modify the code for which they would incur Prolog licensing fees. These factors make it unnecessary to maintain two versions of the program; and as Prolog provides a better development environment, we are phasing out MMTx by freezing its implementation, limiting development to bug fixes.
Background
In order to better understand MetaMap's role in the goal of relating biomedical text to structured sources of biomedical knowledge, it is useful to consider other biomedical mapping programs as well as efforts to assess the degree to which MetaMap succeeds in this goal. That is the purpose of the following sections.
Related work
Over the years several researchers have developed programs to map biomedical text to a knowledge source such as NLM's medical subject headings (MeSH) or, more recently, the UMLS Metathesaurus. Examples of such efforts that predate MetaMap include MicroMeSH,33 CHARTLINE,34 CLARIT,35 and SAPHIRE.36 Each of these systems employed one or more of the following features: lexical analysis, often using a specialized lexicon; syntactic analysis; a mapping procedure accounting for partial matching; and the UMLS Metathesaurus as the target knowledge source, rather than a smaller source such as MeSH. MetaMap's contribution to this environment was that it combined all of these features, emphasizing linguistic principles throughout. Mapping tools developed subsequently to MetaMap include those of Nadkarni et al,37 and KnowledgeMap.38 These recent efforts have been applied to a variety of applications and have achieved varying degrees of success, depending both on how well they solve such NLP problems as parsing, lexical variation and ambiguity resolution, as well as how successfully they have been tailored to specific tasks.
Evaluation of MetaMap
Direct evaluation of MetaMap in which MetaMap's performance is compared with a manually constructed gold standard of mappings to the Metathesaurus has almost never been performed on a realistic scale. However, several indirect evaluations have been performed over the years. Such evaluations consisted of performing a specific NLP task first without MetaMap results and then with them to see if the MetaMap results improved task performance.
As MetaMap was originally developed in the context of MEDLINE citation retrieval, the earliest evaluations consisted of standard information retrieval experiments conducted using MEDLINE test collections. One such experiment used the Metathesaurus concepts found by MetaMap for each document (MEDLINE citation) in a test collection.2 The documents were then indexed without and with these concepts, and the results were compared. This document indexing experiment resulted in a performance improvement (as measured by 11-point average precision) of a modest 4%, a result which is considered to be at best barely discernable.
A second retrieval experiment focused on query expansion instead of document indexing.3 It consisted of processing MEDLINE test collection queries with MetaMap and augmenting the queries with both the resulting Metathesaurus concepts and the text of the phrases discovered early during MetaMap processing. Augmenting each query with the combination of original query text, phrase text and Metathesaurus concepts improved retrieval results, as measured by 11-point average precision over baseline, by 14%, which was almost as strong a result as the best published results at the time of 16%,39 which used document feedback, an automatic form of relevance feedback, on a MEDLINE test collection. An even larger improvement of 20% was obtained using document feedback at the third Text REtrieval Conference (TREC) by the Cornell group.40 Given the power of retrieval feedback, which was not available with the retrieval engine we used for our experiment, it is likely that our 14% result would have improved significantly if we could have augmented our query expansion approach with retrieval feedback.
Moving beyond simple MetaMap-enhanced retrieval, an indexing experiment involving NLM's medical text indexer (MTI),16 18 was performed to compare MTI's indexing effectiveness against official, manually produced MEDLINE indexing.41 (Note that MTI was referred to as the indexing initiative system (IIS) at the time.) The experiment is relevant to evaluating MetaMap as MetaMap is the basis of one of MTI's two foundational indexing methods, the other being a variant of PubMed's related articles facility.18 For the experiment, the documents in several MEDLINE test collections were processed by MTI. Standard retrieval experiments were performed using only the document text as a baseline and then augmenting the document text with either MEDLINE indexing or MTI's indexing. Augmented documents always improved performance over the baseline; and in all but one case, the MEDLINE indexing achieved better performance than MTI indexing. However, the experiment showed that even though MTI indexing does not have the coherence of manual indexing, it can still produce retrieval results almost as good as those obtained with manual indexing.
One example of an experiment comparing MetaMap with another system was performed by Denny et al38 They developed a system, KnowledgeMap, for the purpose of identifying concepts in medical curriculum documents and compared KnowledgeMap's performance with that of MetaMap. They found that on a collection of 10 curriculum documents, KnowledgeMap outperformed MetaMap both on recall (82% vs 78%) and precision (89% vs 85%).
A final, more recent experiment42 compared MetaMap's concept identification ability with that of MGREP, a program developed at the University of Michigan. The task for the experiment was large-scale indexing of online biomedical resources, and the two systems were compared on four document sets ranging in size from 2 K documents to 99 K documents and for two entity types, biological processes and diseases. MGREP is a much simpler program than MetaMap. Rather than using significant linguistic analysis followed by mapping construction from intermediate results, MGREP relies on a comprehensive lexicon of terms of interest to recognize concepts in text. With a couple of exceptions, MetaMap recognized more concepts (occasionally several times more concepts) in text. Not surprisingly, then, MGREP almost always outperformed MetaMap with respect to precision, an exception being that MetaMap slightly outperformed MGREP for recognizing biological processes in http://ClinicalTrials.gov. Recall, for which the more thorough MetaMap would be expected to outperform MGREP, was not reported because of the difficulty in computing it for document collections of the size used in the study. Perhaps just as significantly, MGREP was much faster in performing the indexing task, almost two orders of magnitude faster in one case. Because MGREP clearly outperformed MetaMap for the study, it was chosen as the concept recognition tool for the production indexing system. MetaMap's inability to perform in real-time situations mentioned earlier only confirms the decision.
Research-driven development
Virtually all MetaMap development has been driven by issues arising naturally from research efforts. Some of these issues are theoretically substantial and deep; others are practical and straightforward. They include tokenization issues, output formats, issues relating to text genre or application task, and algorithm tuning. Examples of these issue types are discussed in the following sections.
Tokenization issues
Acronym detection
Most technical domains are heavily laden with acronyms and abbreviations (AA), often accompanied by their definitions or expansions:
The effect of adrenocorticotropic hormone (ACTH) and cortisone on drug hypersensitivity reactions.
In analyzing such text, it is essential that subsequent occurrences of ACTH be analyzed as adrenocorticotropic hormone. We have accordingly implemented AA detection in an algorithm which is fundamentally the same, intuitive algorithm as that described in Schwartz and Hearst.43 We try to match a potential AA, which must be bracketed (eg, (ACTH)) with a potential expansion, which must precede the AA in the same sentence (eg, adrenocorticotropic hormone).
Because of the character-matching required, AA detection can be computationally expensive when applied to long strings. In order to streamline the logic, we have implemented a number of efficiency rules which prevent erroneous attempts to match potential AAs and expansions. Some typical rules include the following:
AA cannot contain more than 20 characters.
Expansions must be longer than their corresponding AA.
Expansions cannot contain parenthesized text.
Single-word AA cannot contain more than 12 characters.
AA cannot begin with such, also or including.
For example, rule 1 prevents costly fruitless attempts to identify an AA in the text bladder contractile dysfunction (bladder decompensation).
Of course some AA do exceed 20 characters, so our algorithm causes some false negatives, but the computational savings outweigh the decrease in recall.
Non-standard input
A common problem for NLP systems is handling non-standard or ill-formed input. In the case of MetaMap, MEDLINE/PubMed citations contain numerous instances of sentence-ending periods not followed by whitespace, which led to numerous false negatives in end-of-sentence detection. For example, the abstract of PMID 18011217 contains a passage with three such instances in succession:
Diphtheria is a disease of childhood. Seventy per cent of all cases and ninety per cent of all deaths occur under 15 years of age.More boys than girls and more women than men have diphtheria.The fatality rate is higher for males than for females.
We discovered over 50 000 such cases in the PubMed database, and implemented a workaround, until the data can be corrected, by assembling a list of the most common words found in this non-standard context and forcing the tokenizer to create sentence breaks in these special contexts.
Output formats
MetaMap's default output is human readable and displays, for each phrase in the input text, an intermediate list of candidate Metathesaurus concepts matching (part of) the phrase, the mappings formed by combining candidates matching disjoint phrase text, and ancillary information, often optional, such as concept CUI and their semantic types.
We have also developed other output formats, some at the behest of our users: MetaMap machine output (MMO), XML output and colorized MetaMap output, each of which is described here.
MMO, the earliest alternative form of MetaMap output, includes a superset of the information in human-readable output and is formatted as Prolog terms. This format enables storing MetaMap output for subsequent postprocessing by other Prolog-based applications, thereby obviating the need for repeated MetaMap analysis of input texts.
Because XML has become somewhat of a lingua franca for internet-based information exchange, we have developed an XML output format that presents the same data as MMO. Consider the simple input text consisting of the single word heart. To provide context, figure 3 contains the human-readable output for heart.
Figure 3.
MetaMap's human-readable output for the input text ‘heart’.
MetaMap's XML output for heart (greatly simplified and compressed for expository conciseness) is shown in figure 4.
Figure 4.
MetaMap's XML output for the input text ‘heart’.
The figure shows that the XML data presented for each concept include:
the concept's (negated) score: −1000;
its CUI: C0018787;
the matching UMLS string and preferred name for the concept: ‘heart’ for both;
the semantic type(s) of the concept: bpoc, the abbreviation for ‘body part, organ, or organ component’;
the UMLS source(s) in which this candidate concept appears: including MSH (for MeSH); and
positional information for utterances, phrases and concepts: a start position of 0 and a span length of 5 for the single utterance, phrase and concept in this simple example. Note that the computation of such positional information was crucial for developing colorized MetaMap output.
Colorized MetaMap output (MetaMap 3-D, or MM3D) is designed to provide visual layers of information for the concepts mapped by MetaMap in a body of text and was specifically developed to display clinical text effectively. The initial version of MM3D highlights all concepts using colors depending on their semantic groupings (eg, concepts from the disorders group are shown in light pink). Names of prescription drugs are also linked to DailyMed.44 In addition, the program denotes syntactic elements such as the head of each phrase using underlining; and phrase boundaries in the text can be turned on or off by the user as desired. Enhancements to MM3D will concentrate on adding UMLS-specific information for each of the mapped concepts, incorporating negation information when available, and possibly adding SemRep and MeSH information when applicable.
Genre and task issues
Many MetaMap features have been developed because of the genre of text to be processed or, more specifically, the tasks to be performed on the text. Several such features are described here. MetaMap options that implement a given feature are shown in parentheses after the feature name.
Term processing (-z)
MetaMap normally expects input in the form of complete sentences which are parsed into phrases and processed independently. If, however, a user already knows the blocks of text that should be analyzed as a unit (vocabulary terms, for example), then term processing can be specified. The parsing analysis still occurs to aid in evaluating results, but lines of input text are not split into multiple phrases for separate processing.
Browse mode (-zogm)
An extension of term processing is MetaMap's browse mode, which is useful for determining how well a set of terms is represented in the Metathesaurus. An example of such a need was the large scale vocabulary test undertaken by NLM and the Agency for Healthcare Research and Quality (AHRQ; formerly, AHCPR) to answer coverage questions and to discover candidate vocabularies for inclusion in the Metathesaurus.45 (Note that a simplified implementation of browse mode called approximate matching was actually used in the large scale vocabulary test study.)
In order to find any Metathesaurus concept even remotely related to an input term, browse mode augments term processing (−z) with overmatches (−o) and concept gaps (−g). Overmatches (eg, mapping medicine to ‘alternative medicine’, ‘medical records’, or even ‘nuclear medicine procedure’) are not normally allowed by MetaMap because the results are generally too semantically distant from the input. Similarly, although MetaMap always allows gaps in the input text (eg, mapping ambulatory heart monitor to ‘ambulatory monitors’), concept gaps (eg, mapping obstructive apnea to either ‘obstructive sleep apnoea’ or ‘obstructive neonatal apnea’) are not normally allowed because gapped concepts are generally too specific. Note that the computation of final mappings (−m) is suppressed for browse mode because of the overwhelming number of final mappings constructible from the large number of intermediate concepts (see candidate identification in figure 1) normally found. Browse mode is computationally expensive even without mapping construction.
Negation (-negex)
Although detecting negated concepts may or may not be useful for ad-hoc information retrieval, it is essential for properly understanding clinical text. In particular, recognizing negated concepts was essential in the context of the medical NLP challenge, sponsored by a number of groups including the Computational Medicine Center (CMC) at the Cincinnati Children's Hospital Medical Center. The challenge consisted of assigning International Classification of Diseases, 9th Revision, clinical modification (ICD-9-CM) codes to clinical text consisting of anonymized clinical history and impression sections of radiology reports. MetaMap was provisionally but successfully modified46 to detect negated predications in these reports using a simplified version of the NegEx algorithm.47 MetaMap now implements an extension of the complete NegEx algorithm.
Word sense disambiguation (-y)
MetaMap's greatest weakness is arguably its inability to resolve Metathesaurus ambiguity, that is, cases in which two or more Metathesaurus concepts share a common synonym. For example, the synonym ‘cold’ (equivalently, ‘cold’ or ‘COLD’) occurs in six distinct concepts such as ‘common cold’, ‘cold temperature’ and ‘cold sensation’. Until recently, we have relied on a manual study of Metathesaurus ambiguity48 to suppress word senses deemed problematical for (literature-centric) NLP usage. For example, the ‘cold’ synonyms in the three remaining cold concepts (‘cold brand of chlorpheniramine-phenylpropanolamine’, ‘cold therapy’ and ‘chronic obstructive airway disease’) are all suppressed for MetaMap processing. Unfortunately, manual suppression only partly solves the ambiguity problem (eg, ‘cold’ remains three-ways ambiguous). Even worse, the problem is growing: the number of cases of ambiguity not already suppressed by Metathesaurus editors rose from 26 084 in the 2007AA release to 36 266 in the 2008AA release, an increase of 39%.
The only long-term solution to an environment with significant ambiguity is to implement some form of WSD. We developed a WSD test collection for Metathesaurus ambiguity49 and have used it to test a WSD algorithm8 based on semantic type indexing, which resolves Metathesaurus ambiguity by choosing a concept having the most likely semantic type for a given context.
Algorithm tuning
Two practical improvements to MetaMap's algorithm include the suppression of word variants that almost always lead to bad mappings and a collection of efficiency modifications. The former improves accuracy while the latter improves throughput.
Variant suppression
A recent improvement to our variant generation logic that has increased precision is the suppression of variants of one and two-character words. For example, in analyzing t-cell, we no longer generate variants that produce the false positive candidates ‘TX’ and ‘TS’ from t. We implemented this change because of the large number of false positives observed and in coordination with a project to filter out Metathesaurus content beyond what MetaMap already does.50 Ex post facto analysis has confirmed that one-character words have five times more variants than the average word, and two-character words have twice as many.
Efficiency modifications
When MetaMap was first implemented, the Metathesaurus contained approximately 440 K concepts. No content-bearing word (such as human, disease and protein) appeared in more than 12 K concepts; consequently the relatively modest size of the Metathesaurus did not impose significant computational constraints. For example, caching results in simple data structures such as linear lists incurred no discernible performance penalty. Since then, however, the number of concepts in the Metathesaurus has grown to almost 2 M, a fourfold increase; and now, over 100 content words each appear in at least 12 K concepts.
Because this increase in the size and complexity of the Metathesaurus was relatively gradual, the accompanying slowdown in MetaMap's throughput was not immediately noticeable. This year, however, the Metathesaurus has grown to such an extent that certain MEDLINE citations took an entire day to analyze, and others failed because they required more physical memory than the 8 GB installed on our workstations. We therefore optimized MetaMap in three significant ways:
caching results in AVL trees (self-balancing binary search trees) rather than linear lists;
expanding the scope of caching from a phrase to an entire citation; and
replacing a straightforward but inefficient call to Prolog's findall/3 all-solutions predicate with pure recursive code.
Our optimizations have met with excellent results: some complex MEDLINE citations now run well over 100 times faster, and none has yet exceeded our memory capacity. One infamous citation, in particular, that used to require over 40 h of processing can now be analyzed in under 2 min. In spite of these dramatic improvements, further optimization will be required in order to handle challenging cases such as the complex phrase mentioned earlier.
Conclusion
This paper has described some of the history, most important features, linguistic basis, architecture, and processing methodology of MetaMap, which has become established as one of the premier applications for the identification of Metathesaurus concepts in biomedical text. Because of our ongoing interaction with our user community, MetaMap has over the years evolved significantly with regard to functionality, implementation, and distribution vehicles since its inception in the mid 1990s, and it is currently used by many groups throughout the world in the biomedical informatics community.
Some of the near-term plans for further MetaMap development include:
improving MetaMap's ability to process clinical text in conjunction with a project which creates UMLS Metathesaurus content views for various biomedical NLP purposes;50
adding chemical name recognition based on Wilbur et al51 to MetaMap's higher-order tokenization capabilities; and
enhancing MetaMap's WSD accuracy by adding more WSD algorithms and basing final ambiguity resolution on a voting mechanism.
Acknowledgments
The authors gratefully acknowledge the contribution by many colleagues to the development of MetaMap over the years. Of particular note, MetaMap's linguistic foundation is due to Thomas C Rindflesch and Allen C Browne. Development support of MetaMap and related tools is shared with project colleagues, James G Mork and Will J Rogers.
Footnotes
Funding: This work was supported by the Intramural Research Program of the National Institutes of Health and the National Library of Medicine.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the metamap program. Proc AMIA Symp 2001:17–21 [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson AR, Rindflesch TC, Browne AC. Exploiting a large thesaurus for information retrieval. Proc RIAO 1994;94:197–216 [Google Scholar]
- 3.Aronson AR, Rindflesch TC. Query expansion using the UMLS Metathesaurus. Proc AMIA Annu Fall Symp 1997:485–9 [PMC free article] [PubMed] [Google Scholar]
- 4.Rindflesch TC, Aronson AR. Semantic processing in information retrieval. Proc Annu Symp Comput Appl Med Care 1993:611–15 [PMC free article] [PubMed] [Google Scholar]
- 5.Rindflesch TC, Aronson AR. Ambiguity resolution while mapping free text to the UMLS Metathesaurus. Proc Annu Symp Comput Appl Med Care 1994:240–4 [PMC free article] [PubMed] [Google Scholar]
- 6.Aronson AR. The effect of textual variation on concept based information retrieval. Proc AMIA Annu Fall Symp 1996:373–7 [PMC free article] [PubMed] [Google Scholar]
- 7.McCray AT, Aronson AR, Browne AC, et al. UMLS knowledge for biomedical language processing. Bull Med Libr Assoc 1993;81:184–94 [PMC free article] [PubMed] [Google Scholar]
- 8.Humphrey SM, Rogers WJ, Kilicoglu H, et al. Word sense disambiguation by selecting the best semantic type based on journal descriptor indexing: preliminary experiment. JASIST 2006;57:96–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aronson AR. MetaMap evaluation. Bethesda MD: National Library of Medicine, 2001. http://skr.nlm.nih.gov/papers/references/mm.evaluation.pdf [Google Scholar]
- 10.http://metamap.nlm.nih.gov
- 11.Masseroli M, Kilicoglu H, Lang FM, et al. Argument-predicate distance as a filter for enhancing precision in extracting predications on the genetic etiology of disease. BMC Bioinformatics 2006;7:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiszman M, Demner-Fushman D, Lang FM, et al. Interpreting comparative constructions in biomedical text. Association for computational linguistics. Proc BioNLP Workshop 2007:137–44 [Google Scholar]
- 13.Ahlers CB, Fiszman M, Demner-Fushman D, et al. Extracting semantic predications from medline citations for pharmacogenomics. Pac Symp Biocomput 2007:209–20 [PubMed] [Google Scholar]
- 14.Demner-Fushman D, Humphrey SM, Ide NC, et al. Combining resources to find answers to biomedical questions. Proc TREC 2007:205–14 [Google Scholar]
- 15.Weeber M, Klein H, Aronson AR, et al. Text-based discovery in biomedicine: the architecture of the dad-system. Proc AMIA Symp 2000:903–7 [PMC free article] [PubMed] [Google Scholar]
- 16.Aronson AR, Bodenreider O, Chang HF, et al. The NLM indexing initiative. Proc AMIA Symp 2000:17–21 [PMC free article] [PubMed] [Google Scholar]
- 17.Kim GR, Aronson AR, Mork JG, et al. Application of a medical text indexer to an online dermatology atlas. Stud Health Technol Inform 2004;107:287–91 [PubMed] [Google Scholar]
- 18.Aronson AR, Mork JG, Gay CW, et al. The NLM indexing initiative's medical text indexer. Stud Health Technol Inform 2004;107:268–72 [PubMed] [Google Scholar]
- 19.Gay CW, Kayaalp M, Aronson AR. Semi-automatic indexing of full text biomedical articles. AMIA Annu Symp Proc 2005:271–5 [PMC free article] [PubMed] [Google Scholar]
- 20.Neveol A, Shooshan SE, Humphrey SM, et al. Multiple approaches to fine-grained indexing of the biomedical literature. Pac Symp Biocomput 2007:292–303 [PubMed] [Google Scholar]
- 21.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat Biotechnol 2006;24:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh Y, Hardardottir GA, Brennan PF. Linguistic analysis: terms and phrases used by patients in e-mail messages to nurses. Stud Health Technol Inform 2004;107:511–15 [PubMed] [Google Scholar]
- 23.Baud RH, Ruch P, Gaudinat A, et al. Coping with the variability of medical terms. Stud Health Technol Inform 2004;107:322–6 [PubMed] [Google Scholar]
- 24.Yetisgen-Yildiz M, Pratt W. The effect of feature representation on MEDLINE document classification. AMIA Annu Symp Proc 2005:849–53 [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou L, Srinivasan P. Concept space comparisons: explorations with five health domains. AMIA Annu Symp Proc 2005:874–8 [PMC free article] [PubMed] [Google Scholar]
- 26.Niu Y, Zhu X, Li J, et al. Analysis of polarity information in medical text. AMIA Annu Symp Proc 2005:570–4 [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz ME. Combining image features, case descriptions and UMLS concepts to improve retrieval of medical images. AMIA Annu Symp Proc 2006:674–8 [PMC free article] [PubMed] [Google Scholar]
- 28.Mougin F, Burgun A, Bodenreider O. Mapping data elements to terminological resources for integrating biomedical data sources. BMC Bioinformatics 2006;7(Suppl 3):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McInnes BT, Pedersen T, Carlis J. Using UMLS Concept Unique Identifiers (CUIs) for word sense disambiguation in the biomedical domain. AMIA Annu Symp Proc 2007:533–7 [PMC free article] [PubMed] [Google Scholar]
- 30.Pakhomov S, Shah N, Hanson P, et al. Automatic quality of life prediction using electronic medical records. AMIA Annu Symp Proc 2008:545–9 [PMC free article] [PubMed] [Google Scholar]
- 31.Fan JW, Friedman C. Word sense disambiguation via semantic type classification. AMIA Annu Symp Proc 2008:177–81 [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson O, Hurdle J. Building a domain information schema using semisupervised machine learning. AMIA Annu Symp Proc 2009:1000 [Google Scholar]
- 33.Lowe HJ, Barnett GO. MicroMeSH: a microcomputer system for searching and exploring the national library medicine's medical subject headings (mesh) vocabulary. Proc Annu Symp Comput Appl Med Care 1987:717–20 [Google Scholar]
- 34.Miller RA, Gieszczykiewicz FM, Vries JK, et al. CHARTLINE: pviding bibliographic references relevant to patient charts using the UMLS Metathesaurus knowledge sources. Proc Annu Symp Comput Appl Med Care 1992:86–90 [PMC free article] [PubMed] [Google Scholar]
- 35.Evans DA, Ginther-Webster K, Hart M, et al. Automatic indexing using selective NLP and first-order thesauri. Proc RIAO 1991;91:624–44 [Google Scholar]
- 36.Hersh WR, Greenes RA. SAPHIRE–an information retrieval system featuring concept matching, automatic indexing, probabilistic retrieval, and hierarchical relationships. Comput Biomed Res 1990;23:410–25 [DOI] [PubMed] [Google Scholar]
- 37.Nadkarni P, Chen R, Brandt C. UMLS concept indexing for production databases: a feasibility study. J Am Med Inform Assoc 2001;8:80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denny JC, Smithers JD, Miller RA, et al. “Understanding” medical school curriculum content using knowledgemap. J Am Med Inform Assoc 2003;10:351–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasan P. Retrieval feedback in MEDLINE. J Am Med Inform Assoc 1996;3:157–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harman D. Overview of the third text retrieval conference (TREC-3). In: Harman D, ed. TREC-3: proceedings of the third text retrieval conference. Washington, DC: Government Printing Office, 1994:1–19 [Google Scholar]
- 41.Kim W, Aronson AR, Wilbur WJ. Automatic MeSH term assignment and quality assessment. Proc AMIA Symp 2001:319–23 [PMC free article] [PubMed] [Google Scholar]
- 42.Shah NH, Bhatia N, Jonquet C, et al. Comparison of concept recognizers for building the open biomedical annotator. BMC Bioinformatics 2009;10(Suppl 9):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz AS, Hearst MA. A simple algorithm for identifying abbreviation definitions in biomedical text. Pac Symp Biocomput 2003:451–62 [PubMed] [Google Scholar]
- 44.http://dailymed.nlm.nih.gov/dailymed/about.cfm
- 45.Humphreys BL, McCray AT, Cheh ML. Evaluating the coverage of controlled health data terminologies: report on the results of the NLM/AHCPR large scale vocabulary test. J Am Med Inform Assoc 1997;4:484–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aronson AR, Bodenreider O, Demner-Fushman D, et al. From indexing the biomedical literature to coding clinical text: experience with mti and machine learning approaches. Proc BioNLP Workshop 2007:105–12 [Google Scholar]
- 47.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 48.Shooshan SE, Aronson AR. Ambiguity in the UMLS Metathesaurus: 2008 edition. Bethesda MD: National Library of Medicine, 2008. http://skr.nlm.nih.gov/papers/references/ambiguity08.pdf [Google Scholar]
- 49.Weeber M, Mork JG, Aronson AR. Developing a test collection for biomedical word sense disambiguation. Proc AMIA Symp 2001:746–50 [PMC free article] [PubMed] [Google Scholar]
- 50.Aronson AR, Mork JG, Neveol A, et al. Methodology for creating umls content views appropriate for biomedical natural language processing. AMIA Annu Symp Proc 2008:21–5 [PMC free article] [PubMed] [Google Scholar]
- 51.Wilbur WJ, Hazard GF, Jr, Divita G, et al. Analysis of biomedical text for chemical names: a comparison of three methods. Proc AMIA Symp 1999:176–80 [PMC free article] [PubMed] [Google Scholar]