Abstract
Modified bases in nucleic acids present a layer of information that directs biological function over and beyond the coding capacity of the conventional bases. While a large number of modified bases have been identified, many of the enzymes generating them still remain to be discovered. Recently, members of the 2-oxoglutarate- and iron(II)-dependent dioxygenase superfamily, which modify diverse substrates from small molecules to biopolymers, were predicted and subsequently confirmed to catalyze oxidative modification of bases in nucleic acids. Of these, two distinct families, namely the AlkB and the kinetoplastid base J binding proteins (JBP) catalyze in situ hydroxylation of bases in nucleic acids. Using sensitive computational analysis of sequences, structures and contextual information from genomic structure and protein domain architectures, we report five distinct families of 2-oxoglutarate- and iron(II)-dependent dioxygenase that we predict to be involved in nucleic acid modifications. Among the DNA-modifying families, we show that the dioxygenase domains of the kinetoplastid base J-binding proteins belong to a larger family that includes the Tet proteins, prototyped by the human oncogene Tet1, and proteins from basidiomycete fungi, chlorophyte algae, heterolobosean amoeboflagellates and bacteriophages. We present evidence that some of these proteins are likely to be involved in oxidative modification of the 5-methyl group of cytosine leading to the formation of 5-hydroxymethyl-cytosine. The Tet/JBP homologs from basidiomycete fungi such as Laccaria and Coprinopsis show large lineage-specific expansions and a tight linkage with genes encoding a novel and distinct family of predicted transposases, and a member of the Maelstrom-like HMG family. We propose that these fungal members are part of a mobile transposon. To the best of our knowledge, this is the first report of a eukaryotic transposable element that encodes its own DNA-modification enzyme with a potential regulatory role. Through a wider analysis of other poorly characterized DNA-modifying enzymes we also show that the phage Mu Mom-like proteins, which catalyze the N6-carbamoylmethylation of adenines, are also linked to diverse families of bacterial transposases, suggesting that DNA modification by transposable elements might have a more general presence than previously appreciated. Among the other families of 2-oxoglutarate- and iron(II)-dependent dioxygenases identified in this study, one which is found in algae, is predicted to mainly comprise of RNA-modifying enzymes and shows a striking diversity in protein domain architectures suggesting the presence of RNA modifications with possibly unique adaptive roles. The results presented here are likely to provide the means for future investigation of unexpected epigenetic modifications, such as hydroxymethyl cytosine, that could profoundly impact our understanding of gene regulation and processes such as DNA demethylation.
Keywords: DNA methylation, dioxygenases, mom, transposons, bacteriophage, AlkB, hydroxymethylcytosine, demethylation, algae, RNA modification, CXXC domain
Introduction
Catalytic modification of bases in nucleic acids is universally observed across the three primary superkingdoms of life and is the basis for a wide range of biological functions.1 Certain modifications of bases in rRNAs and tRNAs, such as methylation, thiouridylation and pseudouridylation, are traceable to the last universal common ancestor of all life and are essential for survival.2,3 Other RNA base modifications are more limited in their distribution. For example, wybutosine is found only in archaeal and eukaryotic tRNAs, whereas certain forms of methylation and thiouridylation show even more narrow phyletic distributions.2,3 In contrast, DNA base modifications are apparently less diverse and more sporadic in their distribution; enzymes catalyzing them are not essential in most lineages of life.4 This difference is potentially attributable to the constraint of needing to maintain double-helical pairing in DNA and protecting the genetic material from the potentially mutagenic effects of base modifications.5
The most common DNA modification in prokaryotes, methylation of cytosine or adenine, is primarily catalyzed by methylases encoded by mobile restriction-modification (RM) systems.4,6 These methylases have a predominantly defensive role in immunizing the host DNA against the activity of the restriction endonucleases, which cleave invading DNA, such as those of bacteriophages.7,8 In certain prokaryotes, DNA methylation additionally supplies an epigenetic mark for DNA repair.9 Eukaryotes too possess several distinct DNA cytosine methylases related to the bacterial RM methylases. These have been shown to have a role in chromatin organization, regulatory gene silencing, repression of selfish DNA elements, and possibly other epigenetic processes in several animals, fungi and plants.10–13 DNA modifications other than methylation are primarily known from caudate bacteriophages and include a spectacular array of modified bases such as 5-hydroxymethylpyrimidines and their mono- or di-glycosylated derivatives, α-putrescinylated or α-glutamylated thymines, sugar-substituted 5-hydroxypentyluracil, and N6-carbamoylmethyl adenines (called Momylation after the mom enzyme of phage Mu).8,14 These atypical modifications are used by phages in countering the host DNA restriction response. Other DNA base modifications have become apparent in eukaryotes. The simplest of these is deamination of cytosine that appears to be mainly involved in diversification of immunity molecules in vertebrates.15–17 Another well-studied eukaryotic modification is the formation of β-D-glucosyl-hydroxymethyluracil or base J from thymine in euglenozoans, including the parasites Trypanosoma and Leishmania.18
While enzymes catalyzing several of the major RNA modifications have been biochemically well-characterized and crystallized, fewer DNA-modifying enzymes have been studied in detail. Of the latter, the well-studied ones are DNA methylases, namely the 5-methylcytosine-generating methylases of both bacteria and eukaryotes and the bacterial N6-methyladenine-generating enzymes.19–22 Additionally, the classic T-even phage modification system comprised of the 5-hydroxymethylcytosine (hmC) synthase and glucosyltransferases that further modify this base have been characterized. These studies have revealed that phage 5-hydroxymethylcytosine and 5-hydroxymethyluracil synthases are derived from the classical thymidylate synthases, which are often encoded by several DNA viruses, including these T-even phages.23 Thus, phage 5-hydroxymethylpyrimidines are not derived by direct DNA modifications but by an incorporation of pre-modified base during viral DNA synthesis. On the other hand phage DNA bases glycosyltransferases (that modify the 5-hydroxymethylpyrimidines) are members of the glycogen synthase/glycogen phosphorylase fold (e.g., alpha-glucosyltransferase and beta-glucosyltransferase) or the Fringe-like glucosyltransferase (e.g., beta-glucosyl-HMC-alpha-glucosyltransferase), which directly modify the hmC in DNA.24,25 Likewise, the phage mu enzyme, Mom, directly modifies adenines in DNA by adding a carbamoylmethyl or a related adduct, and was recently shown to belong to the GCN5-like acetyltransferase fold.26 Other recent studies have shown that the first step in the synthesis of base J in trypanosomes, i.e., oxidation of the methyl group on thymine to generate 5-hydroxymethyluracil, occurs in situ in DNA. This reaction is catalyzed by JBP1 and JBP2, enzymes of the 2-oxoglutarate- and iron(II)-dependent dioxygenase (2OGFeDO) superfamily, and represent the first example of in situ oxidative modification of methylpyrimidines, in contrast to the T-even phage hmC generation pathways in which premodified bases are incorporated into DNA.27,28
Other enzymes of the 2OGFeDO superfamily catalyze a variety of oxidative reactions such as:
(1) Oxidation of carbons in an aromatic ring to generate phenolic groups; e.g., hydroxylases in flavonoid synthesis.29 (2) Oxidation of aliphatic and alicyclic carbons e.g., amino acid modifications in proteins, namely hydroxylysine and hydroxyproline catalyzed respectively by lysyl and prolyl hydroxylases.30 (3) Ring opening/closing via C-N and C-S bond formation; e.g., isopenicillin synthase.31 (4) Oxidation of C-C bond in side-chains linked to an aromatic ring; e.g., thymine-7-hydroxylase, which oxidizes thymine to carboxyuracil in the thymidine/uridine salvage in fungi and bacteria.32 Trypanosome JBP1 and JBP2 also catalyze this class of reaction, albeit on DNA rather than the free base.27,28 (5) Demethylation of N-CH3 side chains linked to heterocyclic aromatic rings. This is typified by the AlkB family that functions in DNA repair by reversing methyl adducts on bases (e.g., N6-methyladenine) produced by DNA alkylating agents via complete oxidation of the methyl group to formaldehyde.33,34 Although both AlkB and JBP1/2 operate on methyl groups on bases in DNA their catalytic domains are only distantly related. This suggested that there might be as yet undetected enzymes that catalyze the oxidative modification of DNA in this superfamily. Most enzymes of this superfamily that act on low-molecular weight substrates are standalone proteins with compact dioxygenase domains. However, those that act on biopolymers like nucleic acids and proteins are frequently fused to other nucleic-acid- or protein-interacting domains (e.g., Swi2/Snf2 ATPase module in JBP2,35 and the MYND finger in Egl-9 like prolyl hydroxylases34). Alternatively, they contain peculiar conserved inserts within the catalytic domain that help in binding their biopolymer targets (e.g., AlkB).36 We accordingly hoped to utilize these features as contextual information in a computational protocol to identify potentially novel members of the 2OGFeDO superfamily that catalyze in situ oxidative modifications of nucleic acids.
Results and Discussion
Prediction and classification of novel nucleic-acid-modifying 2OGFeDO domains
We first identified novel 2OGFeDO domains by means of iterative profile searches with the PSI-BLAST program using several seeds including versions of these domains from JBP1 and JBP2, AlkB, prolyl hydroxylases and several low molecular weight compound dioxygenases, such as the thymine-7-hydroxylase and isopenicillin synthase. In some cases these profile searches converged rather rapidly; hence, we improved the profiles via further searches of the protein sequence database of uncultured microbes from environmental samples. For example, a search of the NCBI non-redundant (NR) database using the 2OGFeDO domains of JBP1 and JBP2 converged within 3 iterations. However, upon searching the NCBI environmental sample database, we identified numerous homologous proteins potentially derived from uncultured marine organisms. Including these hits in the profile for a renewed search of the NR database resulted in the detection of homologous oxygenase domains in the gp2 proteins from the mycobacteriophages Cooper and Nigel and a related prophage integrated into the genome of Frankia alni (e-values < 10−4). Further iterations of these searches recovered homologous regions in the 3 paralogous human oncogenes Tet1 (CXXC6), Tet2 and Tet3,37,38 and their orthologs found throughout metazoa (e < 10−5). These searches also recovered a vast expansion of homologous domains from the mushrooms Laccaria bicolor and Coprinopsis cinerea, smaller expansions in the chlorophyte algae Chlamydomonas reinhardtii and Volvox carteri with significant e-values (e < 10−5). Searches against a panel of eukaryotic proteomes using the profile generated from the above search also recovered few representatives from the heterolobosean amoebofla-gellate Naegleria, the stramenopile algae Aureococcus, Emiliania, Phaeodactylum and Thalassiosira, and the chlorophyte algae Ostreococcus and Micromonas. In reciprocal PSI-BLAST searches these versions consistently recovered each other prior to recovering any other member of the 2OGFeDO superfamily, suggesting that they formed a distinctive family comprised of JBP1/2, the animal Tet proteins and their homologs.
Likewise, profile searches with the other queries also recovered a large number of previously undetected 2OGFeDO domains. To identify versions amongst these, which potentially act on nucleic acids, we used a library of sequence profiles for domains involved in nucleic acid metabolism and chromatin function and scanned all the newly detected 2OGFeDO domain-containing proteins for fusions to any of these domains. As result we identified conserved fusions to different DNA-associated domains such as SAD(SRA), R3H, DNA glycosylase, Swi2/Snf2 ATPase and TAM(MBD),11 and also several RNA-associated domains such as the RRM, pseudouridine synthase, pyrmidine carboxylase fold and RNA methylase domains.2 Additionally, some of the proteins with 2OGFeDO domains more closely related to JBP1/2 were linked in the same polypeptide to the DNA-binding CXXC domain and the chromatin-associated chromodomain. Additional evidence for a possible role in nucleic acid modification was also obtained through systematic analysis of gene neighborhoods and genomic organization (see below for details).
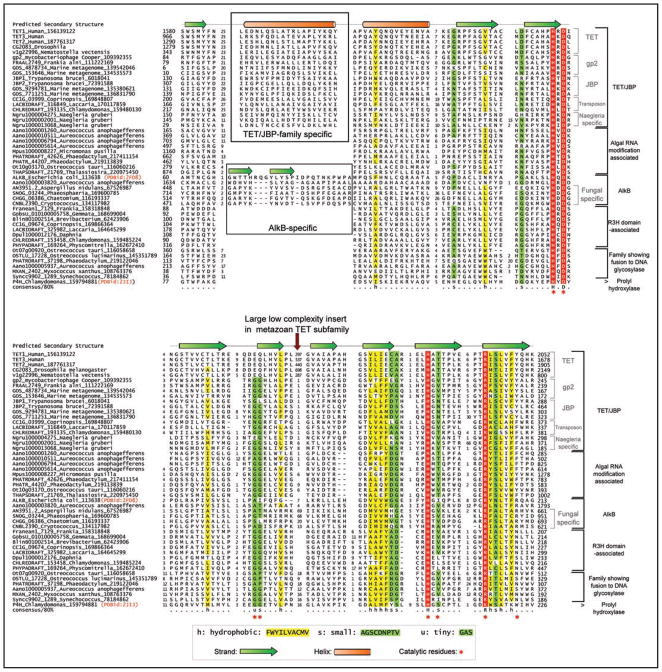
We then clustered these proteins using the BLASTCLUST program and further refined these clusters based on conserved, shared sequence signatures and predicted structure features, and domain architectures to identify 5 distinct families. We aligned each of these families and an examination of their conservation patterns (Fig. 1) showed that they typically conserved: (1) The HxD signature (where ‘x’ is any amino acid), which chelates Fe(II) and is associated with the extended region after the first core strand. (2) A pair of small residues at the end of the strand immediately downstream of the HXD motif, which helps in positioning the active site arginine. (3) The HXs (where ‘s’ is a small residue) in the penultimate conserved strand, in which the H chelates the Fe(II) and the small residue contacts the 2-oxo acid cofactor. (4) The RX5a/R signature (where ‘a’ is usually an aromatic residue: F, Y, W) in the last conserved strand of domain. The first R in this motif forms a salt bridge with the 2-oxo acid and the aromatic residue or second arginine helps in positioning the first metal-chelating histidine (Fig. 1).34 We also generated HMMs for each of the families using their multiple alignments and performed a profile-profile comparison of these HMM against a library of HMMs generated for all structurally characterized domains (i.e., domains from PDB) using the HHpred program. These searches uniformly recovered known 2OGFeDO structures (e.g., prolyl hydroxylase, 2jij or AlkB, 2fd8) with significant p-values (p < 10−12) and an alignment spanning all the key catalytic residues (Fig. 1). Together, these observations indicated that we had indeed identified several novel 2OGFeDOs predicted to oxidatively modify nucleic acids.
Figure 1.
Multiple alignment of selected examples of the newly predicted families of the nucleic-acid-modifying 2-oxoglutarate- and iron(II)-dependent dioxygenase superfamily. Protein sequences are represented by their gene names, species names and GenBank index numbers (where available). Temporary gene names were assigned for predicted proteins from Naegleria, Aureococcus, Daphnia and Micromonas. The full length protein sequences from these are available in the Supplementary material. The coloring scheme and consensus abbreviations are shown in the key. Family names are shown to the right of the alignment. The distinct inserts of the TET/JBP and the AlkB families are shown within boxes. The key conserved residues defining the 2OGFeDO protein have been marked below the alignment. The consensus secondary structure derived from crystal structures of characterized members of the superfamily is shown above.
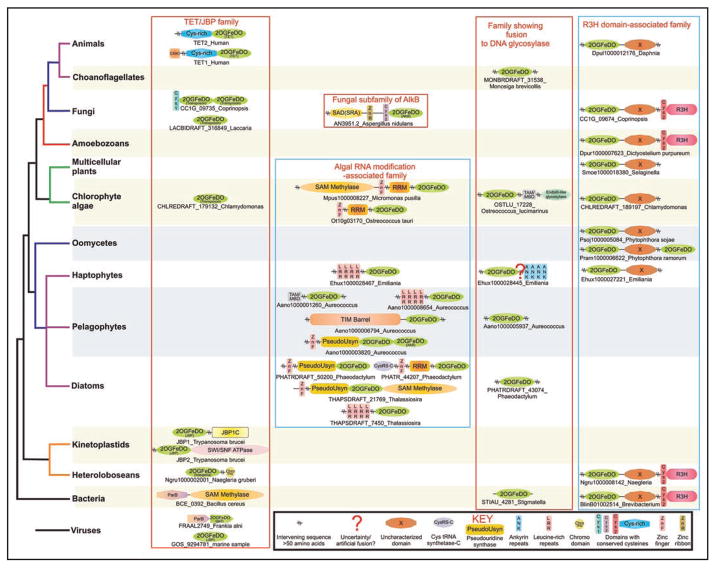
We outline below the 5 sub-families along with their inferred evolutionary histories and predicted functional features based on domain architectures and other forms of contextual information such as genomic organization and gene neighbors (Table 1).
Table 1.
2-OG-Fe(II)-Dependent dioxygenase families predicted to be involved in nucleic acid modification.
| Family; subfamily | Phyletic patterns | Comments |
|---|---|---|
| Tet/JBP family; | Kinetoplastids; | JBP2 is fused to a Swi2/Snf2 ATPase module. |
| JBP subfamily | Uncharacterized versions in marine microbes | JBP1 has an uncharacterized conserved C-terminal domain with absolutely conserved polar residues. |
| Tet/JBP family; TET subfamily | Metazoans | Contains an extension with 8 conserved cysteines and 1 histidine just N-terminal to the core double stranded β-helix of the 2OGFeDO catalytic domain. The vertebrate Tet1 and Tet3 and their orthologs from all other animals show a fusion of the 2OG-Fe(II) oxygenase domain with a N-terminal CXXC domain, a binuclear Zn chelating domain with 8 conserved cysteines and 1 histidine. Contain a large low complexity insert within the catalytic domain predicted to have a predominantly unstructured conformation. |
| Tet/JBP family; transposon-associated subfamily | Basidiomycete fungi: Laccaria, Coprinopsis, Postia, Moniliophthora Chlorophytes: Chlamydomonas, Volvox |
Encoded by a transposons that also contain genes encoding a distinctive transposase and HMG domain protein related to the HMG domain of Maelstrom and another uncharacterized ORF. Some versions from Coprinopsis are fused to or occur adjacent to a gene encoding a novel N-terminal cysteine cluster. |
| Tet/JBP family; Naegleria-specific subfamily | Naegleria | A group of 8 paralogous proteins of which one shows an N-terminal fusion to a chromo domain. |
| Tet/JBP family; bacteriophage gp2 subfamily | Actinophages Cooper and Nigel of Mycobacteria and Frankia prophage; several uncultured viruses from marine samples | gp2 shows a conserved gene neighborhood association with a gene encoding the chromosome segregation associated ParB protein close to the origin of replication of the bacteriophage chromosome. |
| The algal RNA-modification associated family | Stramenopiles: Aureococcus, Phaeodactylum, Thalassiosira, Emiliania Chlorophytes: Ostreococcus, Chlorella, Micromonas |
Fusions to RRM, classical zinc finger with CCHC signature, cysteinyl tRNA synthetase C-terminal domain, pseudouridine synthase, RNA methylase, TIM-barrel hydrolase/decarboxylase, AlkB family 2OGFeDO, TAM(MBD) and Leucine-rich repeats domains. |
| AlkB family; Fungal subfamily | Fungi | Fused to N-terminal SAD(SRA), Zinc ribbon and a distinct cysteine-rich Zn-chelating domain. |
| R3H domain-associated family | Bacteria: actinobacteria, planctomycetes, proteobacteria Eukaryotes: heterolobosea, oomycetes, haptophytes, chlorophyte algae, mosses, amoebozoa, fungi, crustaceans |
Members of this family are usually fused to an uncharacterized domain immediately C-terminus to the 2OGFeDO domain. Many members of this family show a further C-terminal fusion to a cysteine-rich CXCXXC motif followed by a version of the R3H domain. |
| Family showing domain fusion to DNA glycosylases | Eukaryotes: Monosiga, Ostreococcus, Chlorella, Micromonas, Phaeodactylum, Aureococcus, Emiliania, Bacteria: Synechococcus, Myxococcus, Stigmatella | In Ostreococcus there is a fusion to the MBD4 ortholog comprising of a methylated-DNA binding TAM(MBD) domain and a DNA glycosylase domain of the EndoIII superfamily. This family is unified by a “WW” motif found in the strand just N-terminal to the HXD motif of the core catalytic domain. |
The Tet/JBP family
This family is defined by all 2OGFeDOs that are closer to the kinetoplastid JBP proteins and the metazoan Tets than any other family of dioxygenases. They are characterized by a shared derived character (synapomorphy) in the form of an extended α-helix just N-terminal to the first core strand (Fig. 1). This long α-helix appears to be kinked in most members of the Tet/JBP family by a conserved proline in the middle of the helix. This family can be divided into 5 distinct subfamilies (Table 1). At least a subset of members of each of the families shows either fusions to DNA-binding or chromatin-associated protein domains, or gene-neighborhood/genome-context associations with known DNA-binding domains (Table 1, Fig. 2). This strongly supports a role for most members of the family in modifying DNA. In the first experimentally studied subfamily of this group, the JBP subfamily, JBP2 is fused to a Swi2/Snf2 ATPase module, which is consistent with the role for ATP-dependent chromatin reorganization in synthesis of the J base in kinetoplastid DNA.35 The kinetoplastid JBP1 is fused to a previously uncharacterized C-terminal domain, which is also present as a solo version in other uncharacterized kinetoplastid proteins. Its predicted secondary structure indicates an α + β topology and it contains several strongly conserved polar residues including an absolutely conserved GGTRY motif (Suppl. material). This implies that it could possess an uncharacterized enzymatic activity or could constitute a specific base J-binding domain.
Figure 2.
Representative domain architectures of the newly identified versions of nucleic-acid-modifying 2OGFeDO proteins. Architectures are arranged by their phylogenetic and family affinities, and are labeled by their gene and species names. Domain architectures within a family are boxed. Domains are typically denoted by their standard names. Non-standard domain nomenclatures are clarified in the inset key at the bottom of the figure. Operons are shown as arrows where the arrow head points from the 5′ to the 3′ direction of the coding frame of the gene. Gene neighborhoods are labeled by the gene coding for the 2OGFeDO protein.
The DNA-binding CXXC domain found in the Tet subfamily proteins also occurs in several chromatin proteins, including the animal DNA methylase DNMT1 and the methylated DNA-binding protein MBD111,39,40 (Fig. 2). Given the domain architectural parallel to the DNMT1 methyltransferase and the precedence of the pyrimidine modification catalyzed by the related JBP subfamily we proposed that this subfamily might catalyze oxidative modification of 5-methylcytosines in animal DNA. Further experimental characterization of the human Tet1 protein showed that it indeed catalyzes this reaction to generate 5-hydroxymethylcytosine both in vitro and in cells. Studies based on overexpression of Tet1 in cultured human cells support its role in potential demethylation of 5-methylcytosines directly or indirectly via this oxidative intermediate.38 Based on the crystal structure of the AlkB protein (PDB: 2fd8) we observed that the unique cysteine-rich extension found at the N-terminus of the Tet subfamily 2OGFeDOs (Table 1) is likely, in part, to occupy a position similar to the N-terminal DNA-binding extensions of the AlkB protein.36 Hence, we speculate that this domain might similarly be involved in forming a DNA-recognition surface. The low complexity insert in the core double stranded β-helix of the Tet subfamily is exactly in the same position as an unstructured insert seen in the prolyl hydroxylases (PDB: 2JIJ, Fig. 1) and is inferred to be located on the exterior surface on one side of the 2OGFeDO catalytic domain. Its persistence across the entire Tet subfamily, despite lack of sequence conservation, suggests that it might form a generalized protein-protein interaction surface. In most members we also identified one or more high confidence sumoylation sites in this insert suggesting that the Tet family might be regulated through this protein modification.
In the gnathostome lineage, after the divergence of agnathan vertebrates, the Tet subfamily underwent a triplication to spawn the Tet1, Tet2 and Tet3 genes, which are conserved in all gnathostomes. Of these Tet1 and Tet3 retained the ancestral CXXC domain, whereas Tet2 appears to have lost the CXXC domain. However, analysis of the chromosome neighborhood shows that a standalone CXXC domain protein (CXXC4) is encoded in the same chromosome as a neighboring gene usually in the opposite orientation. This suggests that in Tet2 a local chromosomal inversion detached the ancestral CXXC domain-encoding exon to form a separate gene. In light of this we speculate that CXXC4 could possibly function as an independent protein interacting with Tet2 in a complex. Given the regulation of CXXC4 by the Wnt pathway,41 it would be interesting to investigate if this might constitute a specific regulatory mechanism feeding into Tet2 enzymatic action. The exon-intron structure of the Tet family is also largely retained across animals. Except Tet2, in all cases the first conserved coding exon encodes the CXXC domain, which probably represents the ancestral gene bearing the CXXC domain that was fused to the 2OGFeDO domain-coding segment. Thus, the Tet progenitor appears to have been acquired prior to the divergence of extant metazoans and underwent a gene-fusion event with the progenitor of the N-terminal exon encoding the CXXC domain. The sequence between the CXXC and the 2OGFeDO domains is a large low complexity region, which is extremely fast evolving and shows poor conservation of exon-intron boundaries across metazoa. This region is also subject to insertions of microsatellite DNA repeats as seen in Tet1 of the platypus, where it appears to have been incorporated into the coding sequence (Suppl. material). In zebrafish Tet3 this region shows a large insertion of an integrated retrovirus into the intron following the CXXC exon. These observations suggest that this intervening low-complexity region between the CXXC domain and the catalytic module appears to be under little evolutionary constraint. The catalytic module comprising of the cysteine-rich extension and the 2OGFeDO domain are encoded by 7 highly conserved exons. However, within the low complexity insert in the 2OGFeDO domain there is considerable variability in exon-intron boundaries and exon numbers.
The expansions of the Tet/JBP homologs in mushrooms and algae define a distinct subfamily where, with a few exceptions, most representatives are standalone proteins (Table 1, Figs. 2 and 3). However, their genomic context suggests that they are genes within a novel DNA transposon, which appears to have proliferated in some of these organisms (See below). The bacteriophage gp2 subfamily (Table 1) is found close to the viral origin of replication associated with a gene encoding the ParB protein, which belongs to a superfamily of DNA-binding proteins implicated in bacterial and phage chromosome segregation.42 Typically, the bacteriophage chromosome origins are enriched in genes related to chromosome-segregation, partitioning and packaging, suggesting that gp2 might interact with the ParB protein in these functions. In other phages the ParB protein shows fusions with DNA methylases, and these enzymes have been implicated in regulation of replication or chromosome partitioning in enter-obacteriophages such as P1.43 Hence, the actinophage gp2 might modify methylated bases or reverse their methylation to regulate chromosome partitioning in these viruses. Given the presence of 5-hmC in DNA of other phages, it is possible that, like Tet1, these phage proteins oxidize 5-methylcytosine to 5-hmC directly on DNA. On the whole the Tet/JBP family shows a sporadic distribution: in prokaryotes it is restricted to certain bacteriophages of the caudoviral group or their prophage derivatives integrated in various genomes. The bacteriophage gp2s are the smallest versions of these proteins and represent more-or-less the minimal 2OGFeDO catalytic domain. Hence, they could potentially be the ancestral versions, which spawned the different eukaryotic versions through lateral transfer. The four remaining eukaryotic subfamilies of the Tet/JPB family are not particularly closely related to each other in terms of sequence similarity or domain architectures and have very patchy phyletic patterns (see Table 1). Thus, the eukaryotic versions could have emerged either via multiple transfers from the bacteriophage/bacterial source and might have also disseminated via cross-species transposition within eukaryotes (see below).
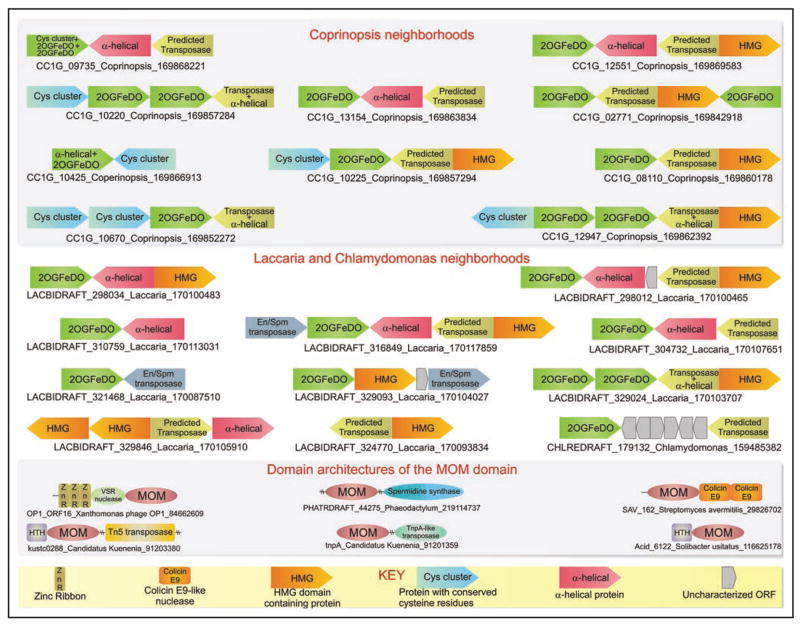
Figure 3.
Genomic organization and domain architectures of predicted transposons encoding DNA-modifying enzymes. Genes are depicted as arrows with the arrow head pointing from the 5′ to the 3′ direction of the coding sequence. Gene neighborhoods of the predicted transposase are typically labeled with the gene name of the 2OGFeDO containing protein, the species name and the gi number. In potential fragmentary elements where the 2OGFeDO is absent, the gene neighborhood is labeled with the gene name of the predicted transposase-containing gene. The key at the bottom of the figure explains non-standard domain and gene names, while other gene and domains names are as commonly used in literature.
The algal RNA-modification associated family
This family is defined by the presence of a synapomorphic tryptophan just N-terminal to the strand prior to the first helix of the core catalytic domain (Suppl. material). In sequence searches, these proteins also tend to recover the Tet/JBP family prior to any other version of the 2OGFeDO superfamily, suggesting that there might be distant relationship between the two families. This family is currently found only in two phylogenetically distinct groups of algae, namely chlorophytes and stramenopiles such as diatoms, pelagophytes and haptophytes. This phyletic pattern suggests that they probably emerged in the primary endosymbiotic photosynthetic chlorophytes and were transferred to stramenopiles during the one or more endosymbiotic associations with the primary photosynthetic lineages. Almost all of these proteins show fusions of the 2OGFeDO domain to domains related to RNA-binding or enzymatic domains involved in RNA metabolism (Table 1 and Fig. 2). The two independent fusions of these 2OGFeDO domains to RNA methylases (Fig. 2) suggests that, like the JBP/Tet family, they might also catalyze further modification of methylated bases, possibly generating a hydroxymethyl derivative like hmC or hmU. Further, fusions to the cysteinyl tRNA synthetase C-terminal domain, which recognizes the anticodon of tRNACys44 and the pseudouridine synthase which modifies tRNA45 suggests that these enzymes catalyze the formation of hydroxylated bases unique to the tRNAs of algae. 2OGFeDO domains of this family are also fused to a TIM-barrel domain, which belongs to a superfamily that includes decarboxylases and amidohydrolases.46 This enzyme could hence potentially act in conjunction with the 2OGFeDO domain in catalyzing removal of a methyl group on a base through an oxidation-decarboxylation mechanism that was proposed earlier for the thymine-7 hydroxylase.32 A member of this family from the pelagophyte Aureococcus contains an interesting C-terminal fusion to a second 2OGFeDO domain, which is however of the AlkB family (see below), suggesting that it might catalyze two distinct oxidative modifications. Members of this family in haptophytes, pelagophytes and diatoms are also fused to a distinctive leucine-rich repeat domain (Fig. 2). The domain architectural diversity of this family (Fig. 2) suggests that it has radiated to catalyze a range of unique RNA modifications, which might have a distinctive adaptive role unique to these algae. In contrast to the majority of these fusions, a representative of this family from Aureococcus (Fig. 2, Suppl. material) is fused to the methylated-DNA binding TAM(MBD) domain. This fusion implies that like Tet1, this protein might catalyze the synthesis of hmC from methylated cytosine in DNA.
The AlkB family
The AlkB family has been previously described and has been extensively characterized both in biochemical and structural terms.34,47 In addition to the versions acting on DNA, we had also described versions from RNA viruses and eukaryotes that are likely to act on RNA. Representatives of the latter group from animals are fused to RNA methylase domains.2 This family is defined by the synapomorphy in the form of a conserved arginine in place of the usual aromatic residue at the end of the C-terminal strand of the core domain34 (Fig. 1). Members of this family also have a unique β-hairpin insert, just N-terminal to the first helix of the core catalytic domain, which interacts with nucleic acids36 (Fig. 1). In this study we discovered a novel subfamily of AlkB proteins in fungi. While the classical AlkB proteins, which act on DNA are not combined with any additional specific DNA-binding domains, this fungal subfamily is typified by a remarkable fusion to multiple N-terminal domains (Table 1 and Fig. 2), including a SAD(SRA) domain.48 Some exemplars of the SAD(SRA) domains have been shown to specifically recognize DNA with methylated cytosines;49 however, representatives of this AlkB subfamily are found in fungi, such as Cryptococcus neoformans, which apparently lack any known DNA cytosine methylation system (Suppl. material). Further, all characterized representatives of the AlkB family appear to function on N-methylated bases rather than C-5 methylated pyrimidines.47 Hence, this version of the SAD(SRA) domain might not recognize 5-methylcytosine, but perhaps some other methylated base. The presence of additional N-terminal DNA-binding domains in this subfamily suggests that, unlike classical AlkBs, it might bind either specific DNA sequences or distinctive DNA structures. Hence, unlike the regular AlkB proteins which repair methylated-DNA in a non-specific manner, members of this subfamily might be involved in a localized DNA repair via recognition of specific sequences or structures of DNA. In evolutionary terms, this fungal AlkB subfamily appears to have been derived through duplication and divergence of the ancestral fungal AlkB, prior to the divergence of ascomycetes and basidiomycetes.
The R3H domain-associated family
This novel family identified in the current study is defined by a RxxW signature at the N-terminus of the first helix of the core catalytic domain and an additional conserved domain just C-terminal to the 2OGFeDO domain (Fig. 2, Suppl. material). This conserved C-terminal domain contains a strongly-conserved signature in the form of HxY and GxD motifs at the N-terminus and a GNxG motif followed by a conserved tyrosine at the C-terminus. This strongly conserved pattern suggests that the domain is likely to be enzymatic. These two N-terminal domains are usually further linked in the same polypeptide to a C-terminal cysteine cluster, predicted to chelate Zn, and a R3H domain (Table 1). R3H domains have been shown to bind both single stranded DNA and RNA,50 indicating that this family is likely to act on bases in single stranded nucleic acids, probably in conjunction with the unknown activity catalyzed by the second conserved domain. Interestingly, this family shows a very patchy phyletic pattern, being found in several phylogenetically distant eukaryotes and bacteria (Table 1). For example, it is only found in Daphnia amongst animals or only in Phytophthora among stramenopiles, but none of their close sister groups among completely sequenced genomes. However, it is fairly widespread in the fungi suggesting that it was at least present in the common ancestor of ascomycetes and basidiomycetes. On the whole the phyletic pattern suggests that the family has either undergone extensive lateral transfer between certain lineages and/or multiple instances of gene loss. This family is also notable for lineage-specific expansions in certain lineages, such as the crustacean Daphnia, the mushroom Coprinopsis, the heterolobosean amoeboflagellate Naegleria and the moss Selaginella (Table 1). Multiple copies have often emerged via local gene-duplications and show no evidence for any association with a conserved transposase or transposon encoded proteins. Such expansions are typical of families that provide an adaptive value by being present in multiple diversified copies, usually in the context of counter-pathogen strategies or detoxification of diverse environmental compounds.51 Hence, a possible role for these proteins could be in the defense against viral nucleic acids or genomic parasites via a novel oxidative modification of nucleic acids.
The DNA glycosylase associated family of 2OGFeDO domains
This family is prototyped by proteins from species of the chlorophyte alga Ostreococcus (e.g., OSTLU_17228), which combine a novel version of the 2OGFeDO domain with a C-terminal DNA glycosylase module. The DNA glycosylase module is orthologous to the animal MBD4,52 and like it combines a EndoIII-superfamily DNA glycosylase domain with a divergent TAM(MBD) domain (Fig. 2). However, the fusion with the 2OGFeDO domain appears to be a lineage-specific one that is not represented in multicellular plants. The domain architecture suggests that the 2OGFeDO domain might function in conjunction with the DNA glycosylase domain, with the former domain oxidatively modifying a base and the latter probably carrying out excision of the modified base. Given that members of this family are also found in photosynthetic stramenopile algae and cyanobacteria (Table 1, Fig. 3), it is possible that they were first acquired by the primary endosymbiotic plant lineages from the cyanobacteria and subsequently transmitted to stramenopiles. However, these versions are usually small proteins that do not show the fusions to the DNA glycosylase domain, making it unclear if they actually modify bases in nucleic acids.
Evidence for oxidative and other DNA-modification activities encoded by transposons
We were struck by the unusual pattern of the lineage-specific expansions and chromosomal distributions of the subfamily of Tet/JBP family from mushrooms and algae (Table 1). These lineage-specific expansions, particularly in mushrooms like Coprinopsis (~40 copies) and Laccaria (~60 copies), are characterized by closely related or even identical copies, with paralogs from the same organism usually being closer to each other than their cognates from other organisms. The different copies are distributed throughout the genome rather than as few loci of multiple tandem repeats. In both Coprinopsis and Laccaria, we observed that the majority of copies of the gene encoding the Tet/JBP family 2OGFeDO protein co-occurred in a tightly-linked genomic neighborhood with either or both of two distinct ORFs; a smaller subset of these neighborhoods also included a further 3rd conserved co-occurring ORF (Fig. 3). In some cases the identical copies of the Tet/JBP family are also found linked to identical copies of one or more of these ORFs at different chromosomal locations in these fungi (Suppl. material). These ORFs also showed a strongly preserved relative orientation with respect to each other (Fig. 3, see below). In computational experiments, the probability of these genes being neighbors in a particular preferred orientation so frequently by chance alone was found to be less than 10−19. Such conserved repetitive gene neighborhoods, which are widely dispersed over the genome in eukaryotes, are only found in the case of transposable elements or integrated viruses. Interestingly, in multiple cases these conserved gene neighborhoods are embedded in what appear to be chromosomal hotspots for transposon integration, as evidenced by the En/Spm transposons or retroelements53 found in their vicinity (Fig. 3). Taken together, these observations strongly indicate that this subfamily of the Tet/JBP family is encoded by a novel active transposable element, which additionally encodes at least the two other ORFs that most frequently co-occur with it in these mushroom genera. Most of the full-length 2OGFeDO genes are predicted to encode active proteins, indicating that they probably function in cis for each copy of the predicted transposable element. However, there are multiple instances in each organism, where one or more of the genes in an element are truncated or disrupted by deletions or mutations, suggesting that they represent non-functional or satellite versions of the parent transposon (Fig. 3, Suppl. information). In the genome of the alga Chlamydomonas, we only found two sufficiently long contigs to investigate the neighborhoods of its representatives of this subfamily of the Tet/JBP family. In both those cases we found linkages with the larger of the co-occurring ORFs found in the above fungal gene neighborhoods (Fig. 3). This suggests that they are indeed likely to be part of a similar transposon even in the alga. The pattern of distribution, which is currently limited to certain fungi and chlorophytes, also implies that these elements are likely to have spread laterally across phylogenetically distant groups.
To obtain a better understanding of this element we performed sequence profile analysis of the ORFs that co-occur in these elements. The smaller of the two most frequently occurring ORFs (Fig. 3) in the predicted transposon was found to contain a specialized version of the DNA-binding HMG domain that is most closely related to HMG domains of animal maelstrom proteins.54 The larger of the two ORFs encodes a protein of 850–1,100 amino acids, which often contains multiple cysteine cluster domains, potentially defining one or more Zn-chelating units (Fig. 4, Suppl. material). However, the core of this ORF contains a highly conserved domain with 6 characteristic sequence motifs (CX1–2H, GE, DXXC, HXXXHXXXC and GEXXE, where h is a hydrophobic residue). We propose that this distinctive conservation pattern defines the catalytic domain of the novel transposase used by these mobile elements. While it appears unrelated to any previously characterized transposase domain, the predicted secondary structure of this conserved domain is not incompatible with the RNAse H fold found in several transposases.53,55 The third ORF that co-occurs with these only in a subset of fungal elements (Fig. 3) is a small predicted α-helical protein with multiple conserved tryptophans and no detectable relationship to characterized protein domains (Suppl. material). Genes encoding the Tet/JBP family protein and the HMG domain protein are always oriented in the same direction with respect to each other, whereas those encoding the predicted transposase protein are oriented in the opposite direction (Fig. 3). Thus, the predicted transposase gene is most often head-to-head with respect to the Tet/JBP family gene and tail-to-tail with the HMG protein-encoding gene. The third uncharacterized ORF if present is almost always oriented in the same direction as the predicted transposase gene. It is conceivable that the strictly maintained pattern opposite orientation of these two genes with respect to the predicted transposase might provide a means of differentially regulating their expression, possibly in a mutually exclusive fashion. Studies in the model mushroom Coprinopsis have suggested that RNA-targeted DNA cytosine methylation might play a role in gene silencing.56 In animals, the maelstrom protein, which contains an HMG domain related to the version encoded by these novel transposons, has been implicated in the repression of transposons via cytosine methylation and is part of the RNA-binding nuage complex.57 In this light, we speculate that the expression of the transposon encoded Tet/JBP-related 2OGFeDO protein might result in oxidation of methylated cytosines on the transposon to hmC, which in conjunction with the HMG domain protein, could help in regulating gene expression of the transposon and/or activity of the transposase.
Figure 4.
Multiple alignment of the proposed catalytic domain of the transposase of the novel predicted transposon encoding 2OGFeDO proteins. Protein sequences are represented by their gene names, species names and GenBank index numbers. The predicted secondary structure is shown above the alignment. The coloring scheme and consensus abbreviations are shown in the key. Conserved residues defining the catalytic site of the predicted transposase are marked below the alignment.
While the highly mobile restriction-modification systems encode DNA modification (methylation) enzymes, such DNA-modification systems have not yet been observed in conventional multi-copy number transposons. To our knowledge the above-identified Tet/JBP-family-encoding transposons represent the first case of an apparently active eukaryotic transposable element that encodes its own DNA-modification enzyme with a potential regulatory role. Hence, we were curious to investigate if further examples of such transposons encoding DNA-modification enzymes existed. We according systematically searched transposons identified on the basis of recognized transposase domains53 with a library of profiles of catalytic domains known to modify DNA, such as DNA methylases, 2OGFeDOs and phage Mu Mom-like enzymes. As a result we uncovered two distinct groups of transposons in bacteria such as Kuenenia, Nitrococcus, Acidithiobacillus and Leptospirillum, which showed associations with Mom-like enzymes (Fig. 3, Suppl. material). In these cases a catalytically active Mom domain is respectively fused to transposase domains of either the TnpA or the TN5 family. Given that the Mom family catalyzes addition of carbamoylmethyl or a related adduct to DNA,8,14,26 it is likely that these transposon proteins are “two-headed” enzymes that catalyze both the modification of DNA via the Mom domain and transposition via the transposase domain. Even in these cases we suggest that Momylation by the transposon encoded protein might regulate the transcription or transposition of the mobile elements that encode them. Such elements are found to be particularly expanded in the bacterium Kuenenia stuttgartiensis. In addition to these linkages, we also found a conserved fusion of the Mom domain with a restriction endonuclease-like domain of the very short patch repair nuclease superfamily55 in several bacteria, and to a nuclease of the Colicin E9-like family in Streptomyces (Fig. 3, Suppl. material). These might represent uncharacterized restriction-modification systems, wherein Momylation might take the place of methylation. Thus, the above observations imply that transposons from both eukaryotes and bacteria might encode their own DNA modifying enzymes to regulate their gene expression or transposition.
Material and Methods
The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda) was searched using the PSI-BLAST programs.59 Profile searches using the PSI-BLAST program were conducted either with a single sequence or a sequence with a PSSM used as the query, with a profile inclusion expectation (E) value threshold of 0.01, and were iterated until convergence.59 For all compositionally biased queries the correction using composition-based statistics was used in the PSI-BLAST searches.60 Multiple alignments were constructed using the Kalign program,61 followed by manual correction based on the PSI-BLAST results. The multiple alignment was used to create a HMM using the Hmmbuild program of the HMMER package.62 It was then optimized with Hmmcaliberate and the resulting profile was used to search a database of completely sequenced genomes using the Hmmsearch program of the HMMER package.62 Profile-profile searches were performed using the HHpred program.63 The JPRED program64 and the COILS program65 were used to predict secondary structure. Globular domains were predicted using the SEG program with the following parameters: window size 40, trigger complexity = 3.4; extension complexity = 3.75.66
The Swiss-PDB viewer67 and Pymol programs68 were used to carry out manipulations of PDB files. Reconstruction of exon-intron boundaries was done using the NCBI Splign program69 with the tblastn searches against chromosomes as a guide. Gene neighborhoods were determined using a custom script that uses completely sequenced genomes or whole genome shotgun sequences to derive a table of gene neighbors centered on a query gene. Then the BLASTCLUST program70 is used to cluster the products in the neighborhood and establish conserved co-occurring genes. These conserved gene neighborhood are then sorted as per a ranking scheme based on occurrence in at least one other phylogenetically distinct lineage (“phylum” in NCBI Taxonomy database), complete conservation in a particular lineage (“phylum”) and physical closeness on the chromosome indicating sharing of regulatory—10 and—35 elements.
Evolutionary Implications and General Conclusions
Our prediction of novel enzymes catalyzing the oxidative modification of nucleic acids has notable implications for both the evolution of nucleic acid metabolism and the future study of gene regulation. While oxidative modifications of proteins have been known for a long time, the existence of direct oxidative modifications of nucleic acids was not widely suspected. Our prediction of AlkB as an oxidative DNA-repair enzyme of the 2OGFeDOs superfamily, and its subsequent experimental confirmation, provided the first computational support for such enzymes and the modifications catalyzed by them being more widely prevalent. This was further extended by studies on the biochemistry of the unusual DNA-modification, base J of kinetoplastids.18 In this study we show that there are several such potential enzymes, both in previously well-studied model organisms such as mammals, as well as poorly characterized clades of fungi, algae and various early branching eukaryotes. Strikingly, there is no support for these nucleic-acid-modifying enzymes forming one related sub-group within the 2OGFeDO superfamily, instead they appear to belong to several families, most of which are only distantly related to each other. This would imply that the 2OGFeDO superfamily has been recruited for oxidative modification of nucleic acids on multiple occasions. Within the Tet/JBP family there appears to be a correlation between their spread and the evolution DNA methylation. Of these the Tet subfamily is correlated in animals with the presence of DNA-modifying cytosine methylases DNMT1 and DNMT3 and appears to act primarily in the oxidation of 5-methylcytosine.38 Unlike animals, multicellular plants have a novel DNA glycosylase, which appears to be their primary demethylating enzyme, and accordingly entirely lack enzymes of the Tet/JBP family.58 In contrast, chlorophyte algae, mushrooms and Naegleria, which also encode multiple DNA methyltransferase genes, have members of the Tet/JBP family that might modify the methylated cytosine in these organisms. Further, in addition to acting on RNA, some members of the algal RNA-modification-associated family (Table 1) might operate on methylated cytosine in DNA, as suggested by the fusion to the TAM(MBD) domain (Fig. 2). It is also interesting to note that, like the eukaryotic DNA methyltransferases, even the Tet/JBP family of enzymes might have descended from selfish elements like viruses or transposons found in the bacterial world. In this context is interesting to note that the phage mu MOM-like enzyme has been acquired by stramenopile algae, such as the diatom Phaeodactylum and Emiliania (Suppl. material), suggesting that on multiple, independent occasions DNA-modifying enzymes of prokaryotic selfish elements might have been reused in regulatory contexts by eukaryotes.
Our computational prediction of novel oxidative modifications of nucleic acids opens up new vistas for exploring previously unforeseen aspects of gene regulation in eukaryotes. Experimental analysis of the Tet1 protein shows that it might be a critical regulator of gene expression by oxidatively modifying 5-methylcytosine and possibly facilitating its demethylation.38 The presence of such enzymes across several eukaryotic lineages suggests that oxidized pyrimidine derivatives could provide novel epigenetic marks, or even a means of erasing prior marks in the form of DNA methylation. This study also points to the possibility of novel RNA-modifying enzymes in particular eukaryotic lineages. Experimental investigation of modifications catalyzed by these enzymes might indeed help in elucidating the lineage-specific adaptive value of such modifications.
Supplementary Material
Acknowledgments
L.A. and L.M.I. acknowledge the Intramural research program of the National Library of Medicine, National Institutes of Health, USA for funding their research. M.T. and A.R. are supported by a pilot grant from the Harvard Stem Cell Institute.
Footnotes
After this paper was submitted for review two new publications on the TET2 gene came to our attention (A Tefferi et al., Frequent TET2 mutations in systemic mastocytosis: clinical, KITD816V and FIP1L1-PDGFRA correlates, Leukemia 2009 and A Tefferi et al, TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis, Leukemia 2009). Both these studies show that the TET2 gene is mutated in multiple myeloproliferative neoplasms. The mutations recorded in these studies include those resulting in loss of the catalytic domain as well as point mutations disrupting the second metal-chelating histidine and other conserved residues in the catalytic domain and its cysteine-rich N-terminal extension. These mutations suggest that the Tet2 catalytic activity is likely to be required for its tumor suppressor function and loss of Tet2 activity might be correlated with the hypermethylation reported in these myeloproliferative neoplasms.
References
- 1.Bloomfield VA, Crothers DM, Tinoco I., Jr . Nucleic Acids: Structures, Properties and Functions. Sausalito, CA: University Science Books; 2000. [Google Scholar]
- 2.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–64. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, et al. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:118–21. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:269–70. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–81. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–56. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol Rev. 1993;57:434–50. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren RA. Modified bases in bacteriophage DNAs. Annu Rev Microbiol. 1980;34:137–58. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 9.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev. 2006;4:183–92. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 11.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 13.Selker EU. Genome defense and DNA methylation in Neurospora. Cold Spring Harb Symp Quant Biol. 2004;69:119–24. doi: 10.1101/sqb.2004.69.119. [DOI] [PubMed] [Google Scholar]
- 14.Gommers-Ampt JH, Borst P. Hypermodified bases in DNA. Faseb J. 1995;9:1034–42. doi: 10.1096/fasebj.9.11.7649402. [DOI] [PubMed] [Google Scholar]
- 15.Arakawa H, Hauschild J, Buerstedde JM. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science (New York, NY) 2002;295:1301–6. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 16.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 17.Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–56. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 18.Borst P, Sabatini R. Base J: discovery, biosynthesis and possible functions. Annual review of microbiology. 2008;62:235–51. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 19.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–51. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton JR, Liebert K, Bekes M, Jeltsch A, Cheng X. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J Mol Biol. 2006;358:559–70. doi: 10.1016/j.jmb.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran PH, Korszun ZR, Cerritelli S, Springhorn SS, Lacks SA. Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of streptococcus pneumoniae bound to S-adenosylmethionine. Structure. 1998;6:1563–75. doi: 10.1016/s0969-2126(98)00154-3. [DOI] [PubMed] [Google Scholar]
- 22.Reinisch KM, Chen L, Verdine GL, Lipscomb WN. The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell. 1995;82:143–53. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 23.Song HK, Sohn SH, Suh SW. Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex. EMBO J. 1999;18:1104–13. doi: 10.1093/emboj/18.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm L, Sander C. Evolutionary link between glycogen phosphorylase and a DNA modifying enzyme. EMBO J. 1995;14:1287–93. doi: 10.1002/j.1460-2075.1995.tb07114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morera S, Lariviere L, Kurzeck J, Aschke-Sonnenborn U, Freemont PS, Janin J, Ruger W. High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding. J Mol Biol. 2001;311:569–77. doi: 10.1006/jmbi.2001.4905. [DOI] [PubMed] [Google Scholar]
- 26.Kaminska KH, Bujnicki JM. Bacteriophage Mu Mom protein responsible for DNA modification is a new member of the acyltransferase superfamily. Cell cycle (Georgetown, Tex) 2008;7:120–1. doi: 10.4161/cc.7.1.5158. [DOI] [PubMed] [Google Scholar]
- 27.Vainio S, Genest PA, Ter Riet B, van Luenen H, Borst P. Evidence that J-binding protein 2 is a thymidine hydroxylase catalyzing the first step in the biosynthesis of DNA base. J Mol Biochem Parasitol. 2009;164:157–61. doi: 10.1016/j.molbiopara.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, et al. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35:2107–15. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukacin R, Britsch L. Identification of strictly conserved histidine and arginine residues as part of the active site in Petunia hybrida flavanone 3beta-hydroxylase. Eur J Biochem. 1997;249:748–57. doi: 10.1111/j.1432-1033.1997.t01-2-00748.x. [DOI] [PubMed] [Google Scholar]
- 30.Clifton IJ, Hsueh LC, Baldwin JE, Harlos K, Schofield CJ. Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent oxygenases. Eur J Biochem. 2001;268:6625–36. doi: 10.1046/j.0014-2956.2001.02617.x. [DOI] [PubMed] [Google Scholar]
- 31.Roach PL, Clifton IJ, Fulop V, Harlos K, Barton GJ, Hajdu J, et al. Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature. 1995;375:700–4. doi: 10.1038/375700a0. [DOI] [PubMed] [Google Scholar]
- 32.Smiley JA, Kundracik M, Landfried DA, Barnes VR, Sr, Axhemi AA. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochim Biophys Acta. 2005;1723:256–64. doi: 10.1016/j.bbagen.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–8. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 34.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9 and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:7. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiPaolo C, Kieft R, Cross M, Sabatini R. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol Cell. 2005;17:441–51. doi: 10.1016/j.molcel.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 36.Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–84. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 37.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–41. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 38.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by the MLL fusion partner. TET1. 2009 doi: 10.1126/science.1170116. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cross SH, Meehan RR, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nature Genet. 1997;16:256–9. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 40.Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006;25:4503–12. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hino S, Kishida S, Michiue T, Fukui A, Sakamoto I, Takada S, et al. Inhibition of the Wnt signaling pathway by Idax, a novel Dvl-binding protein. Mol Cell Biol. 2001;21:330–42. doi: 10.1128/MCB.21.1.330-342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes F, Barilla D. Assembling the bacterial segrosome. Trends Biochem Sci. 2006;31:247–50. doi: 10.1016/j.tibs.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Lobocka MB, Rose DJ, Plunkett G, 3rd, Rusin M, Samojedny A, Lehnherr H, et al. Genome of bacteriophage P1. J Bacteriol. 2004;186:7032–68. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hauenstein S, Zhang CM, Hou YM, Perona JJ. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat Struct Mol Biol. 2004;11:1134–41. doi: 10.1038/nsmb849. [DOI] [PubMed] [Google Scholar]
- 45.Ofengand J, Bakin A, Wrzesinski J, Nurse K, Lane BG. The pseudouridine residues of ribosomal RNA. Biochem Cell Biol. 1995;73:915–24. doi: 10.1139/o95-099. [DOI] [PubMed] [Google Scholar]
- 46.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 47.Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol. 2004;5:148–57. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 48.Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–84. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grishin NV. The R3H motif: a domain that binds single-stranded nucleic acids. Trends Biochem Sci. 1998;23:329–30. doi: 10.1016/s0968-0004(98)01258-4. [DOI] [PubMed] [Google Scholar]
- 51.Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–59. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–4. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 53.Galun E. Transposable Elements: A Guide to the Perplexed and the NoviceWith Appendices on RNAi, Chromatin Remodeling and Gene Tagging. Dordrecht: Kluwer Academic Publishers; 2003. [Google Scholar]
- 54.Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development (Cambridge, England) 2003;130:859–71. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 55.Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–32. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walti MA, Villalba C, Buser RM, Grunler A, Aebi M, Kunzler M. Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs. Eukaryot Cell. 2006;5:732–44. doi: 10.1128/EC.5.4.732-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–97. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, et al. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lassmann T, Frings O, Sonnhammer EL. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eddy SR. Profile hidden Markov models. Bioinformatics (Oxford, England) 1998;14:755–63. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 63.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–11. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 65.Gruber M, Soding J, Lupas AN. Comparative analysis of coiled-coil prediction methods. J Struct Biol. 2006;155:140–5. doi: 10.1016/j.jsb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 66.Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput Chem. 1994;18:269–85. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 67.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 68.Delano WL. San Carlos, CA: USA DeLano Scientific; 2002. [Google Scholar]
- 69.Kapustin Y, Souvorov A, Tatusova T, Lipman D. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol Direct. 2008;3:20. doi: 10.1186/1745-6150-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.BLASTCLUST program. ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.