Abstract
The authors developed a computerized algorithm that estimates ‘who transmits to whom’—that is, the likeliest transmission paths during an outbreak of person-to-person transmitted illness. This algorithm uses basic information about natural history of the disease, population structure, and chronology of observed symptoms. To assess the algorithm efficacy, the authors built a simulator with parameters describing the disease and the population to simulate random outbreaks of influenza. The algorithm's performance was compared with three reference methods that simulated how human operators would handle such situations. For any size of outbreak, the algorithm outperformed the reference methods and provided a higher proportion of cases for whom the source subject who transmitted infection was identified. The authors also illustrated applicability of the algorithm for describing outbreaks of influenza in nursing homes. The use of this algorithm to draw transmission maps in investigations of outbreaks with person-to-person transmitted agents could potentially guide public health measures regarding the control of such outbreaks.
Introduction
A timely investigation of an outbreak is critical in order to implement the most appropriate control measures. The investigation is based on a descriptive and analytical phase, which includes describing cases by time, place, and characteristics of infected subjects. Traditionally, the time course of an outbreak is depicted by drawing a histogram of the number of cases by their dates of onset: the epidemic curve. The epidemic curve provides information on the transmission pattern of the outbreak and helps identify the incubation period.1 In outbreaks of person-to-person transmitted agents, a more detailed analysis can be accomplished by drawing a chart where each case is represented in time, together with main related events, including contacts with other cases. This representation is sometimes called a transmission map2 and can be particularly useful when the outbreak occurs in closed environments. A transmission map will reveal when interhuman transmission might have been possible and eventually will help identify individuals still contagious during the time of the investigation.
Computerized algorithms have been developed in the hospital setting to help epidemiologists or infection control specialists in detecting and investigating outbreaks, and included expert systems to detect and report nosocomial infections3 4 and predictive models to identify patients at high risk for colonization or infection with an antibiotic-resistant pathogen.5 In the community setting, a huge amount has been published regarding the use of computerised algorithm to detect and track the spread of infectious diseases.6–11
However, we are not aware of a tool that helps identify ‘who transmits to whom’ during an outbreak and to draw the likeliest pathway of transmission between individuals. This information may have important implications for outbreaks monitoring and control.
Material and methods
Our work aimed at identifying transmissions of an infectious agent during an outbreak and building the chronology of these transmissions. We developed a computerized algorithm (hereafter named the WTW algorithm) that estimates the likeliest transmission path among subjects and the dates of transmission. The WTW algorithm uses basic information about natural history of the disease, and a dataset describing the population structure and the chronology of observed symptoms.
Parameterization of the WTW algorithm
The WTW algorithm is presented in discrete time and the time step is one day.
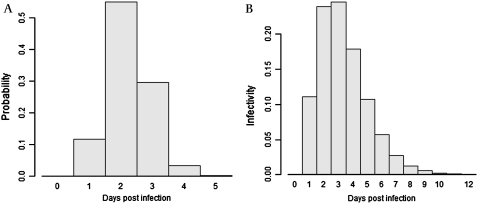
First, we define incubation and infectivity functions for the disease (figure 1). The incubation period is described by a function, incub (i, t), giving the probability for subject i to have been infected t days before the onset of his/her symptoms. Infectivity is described by a function, infec (i, t), giving the daily probability (a hazard function) that the agent would be transmitted from an infectious (i) to a susceptible subject according to the time since infection (t).
Figure 1.
Incubation period and daily infectivity. (A) Incubation period (time from infection to onset of symptoms) distribution. (B) Daily infectivity following infection.
We describe contacts between infected subjects by filling matrices Ci,j (d).
With:
{Subjects}: the set of subjects known to have been infected during the follow-up period
{Sources}: the set including {Subjects} and a dummy element representing infections coming from subjects not listed in the studied population
D: the set of dates describing the follow-up period.
Entries ci,j (d) of these matrices describe the intensity of contact between subjects i and j during date d. Their values weight intensity, frequency, and length of contacts between subjects, during which transmission of the infectious agent can occur. These values vary between 0 (no contact, transmission between subjects excluded) and 1 (maximum intensity contact, optimal condition for a transmission). These values may be collected for each individual distinctly (eg, contact between subjects i and j occurred in a single occasion at a given date), or for groups of individuals (eg, physicians of a ward have an average level of contact with every patient of the same ward during a given period of time).
Then, we define a set T of possible transmissions ti,j,d from subject i to subject j at date d.
A probability of occurrence a priori p0(i, j, d) is assigned to each transmission ti,j,d:
With:
di: the date of infection of subject i
dsj: the date of symptoms onset of subject j
infec(i, d –di), ci,j(d), incub(j, dsj –d), as described previously is set to its maximum value when i is the dummy element representing subjects not listed in the observed case series).
Assuming that k infected subjects have been observed, we developed an algorithm to find the element X* with coordinates , which minimises the function f:
where os is the subject who transmitted infections to subject s at date d.
The function f is applied to a finite set of coordinates, but the number of possibilities is of order k ^ (k×maximum duration of infectivity) and, even for small number of k, a combinatorial method has to be used. The set of candidate solutions X to the minimization of f is obtained by a modified branch and bound algorithm.12 Briefly, the branch and bound algorithm is a general method for finding optimal solutions of various optimization problems (in our case, finding the minimum value of a function f) with a systematic enumeration of all the candidate solutions. It requires a splitting procedure (‘branching’) that, given a set of candidate solutions, returns two or more smaller subsets whose union covers the set, and a second procedure (‘bounding’) that computes upper and lower bounds for the minimum value of f within each subset. The key idea of this algorithm is that if the lower bound for a subset A of candidate solutions is greater than the upper bound for some other set B, then A may be safely discarded from the search. The recursion stops when the current candidate subset is reduced to a single element.
In our study, the result produced can be either a single solution X* or a subset of solutions containing a chosen number of the best solutions. Solutions estimate not only ‘who transmits to whom’ but also the likeliest date of infection for each subject.
Outbreak simulation
To assess the efficacy of the WTW algorithm we built a simulator which used parameters describing the disease and the population to randomly simulate outbreaks.
We used incubation and infectivity functions of influenza13: incubation and infectivity were defined by γ functions (median 2.2 and 3.0 days, SD 0.6 and 1.7 days, respectively).
We simulated 600 scenarios, in a population of 40 subjects split into two to five groups, initially susceptible for the disease and followed up for 50 days. For each pair of groups, a random uniform value between 0 and 1 was drawn to describe average level of contacts within or between groups. For each pair of individuals, a random uniform value between 0 and 1 was drawn to introduce heterogeneity in contacts at an individual level. Entries of the Ci,j(d) matrices were set as the product of these two values. We introduced temporal variations (± 5%, approximately) of these entries between each time step.
Once the population was generated, the propagation of the infectious agent was simulated assuming that all subjects were susceptible. An index subject (defined as the first infected subject in the studied population) was randomly selected then became ill and infectious according to the defined incubation and infectivity functions. At each date d, a probability ptrans(i, j, d) of transmission between each infectious subject i and each susceptible subject j was calculated.
A transmission event was simulated if a random value drawn from a continuous uniform distribution was lower than the calculated probability. Thus, the same initial set of parameters to describe the population could generate several different outbreaks according to the random values generated during the simulation process. The simulation stopped when no infectious subject remained.
Evaluation of the WTW algorithm
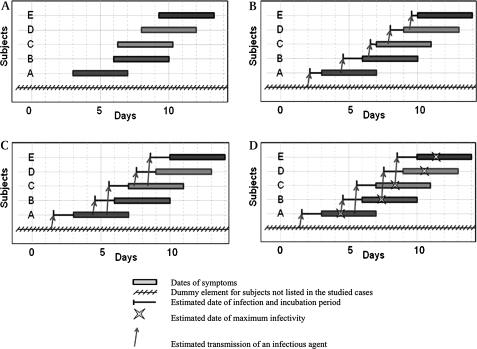
We defined three reference methods (see figure 2):
Figure 2.
Illustration of reference methods used to estimate source subjects and dates of infections. The mean incubation period is 1.5 days. The maximum infectivity is obtained 3 days from infection. (A) Data used by each method consists in dates of onset and end of symptoms for any subject (rectangles). (B) The ‘naive’ method M1 assigned a sequence of transmissions identical to the sequence of cases. Dates of infection are chosen equal to the mean of two consecutive dates. (C) Method 2 integrates as additional constraint the fact that subject i cannot transmit infection during the latent period (equal to the incubation time in this example, 1.5 days). (D) Method 3 takes into account the period between infection and the maximum infectivity of subject (3 days from infection) in addition to the previous method.
The ‘naive’ method (M1) sorted subjects from their dates of onset of symptoms, and assigned a sequence of transmissions identical to the sequence of cases: each subject was supposedly infected by the previous one. Dates of infection were chosen equal to the mean of two consecutive dates of onset of symptoms. Note that this method failed to account for incubation time, varying infectivity, and heterogeneity of contacts.
The second method (M2) integrated as additional constraint the fact that subject i cannot transmit infection before a given amount of time corresponding to the latent period. For the sake of simplicity in our example we assumed that the latent period was similar to the incubation time, so that the subject i could not transmit the infectious agent before onset of his/her symptoms. The subject i at the origin of infection was, thus, among those already symptomatic when j was infected, the most lately infected. Note that this method still failed to account for varying infectivity and heterogeneity of contacts.
The third method (M3) took into account the period between infection and the maximum infectivity of subject in addition to the previous method. After assessing the most probable date of infection according to subject j's onset of symptoms, subject i whose infectivity was the highest on this date (according to the time after infection) was retained as source of transmission. Note that this method did not account for heterogeneity of contacts.
The performances of the WTW algorithm were compared with the three reference methods. The WTW algorithm was run assuming that contacts by groups and at an individual level were known. The first outcome measure was, for each scenario, the proportion of infected subjects for whom the source subject (the subject who transmitted infection) was properly estimated. A second outcome measure was the mean absolute difference (in days) between simulated and predicted dates of infections.
Application to real data
The WTW algorithm was applied to real data collected in a cluster-randomized controlled trial evaluating the effectiveness of staff influenza vaccination on mortality of nursing homes residents.14 In this study, a detailed investigation of each outbreak of influenza-like illness (ILI) was conducted. The presence of type A influenza virus infections was confirmed by rapid diagnostic tests (Quick View Influenza Test, Quidel Corp, San Diego, CA, USA). In order to use the WTW algorithm, contact matrices were parameterized according to characteristics of groups (geographic area of the units and places of dining rooms for residents, main places of work for staff teams) and individual characteristics of residents (mobility dysfunctions, dependency, and importance of daily needs for help from staff, personal habits, and references of the other residents often met, frequency of visits from family and dates of absence) and staff (personal schedule, dates of awayness). The definitions of incubation and infectivity functions used were those previously described (figure 1).
Entries of the contact matrices regarding the dummy element for subjects not listed in the observed cases series were entered differently for members of the staff, who were supposed to spend half of their time outside the nursing home and to meet infectious individuals, and for residents, who had limited contacts with other people outside the nursing home.
Results
Simulated data
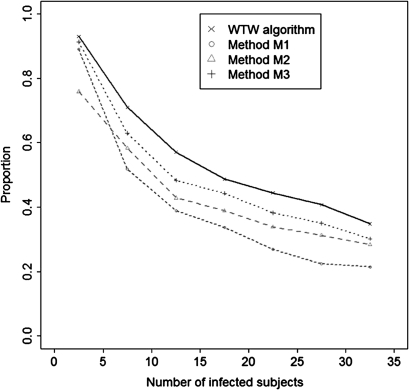
The proportion of infected subjects for whom the source subject was identified was greater with the WTW algorithm than with the other three reference methods (table 1). The proportion decreased with the size of the outbreak, irrespectively of the method used (figure 3). For outbreaks of size 2 to 10, 80% of source subjects were correctly identified with method 1, 72% with method 2, 85% with method 3, and 88% with the WTW algorithm. For larger outbreaks with 11 to 40 cases, the respective values were 27%, 34%, 38%, 43%. The mean absolute differences between simulated and predicted dates of infections were lower in the WTW algorithm and with method 3 than with other methods.
Table 1.
Performances of the WTW algorithm and of other heuristically developed methods (see text and figure 2 for explanation) to retrieve the source subject and the date of infection of cases during simulated outbreaks
| Simulations | Size* | 2–10 subjects | 11–40 subjects | Total |
| 4 (2–6) | 24 (19–31) | 17 (4–27) | ||
| Number | 200 | 400 | 600 | |
| Method M1 | Source† | 0.80 (0.67–1.00) | 0.27 (0.18–0.35) | 0.47 (0.22–0.67) |
| Dates‡ | 1.19 (0.62–1.67) | 2.10 (1.97–2.25) | 1.77 (1.50–2.16) | |
| All§ | 0.52 (0.00–1.00) | 0.00 (0.00–0.00) | 0.19 (0.00–0.00) | |
| Method M2 | Source† | 0.72 (0.50–1.00) | 0.34 (0.26–0.40) | 0.48 (0.29–0.60) |
| Dates‡ | 0.62 (0.42–1.00) | 0.66 (0.57–0.74) | 0.65 (0.50–0.78) | |
| All§ | 0.38 (0.00–1.00) | 0.00 (0.00–0.00) | 0.14 (0.00–0.00) | |
| Method M3 | Source† | 0.85 (0.67–1.00) | 0.38 (0.29–0.45) | 0.55 (0.33–0.75) |
| Dates‡ | 0.35 (0.00–0.50) | 0.39 (0.32–0.47) | 0.38 (0.29–5.00) | |
| All§ | 0.58 (0.00–1.00) | 0.00 (0.00–0.00) | 0.22 (0.00–0.00) | |
| WTW algorithm (best solution) | Source† | 0.88 (0.75–1.00) | 0.44 (0.34–0.51) | 0.60 (0.38–0.82) |
| Dates‡ | 0.37 (0.00–0.52) | 0.44 (0.37–0.51) | 0.41 (0.32–0.50) | |
| All§ | 0.63 (0.00–1.00) | 0.00 (0.00–0.00) | 0.23 (0.00–0.00) | |
| WTW algorithm (10 solutions) | Source† | 0.87 (0.75–1.00) | 0.44 (0.34–0.51) | 0.60 (0.38–0.83) |
| Dates‡ | 0.40 (0.24–0.54) | 0.44 (0.37–0.51) | 0.43 (0.33–0.52) | |
| All (any sol.)¶ | 0.93 (0.97–1.00) | 0.48 (0.37–0.56) | 0.64 (0.42–1.00) | |
| All (same sol.)** | 0.73 (0.00–1.00) | 0.09 (0.00–0.00) | 0.27 (0.00–1.00) |
Number of infected subjects for each simulation: mean (IQR).
Proportion of infected subjects for whom the source subject was identified: mean (IQR).
Mean absolute difference between simulated and predicted dates of infections (in days): mean (IQR).
Proportion of simulations for which all the source subjects were identified: mean (IQR).
Proportion of simulations for which all the source subjects were identified at least once among any of the 10 best solutions proposed by the WTW algorithm: mean (IQR).
Proportion of simulations for which all the source subjects were simultaneously identified at least once among the 10 best solutions proposed by the WTW algorithm: mean (IQR).
Figure 3.
Mean proportion of infected subjects for whom the source subject was properly estimated, according to the number of infected subjects in the simulations (by range of five subjects).
When the 10 likeliest sequences of transmissions were selected using the WTW algorithm, the source subject was properly estimated at least once among this set of 10 solutions in 66% of simulated infections on average and the source subject and dates of infection were simultaneously adequately identified in 40% of simulated infections.
Application to real data
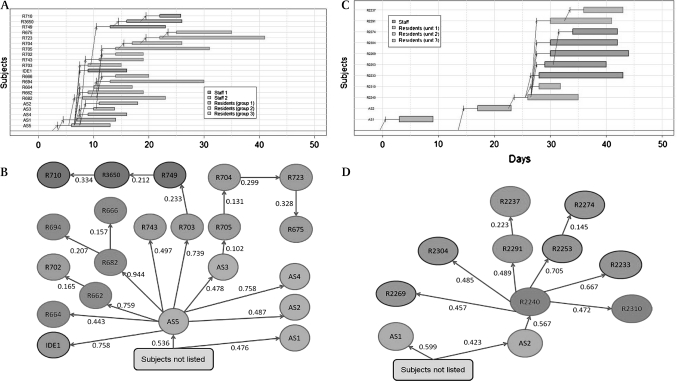
The transmission maps of two out of the four outbreaks investigated during the trial of influenza vaccination in nursing home staff are shown in figure 4. These ILI clusters were reported in two nursing homes between February 8 and March 13, 2007, in the control arm. In nursing home A, the index patient was a staff member (AS5). He/she was presumably the source of spreading of ILI among residents in units 3 (eg, resident R682–probability 0.996) and 2 (eg, resident 703–probability 0.962) and other staff members (eg, member AS4–probability 0.987), whereas the source of spreading in unit 1, was presumably the infection of resident R749 by resident R703 (probability 0.454). Some other transmissions were more difficult to identify: among the 10 likeliest solutions retrieved by the WTW algorithm, infection of resident R705 by staff member AS3 (probability 0.198) was retained five times (including in the likeliest solution), while infection by R682, R703, or R662 (probabilities 0.168, 0.124, 0.164, respectively) was retained in 3, 1, and 1 solution(s), respectively. According to the likeliest estimate, the probabilities for each transmission retained in the transmission map of nursing home A varied from 0.102 to 0.759, with a mean value of 0.405.
Figure 4.
Illustrations of the WTW algorithm: transmission maps and estimated probability for each transmission of influenza between cases during outbreaks of influenza in two nursing homes. (A) Transmission map in nursing home A. (B) Estimated probability for each transmission (at dates displayed in figure 4A) in nursing home A. (C) Transmission map in nursing home B. (D) Estimated probability for each transmission (at dates displayed in figure 4C) in nursing home B.
In nursing home B, the index patient was a staff member but was very unlikely to have transmitted infections to another observed case (probabilities that AS2 was contaminated by AS1 or by someone not listed in the observed case series: 14.0% and 86.0%, respectively).
Discussion
The WTW algorithm may be a helpful tool for investigating outbreaks. Its relative performances may however depend on the natural history of the disease under study. Although we chose to simulate influenza outbreaks, with a latent period and an incubation time that almost completely overlapped, we believe that the WTW algorithm should be even more powerful in investigating outbreaks of diseases with a latent period shorter than the incubation time (eg, measles, chickenpox…). In this case, it is likely that a human operator would have a greater difficulty for estimating a transmission map from data on observed symptoms only. Moreover, the WTW algorithm includes parameters describing encounters between subjects, which are major determinants in the sequence of transmissions in diseases with long periods of incubation or infectivity (eg, AIDS, viral hepatitis).
A critical question is how the algorithm would deal with missing data and how robust it is to the degree of ‘missingness’ and the type of missing data. When precise information is lacking on contacts, the algorithm can be easily adapted by introducing uninformative constant values as entries in the contact matrix. In a worst-case scenario in which no data on contacts would be available for any subject, the results given by the WTW algorithm would be close to those given by method 3. When unobserved cases are potential sources of transmissions, the algorithm will attribute these potential sources to the ‘dummy element’ that represents subjects not listed in the studied population.
The WTW algorithm can provide other estimates of the natural history of the disease under study. Once a solution has been obtained in terms of transmission path, calculation of the serial interval—that is, the average time between onsets of symptoms in index case and secondary cases, is straightforward. By conducting a sensitivity analysis on incubation time and infectivity, one could explore how departures from baseline hypotheses regarding these parameters modify the likeliest transmission path.
Finally, the WTW algorithm provides information on the dynamics of an outbreak, and can help identify sources of infection in order to take appropriate preventive measures. The algorithm can be generalized to all contexts in which the three components––natural history parameters, contacts, and observed dates of events—can be provided. In the field of human medicine, users of the algorithm should be hygienists, and epidemiologists devoted to outbreak investigations. It can be used even with scarce data at the beginning of an outbreak to give approximate estimations, whose accuracy will improve throughout the outbreak, as new data become available. This ‘real-time’ use may contribute to identify spreaders or groups at higher risk of transmission (according to previous infections). If interventions are implemented to mitigate spreading (eg, isolation of infected subjects), they can easily be introduced in the algorithm (by varying matrix entries for these subjects) to help real-time identification of new sources of infection. We believe that our algorithm could help monitor outbreaks and therefore complete previously existing computerized surveillance tools or expert systems devoted to infections tracking.
The WTW algorithm could help guide public health decisions during an outbreak, whether it occurs in a closed setting or in the community. This algorithm uses data and gives estimations individually for a limited number of subjects, and would not be applicable in a wide community epidemic context. However, it would be all the more useful as the outbreak occurs in a closed population, where contacts with several potential sources of infection could occur (and where the pathway of transmission is difficult to derive manually), whereas, in the community, contacts between a source and a susceptible subject are often resumed to a binary indicator (yes/no) and the pathway of transmission straightforward.
We encourage using such an algorithm to draw transmission maps in investigation of outbreaks of person-to-person transmitted agents. To help use the WTW algorithm, we provide an online appendix with a step-by-step example.
Supplementary Material
Acknowledgments
We thank Dr Rebecca Freeman-Grais and Pierre-Loïc Assayag for their helpful comments and careful reading of the manuscript, and Magali Lemaitre and Pierre-François Kouyami for their help in collecting the data in the nursing homes.
Footnotes
Funding: The Nursing Home Trial was supported by a grant from Ministere de la Sante et la Direction des Hopitaux (PHRC 2005, AOM#05050) and was sponsored by Assistance Publique des Hopitaux de Paris. Other funders: Ministere de la Sante et la Direction des Hopitaux, Assistance Publique des Hopitaux de Paris.
Competing interests: None.
Provenance and peer review: Not commissioned–externally peer reviewed.
References
- 1.Reingold AL. Outbreak investigations—a perspective. Emerg Infect Dis 1998;4:21–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Boer MGJ, Bruijnesteijn van Coppenraet LES, Gaasbeek A, et al. An outbreak of Pneumocystis jiroveci pneumonia with 1 predominant genotype among renal transplant recipients: interhuman transmission or a common environmental source? Clin Infect Dis 2007;44:1143–9 [DOI] [PubMed] [Google Scholar]
- 3.Evans RS, Larsen RA, Burke JP, et al. Computer surveillance of hospital-acquired infections and antibiotic use. JAMA 1986;256:1007–11 [PubMed] [Google Scholar]
- 4.Kahn MG, Steib SA, Spitznagel EL, et al. Improvement in user performance following development and routine use of an expert system. Medinfo 1995;8(Pt 2):1064–7 [PubMed] [Google Scholar]
- 5.Evans RS, Wallace CJ, Lloyd JF, et al. Rapid identification of hospitalized patients at high risk for MRSA carriage. J Am Med Inform Assoc 2008;15:506–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klompas M, Haney G, Church D, et al. Automated identification of acute hepatitis B using electronic medical record data to facilitate public health surveillance. PLoS One 2008;3:e2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meurer WJ, Smith BL, Losman ED, et al. Real-time identification of serious infection in geriatric patients using clinical information system surveillance. J Am Geriatr Soc 2009;57:40–5 [DOI] [PubMed] [Google Scholar]
- 8.Moore K. Real-time syndrome surveillance in Ontario, Canada: the potential use of emergency departments and telehealth. Eur J Emerg Med 2004;11:3–11 [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention (CDC) Automated detection and reporting of notifiable diseases using electronic medical records versus passive surveillance–Massachusetts, June 2006-July 2007. MMWR Morb Mortal Wkly Rep 2008;57:373–6 [PubMed] [Google Scholar]
- 10.Ginsberg J, Mohebbi MH, Patel RS, et al. Detecting influenza epidemics using search engine query data. Nature 2009;457:1012–14 [DOI] [PubMed] [Google Scholar]
- 11.Temime L, Opatowski L, Pannet Y, et al. Peripatetic health-care workers as potential superspreaders. Proc Natl Acad Sci USA 2009;106:18420–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silver E. An overview of heuristic solution methods. J Oper Res Soc 2004;55:936–56 [Google Scholar]
- 13.Carrat F, Vergu E, Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol 2008;167:775–85 [DOI] [PubMed] [Google Scholar]
- 14.Lemaitre M, Meret T, Rothan-Tondeur M, et al. Effect of influenza vaccination of nursing home staff on mortality of residents: a cluster-randomized trial. J Am Geriatr Soc 2009;57:1580–6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.