INTRODUCTION
Synthetic biology is the application of engineering and mathematical principles to develop novel biological devices and circuits. What separates synthetic biology from traditional molecular biology is the development of standardized interchangeable DNA “parts,” just as advances in engineering in the nineteenth century brought about standardized railroad gauges and screw threads ( Campbell [2005] offers a brief review of the emergence of synthetic biology as a distinct field). Through the use of these “parts,” laboratories around the world can easily exchange and combine biological constructs to develop novel “machines” for a variety of applications ( Ferber, 2004, Purnick and Weiss, 2009), including medicinal chemistry ( Martin et al., 2003; Tsuruta et al., 2009) and genetic engineering ( Hasty et al., 2002; Sprinzak and Elowitz, 2005).
While synthetic biology can trace its roots to large research universities, over the last five years the field has emerged as an important area for collaborations among science, technology, engineering, and mathematics (STEM) disciplines at primarily undergraduate institutions as well. Undergraduate student research has become an integral part of synthetic biology, in large part because of the National Science Foundation–sponsored undergraduate student research Jamboree in 2004 ( Campbell, 2005). This meeting evolved into the annual International Genetically Engineered Machine (iGEM) competition held annually at the Massachusetts Institute of Technology (MIT), which attracts undergraduate student researchers from around the world to design biological devices that address a wide range of problems related to biology, agriculture, energy, the environment, and mathematics (http://2010.igem.org/Main_Page).
A common problem faced by primarily undergraduate institutions is the lack of funding and material support needed to adequately expose students to modern biology, including synthetic biology. To help alleviate this problem, the Genome Consortium for Active Teaching (GCAT) was founded in 2000 by Malcolm Campbell at Davidson College to bring genomics into the undergraduate curriculum. GCAT's first tangible activity was to serve as a central clearinghouse both for the purchase and reading of DNA microarrays and for information on how to execute genomics experiments at undergraduate institutions. In response to the evolution of molecular biology in the last decade, Campbell, along with Davidson colleague Laurie Heyer and collaborators Todd Eckdahl and Jeff Poet of Missouri Western State University, organized a Howard Hughes Medical Institute (HHMI)-sponsored GCAT workshop at Davidson in July of 2010. This workshop explored how faculty from multiple disciplines could work together to bring synthetic biology to the undergraduate classroom and laboratory. The workshop was attended by a biologist and nonbiologist pair of faculty from 15 primarily undergraduate institutions, with the goal that each pair could begin the preparations necessary to bring multidisciplinary synthetic biology activity to their respective institutions (see Figure 1 for the organizers and participants). The nonbiologists attending the meeting had expertise in a diverse group of STEM fields including chemistry, biochemistry, physics, mathematics, statistics, computer science, and engineering.
Figure 1.
GCAT Synthetic Biology Workshop participants, organizers, and HHMI representatives. Top row (left to right): Laurie Heyer, Jeff Poet, Jeff Matocha, Nathan Reyna (back), Malcolm Campbell, Qiang Shi, and Kathy Ogata; second row: Nighat Kokan, Robert Jonas, Santiago Toledo, Vidya Chandrasekaran, Valerie Burke, Yixin Yang, Andrea Holgado, and Anil Pereira; third row: Todd Eckdahl, Susmita Acharya, Consuelo Alvarez, Paul Hemler, Michael Wolyniak, Libby Shoop, and Paul Overvoorde; fourth row: Nathan Reyna, Matthew Tuthill, Carl Salter, Chris Jones, Robert Morris, Tom Twardowski, Joyce Stamm, and Talitha Washington; bottom row: all attendees, Theresa Grana, Leo Lee, Jodi Schwarz, and Teresa Garrett.
MEETING EVENTS AND THEMES
The workshop started with discussions on the fundamental principles of synthetic biology, the suitability of synthetic biology for multidisciplinary research with undergraduate students, the connection and communication between biology and collaborating disciplines necessary for success, and the elements that make up a successful synthetic biology project. The discussions were followed by an introduction to the Biobrick standard assembly scheme as the foundation of synthetic biology and the Registry of Standard Biological Parts, a community resource maintained at MIT in which developed or developing parts are listed (http://partsregistry.org/Main_Page). To apply the lessons of these discussions, each pair of faculty worked with the Registry to design a “lava lamp,” a DNA construct that would allow bacteria to fluoresce and float in response to chemical inputs.
Some of the workshop sessions were broken up into biologist and nonbiologist groups to provide an opportunity for discussion among faculty from common disciplines. In these discussions, strategies for identifying areas of common interest and expertise from two different disciplines and developing successful interdisciplinary communication were emphasized.
The ethical issues of designing synthetic biology systems were briefly discussed by both the whole group and in small sessions over lunch with the workshop organizers as well as bioethicists from Davidson College. Given that this session was coincidentally held at the same time that congressional hearings on this very subject were being held on Capitol Hill, the topic was especially relevant to the future of synthetic biology and will no doubt become more significant as the general public becomes more aware of the field.
“Wet lab” time was also provided in which basic PCR and oligonucleotide assembly projects were conducted to provide a taste of the workbench “nuts-and-bolts” of synthetic biology work. This portion of the meeting was especially valuable to the foundation of collaboration between faculty of different disciplines, because the biologist member of each pair was charged with explaining the principles and practice of the techniques to the nonbiologist, and the nonbiologist, in turn, provided context about how such techniques could possibly be used in the construction of a biological project that could also address their particular research interests.
The remainder of the meeting was dedicated to the development of potential synthetic biology project ideas for undergraduate research and classroom purposes based on the interests and expertise of each faculty pair as well as the skills learned from the workshop. A wide range of projects was proposed for classroom and laboratory-based synthetic biology work that may be explored in the online listing of GCAT Synthetic Biology Workshop Results/Project Presentations (www.bio.davidson.edu/projects/gcat/workshop_2010/workshop_2010_results.html).
The proposed projects reflected the diversity of academic interests among meeting participants as well as their efforts to pursue ideas that would be stimulating to both the biologist and the nonbiologist. For instance, one team made up of a chemist and a biologist proposed exploring environmental problems and issues, specifically water contamination by heavy metals. Another team with a computer scientist developed a project to engineer a biological “wire” capable of propagating a chemical signal down its length. A third group with a mathematician would like to develop mathematical models of a system in which bacteria express varying levels of a signaling molecule. The versatility of synthetic biology, as well as its many areas of applications that remain under development, proved appealing to all of the nonbiologists in attendance.
Several key themes emerged repeatedly throughout the workshop. First, synthetic biology requires a new way of thinking for most scientists. It requires one to view problems through the lens of an engineer and consider issues of standardization, modularity, abstraction, and modeling and their applicability to the problem at hand. This field is an excellent pedagogical and research tool to foster interdisciplinary work and learning. However, successful interdisciplinary work requires the collaborators to have frequent communication, to learn how to speak the language of each others' disciplines, and to think about problems in novel ways.
Synthetic biology is incredibly flexible. That is, almost any scientific interest can be accommodated within the field, making it relatively easy to find projects that require contributions from all members of a multidisciplinary team. This is essential because if any member of the team feels that their role in the overall project is not important, the project ultimately may not work. This flexibility also makes synthetic biology particularly accessible and stimulating for faculty and undergraduate students at small institutions. In most cases, individual faculty members are the sole representative of their academic specialty in the department. Having a way to leverage the interests, knowledge, and excitement of colleagues with whom there would normally be little to no communication provides a stimulating opportunity for new directions in research and teaching. This causes faculty and students to be forced out of intellectual comfort zones. Exposing students to other approaches and ways of thinking as part of a hands-on research experience will give students an unparalleled appreciation of the value and realities of interdisciplinary thinking, an asset that will be increasingly valuable to them in the future.
Over several years and after leading several cohorts of undergraduates in the successful generation of iGEM projects, the workshop organizers developed an overall set of goals for the projects they developed with their students:
Everyone (faculty and students) should learn new things.
Everyone should have fun.
The project should contribute to the knowledge base in synthetic biology.
As mentioned more than once during the workshop, these goals are in listed in order of importance. If project participants are learning and enjoying what they are doing, then they are succeeding. Based on the success that combined Davidson/Missouri Western teams have had in preparing projects for the annual iGEM jamboree, it seems that their focus on the first two aforementioned goals has enabled them to consistently achieve the third as well.
RESOURCES FOR THE DEVELOPMENT OF SYNTHETIC BIOLOGY PROJECTS
To facilitate the transition for international scientists between ongoing traditional research and synthetic biology multidisciplinary research, a number of resources were presented at the meeting:
GCAT Listserv
The GCAT Listserv (known as GCAT-L) is an email discussion list that connects faculty members using genomic methods in their undergraduate courses. Messages sent to GCAT-L by any one of its subscribers are distributed to all other subscribers. Information on how to subscribe and use GCAT-L is posted at www.bio.davidson.edu/projects/gcat/GCAT-L.html.
Wiggio/Wiki
To facilitate communication between collaborators on different campuses, Wiggio.com is an online toolkit that is freely available on the Internet and that allows file sharing and editing, management of group calendars, posting of links, setting up conference calls, online chat, and sending text, voice, and email messages to group members. Group members can define how each is informed about upcoming group activity. The toolkit can be accessed at http://wiggio.com. In addition, the GCAT community Wiki has been set up for use by the GCAT community and is maintained by its users; it can be accessed at http://gcat.davidson.edu/GcatWiki/index.php/Main_Page.
GCAT Mini-Registry
A mini-registry was provided to workshop participants that contained 10 DNA constructs that are present in the Registry of Standard Biological Parts. The constructs included promoters, ribosome binding sites (RBS), double terminators, red fluorescent protein (RFP), and green fluorescent protein (GFP).
GCATalog
This web-based catalog, developed by Bill Hatfield at Davidson along with Laurie Heyer and Malcolm Campbell, is freely available and is optimized for use in synthetic biology applications. Using GCATalog (http://gcat.davidson.edu/GCATalog/), synthetic biology users can generate a publicly accessible freezer inventory that allows the synthetic biology community to share commonly used Biobricks as well as other biological constructs.
Open WetWare
An Open WetWare page was created to promote sharing of unpublished work and protocols between scientists engaged in biological engineering (http://openwetware.org/wiki/Main_Page). This page has information related to labs working in synthetic biology, course and teaching resources, protocols, and a continually updated blog. Another related website highlighted as a repository of synthetic biology tools was that of Drew Endy's laboratory in the Department of Bioengineering at Stanford University (http://openwetware.org/wiki/Endy_Lab). This Open WetWare-linked laboratory website has information on ongoing research, publications, and detailed descriptions of projects and tools generated.
Registry of Standard Biological Parts Help Page
As previously mentioned, a registry of standardized parts to build synthetic biology devices is available at http://partsregistry.org/Main_Page. New users may wish to take advantage of the Help page available from the Registry in which detailed instructions related to Biobricks, including their assembly and the registration of newly developed parts, are explained. (http://partsregistry.org/Help:Contents).
WORKSHOP ASSESSMENT
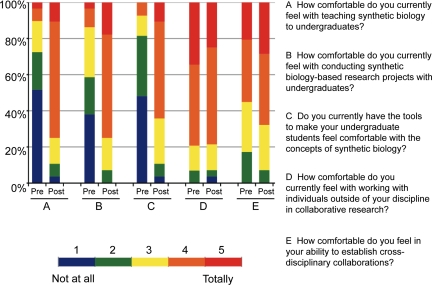
To gauge the effectiveness of the workshop in providing the resources and creative sparks necessary for developing synthetic biology projects in the classrooms and laboratories of its participants, anonymous pre- and postassessment surveys were conducted. The two surveys compared the participants' pre- and postworkshop perceptions of synthetic biology as a distinct field, as an area for multidisciplinary collaboration, and as a viable option for their classroom and research programs. The results revealed a general feeling of excitement and improved understanding about synthetic biology (see Figure 2), with the majority agreeing with the comment from one participant who stated that “I now feel confident that I understand the basic ins and outs of synthetic biology—what it is and isn't—as well as how I can implement projects in this area with my students.”
Figure 2.
Comparison of preassessment and postassessment of the GCAT workshop by its participants.
Interestingly, although 75% of workshop attendees had previous experience in basic molecular biology laboratory work, data analysis, and experimental design, only 25% had previously engaged in a multidisciplinary collaboration. This trend was reflected in the postassessment of the meeting as well, as many felt that the forging of collaborative projects that satisfy the intellectual curiosities of faculty from disparate disciplines stood as the most significant challenge for the successful implementation of synthetic biology projects. Several participants noted that the meeting was, in the words of one attendee, “important to meet and establish a network of colleagues,” and it became apparent to most participants that the establishment of collaborations between different disciplines and between different institutions of all sizes will be essential to successfully develop synthetic biology projects that expose students to the collaborative and multidisciplinary nature of modern scientific research. Indeed, the pursuit of multidisciplinary collaborations engendered by this workshop reflects the need for academics from different disciplines to join forces to share ideas and resources in their educational endeavors as obstacles including scarcity of individual college resources and increased competition for research funding threaten to curtail the development of scientific educational opportunities for students ( Dodson et al., 2010).
INCORPORATING SYNTHETIC BIOLOGY IN RESEARCH
Research in synthetic biology is an excellent way to bridge the STEM disciplines in a way that is accessible to students. This type of research is well suited for undergraduate involvement as evidenced by the increasing number of teams competing at iGEM (129 teams registered for 2010 as of July versus 112 teams in 2009 and 84 in 2008). While the majority of these teams are from research universities, primarily undergraduate institutions such as Davidson College and Missouri Western State University have been successful at the competition and have also published some of their work in peer-reviewed journals ( Haynes et al., 2008; Jordan et al., 2009). Also, many undergraduate institutions are already well positioned to pursue projects in synthetic biology because only basic computer and molecular biology resources are needed and the necessary reagents are relatively inexpensive. While iGEM participation allows access to the entire library of available Biobricks, participation in this program may be cost-prohibitive for many institutions. The GCAT community is currently exploring strategies to address this issue but notes that in the meantime it is relatively simple to construct plasmids using Biobrick strategies.
There are a number of approaches that may be taken to develop research programs in synthetic biology, which range from student-generated and self-contained summer research projects to long-term faculty-developed projects designed for student involvement over a number of years. To develop bona fide interdisciplinary projects, it is important to create the time and space for idea generation. If possible, the organizers suggested attending iGEM as an observer to see first-hand the wide range of approaches to synthetic biology. Closer to home, exploring previous iGEM projects online and the synthetic biology resources GCAT has to offer can also provide a good starting point.
A successful interdisciplinary synthetic biology collaboration depends on the communication achieved by scheduling regular meetings between partners. The Davidson/Missouri Western team achieves this by holding weekly biology/math colloquia with interested students via Wiggio.com, which connects the campuses as mentioned above. Over time, professors and students gain an appreciation for the interconnectedness of different disciplines and, most importantly, have fun in the collaborative process.
INCORPORATING SYNTHETIC BIOLOGY IN THE CLASSROOM
The presentations and exercises from the workshop sparked many conversations about ways to use this material in the classroom. The accessibility of parts and the willingness of the synthetic biology community to share knowledge and, potentially, resources with beginners have most participants preparing lab experiences for the coming academic year and beyond.
Brainstorming about ways to make a bacterial “lava lamp” got most of the workshop participants excited to think of the variety of tasks that bacteria can be induced to perform. The designing of a classroom project with a long-range engineering goal in mind could be a very good way to enlist students' creative talents. Whether or not it ever got built, contemplating the feasibility of such a project may elicit significant student buy-in of synthetic biology concepts.
Once students have taken ownership of the overall picture and seized upon a long-term goal, there are innumerable things to try, from straightforward to Byzantine. For example, one might examine the dose response of an inducible promoter by developing a promoter + ribosome binding site + GFP plasmid and measuring fluorescence as a function of inducer concentration. In parallel, students could develop a mathematical model that would predict the promoter's effects over a hypothetical range of inducer concentrations. Cis-trans effects could be examined by using two plasmids, one carrying kanamycin resistance as well as one structure of interest and another with ampicillin resistance and the other structure. Other variables could include bacterial strain, ribosome binding site, distance between the various components, or promoters, and all of these variables invite the introduction of mathematical modeling and analysis to gain further insight from these studies. Much new learning could occur with the available Biobricks, or students could synthesize new ones to test their pet ideas. It seems that the only limitation is the imagination of the student. Any new constructs could be entered into the Registry of Standard Biological Parts and the GCATalog for sharing with the community at large.
SUMMARY
Synthetic biology is a newly emerging field in which costs are relatively low and the value of student input can be high. It rewards tackling the sort of interdisciplinary problems that are increasingly important for students' professional futures but are often difficult to undertake from traditional disciplinary towers. Undergraduate students have shown both interest and ability in pursuing this research: The results of 5 years of iGEM jamborees amply testify to the potential for undergraduate success in this endeavor.
The organizers are considering the possibility of offering additional faculty workshops in the future. The participants of the 2010 meeting found this experience to be a focused, effective, and fun introduction to synthetic biology for those who want to explore this exciting new field. Hopefully, these same experiences will continue to be shared through future workshops as an effort to create opportunities in synthetic biology for faculty and their students.
Supplementary Material
ACKNOWLEDGMENTS
In addition to the organizers and participants, two representatives from the HHMI Science Education Alliance, Grants and Special Programs Office were present as observers during the entire workshop. They were the Director, Tuajuanda C. Jordan, and the Program Officer, Lucia P. Barber. Furthermore, Cathryn Westra, a Davidson student majoring in Science Writing, observed the meeting and created a blog of her observations at http://gcat2010.wordpress.com/. The pre- and postassessments conducted for this report were approved before their administration by the Human Subject Review Committees of Hampden-Sydney College and Longwood University. We thank the organizers of the 2010 GCAT Synthetic Biology workshop, Malcolm Campbell and Laurie Heyer from Davidson College and Todd Eckdahl and Jeff Poet from Missouri Western State University. This workshop was possible due to HHMI grant % 52006292.
REFERENCES
- Campbell, 2005.Campbell M. A. Meeting report: synthetic biology jamboree for undergraduates. Cell Biol. Educ. 2005;4:19–23. doi: 10.1187/cbe.04-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson, 2010.Dodson M. V., et al. Perspectives on the formation of an interdisciplinary research team. Biochem. Biophys. Res. Commun. 2010;391:1155–1157. doi: 10.1016/j.bbrc.2009.11.040. [DOI] [PubMed] [Google Scholar]
- Ferber, 2004.Ferber D. Microbes made to order. Science. 2004;303:158–161. doi: 10.1126/science.303.5655.158. [DOI] [PubMed] [Google Scholar]
- Hasty et al., 2002.Hasty J., McMillen D., Collins J. J. Engineered gene circuits. Nature. 2002;240:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- Haynes, 2008.Haynes K. A., et al. Engineering bacteria to solve the Burnt Pancake Problem. J. Biol. Eng. 2008;2:1–12. doi: 10.1186/1754-1611-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, 2009.Jordan B., et al. Solving a Hamiltonian Path Problem with a bacterial computer. J. Biol. Eng. 2009;3:11. doi: 10.1186/1754-1611-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin et al., 2003.Martin V. J., Pitera D. J., Withers S. T., Newman J. D., Keasling J. D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Purnick and Weiss, 2009.Purnick P. E. M., Weiss R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- Sprinzak and Elowitz, 2005.Sprinzak D., Elowitz M. B. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- Tsuruta, 2009.Tsuruta H., et al. High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS ONE. 2009;4:e4489. doi: 10.1371/journal.pone.0004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.