Abstract
The natural killer gene complex (NKC) on chromosome 6 contains clusters of genes that encode both activation and inhibitory receptors expressed on mouse natural killer (NK) cells. NKC genes, particularly belonging to the Nkrp1 and Ly49 gene families, display haplotype differences between different mouse strains and allelic polymorphisms of individual genes, as previously revealed by conventional analysis in a small number of inbred mouse strains. Herein we used array-based comparative genomic hybridization (aCGH) to efficiently compare the NKC in 21 mouse strains to the reference C57BL/6 strain. By using unsupervised clustering methods, we could sort these variations into the same groups as determined by previous RFLP analyses of Nkrp1 and Ly49 genes. Prospective analyses of aCGH and RFLP data validated these relationships. Moreover, aCGH data predicted monoclonal antibody reactivity with an allospecific determinant on molecules expressed by NK cells. Taken together, these data demonstrate the structural variation in the NKC between mouse strains as well as the usefulness of aCGH in analysis of complex, polymorphic gene clusters.
Keywords: Natural killer (NK) cells, natural killer gene complex (NKC), array-based comparative genomic hybridization (aCGH), RFLPs
Introduction
Natural killer (NK) cells are lymphocytes initially described for their “natural” ability to kill tumor targets. They are now appreciated as also having critical roles in innate immune responses to infections. In addition to their responses to cytokines, they can specifically recognize and kill their cellular targets which can also stimulate their capacity to secrete cytokines, such as interferon-γ (IFNγ)1. Whether or not an NK cell is triggered by a cellular target is dependent on input from two functional types of receptors: 1) Activation receptors that directly recognize target surface ligands and stimulate NK cell functions; and 2) inhibitory receptors recognizing surface ligands representing self, such as major histocompatibility complex (MHC) class I molecules. In general, the activation receptors contain charged transmembrane residues that facilitate association with signal transducing transmembrane molecules. In contrast, inhibitory receptors lack such transmembrane residues and instead contain immunoreceptor tyrosine-based inhibitory motifs (ITIM) in their cytoplasmic domains. Simultaneous engagement of both types of receptors generally results in no NK cell activation whereas down-regulation of otherwise ubiquitously expressed MHC class I molecules releases NK cells from inhibition, allowing NK cell stimulation. The latter scenario explains the “missing-self” hypothesis which proposes that down-regulation of MHC class I to avoid T cell attack, such as in viral infections, would unleash NK cells. Other NK cell inhibitory receptors recognize non-MHC ligands that may also regulate NK cell functions.
In the mouse, the predominant NK cell receptors are type II integral membrane proteins with lectin-like external domains, and are encoded in the NK gene complex (NKC) on distal mouse chromosome 62; 3. The mouse NKC itself can be divided into three major regions. A centromeric region contains genes for the Nkrp1 family of receptors specific for Clr molecules, structurally related glycoproteins encoded by genes intermingled among the Nkrp1 genes4-6. The telomeric Ly49 region primarily encodes inhibitory NK cell receptors involved in self-non-self discrimination by recognition of MHC class I1; 6; 7. Between the Nkrp1 and Ly49 regions is a region containing genes for CD94 and the NKG2 family of molecules that are also present in humans8. CD94 heterodimerizes with NKG2 family members to form receptors for an invariant MHC molecule (mouse Qa1 and HLA-E). Whereas the CD94 and NKG2 genes appear to display relatively little polymorphism, there is some evidence for polymorphism in the mouse Nkrp1 and Ly49 gene families.
Polymorphisms of the NKC have been described at two levels, structural genomic variations and allelic polymorphisms of individual genes. With respect to structural variation in the NKC, previous studies have revealed restriction fragment length polymorphic (RFLP) variants among inbred mouse strains. For Nkrp1, mice can be grouped according to 4 RFLP variants2. Limited analysis indicates that some of these RFLP variants are associated with allelic polymorphisms of individual Nkrp1 genes9-11. The intermingling of genes for Nkrp1 receptors and their Clr ligands suggests a genetic mechanism to protect receptor-ligand pairs important in host preservation of self, as seen in other species including plants. Moreover, there is evidence indicating genetic protection of this region because of infrequent recombination events in the Nkrp1 region, suggesting that structural variation in this region of the NKC may be limited.
In contrast, it would not be surprising if the Ly49 region itself demonstrates relatively profound genetic diversity since Ly49 receptors bind highly polymorphic major histocompatibility complex (MHC) molecules encoded on chromosome 17. Indeed, this is supported by limited analysis. Ly49 RFLP variants show 5 patterns12. In detailed analyses of bacterial artificial chromosomes derived from 4 mouse strains (C57BL/6J, 129S1/SvImJ, BALB/cByJ, and NOD/ShiLtJ), each representing a different RFLP group, there were differences in Ly49 gene content13-16. In addition, the strains display allelic polymorphism in individual Ly49 genes. Thus, there is structural variation in the NKC, including RFLPs that correspond to differences in gene content and allelic polymorphisms but prior analysis has been restricted to a small number of inbred strains.
In a previous study, we used array-based comparative genomic hybridization (aCGH) with in a genome-wide analysis of a panel of inbred mouse strains to efficiently evaluate structural variations in large genomic regions 17. In aCGH, DNA from a test and reference genome is hybridized to a microarray containing probes, typically oligonucleotides, representing defined chromosomal locations spaced more or less evenly across the genome. In this assay, structural variation is apparent when there are differences in hybridization intensity as compared to a reference genome18; 19. This structural variation is often thought to reflect variable numbers of copies of DNA segments, termed copy number variation (CNV), although allelic polymorphisms of genes could also theoretically affect hybridization. Moreover, the application of aCGH to polymorphic gene clusters has not been well studied. In our previous analysis (388, 352 unique probes with median spacing of 5 kb), we found structural variations in several genomic regions containing genes categorized as being involved in immune response by gene ontology lists17, including the NKC8.
Herein, we used high resolution aCGH (2.1 million unique probes, median spacing 1 kb) to analyze multiple inbred mouse strains to yield further insight into structural variation of the polymorphic NKC. By clustering analysis, the aCGH data could be precisely correlated with overall genome structural variation of the NKC as detected by RFLP variants in retrospective and prospective analyses. Moreover, we determined that strains related by aCGH and RFLP analysis share expression of allotypic determinants on NK cell surface molecules as detected by monoclonal antibody reactivity.
Results
Identification of the NKC as a variable region on chromosome 6
Conventional analysis previously demonstrated structural variation in the NKC but detailed analyses have been limited to the C57BL/6J, 129S1/SvImJ, BALB/cByJ, and NOD/ShiLtJ inbred mouse strains and primarily for the Ly49 gene cluster13-16. Here we used aCGH to further analyze the NKC on distal mouse chromosome 6, a chromosomal region that we previously showed was among the most variable regions of the mouse genome17.
Consistent with known variability in this region by conventional physical mapping and sequencing studies13-16, we determined by aCGH that the NKC region (128,574,123 bp to 130,348,416 bp positions) was a variable region on chromosome 6 in the 129S1/J, BALB/c and NOD strains as compared to C57BL/6 reference strain (Figure 1, Supplemental Figures S1 and S2). By contrast, variability was less evident along most of the entire length of chromosome 6. Thus, the NKC region appears to be one of the more variable regions on chromosome 6 in aCGH comparison of 4 mouse strains known to have variability in this genomic region.
Figure 1. Index of variability along chromosome 6.
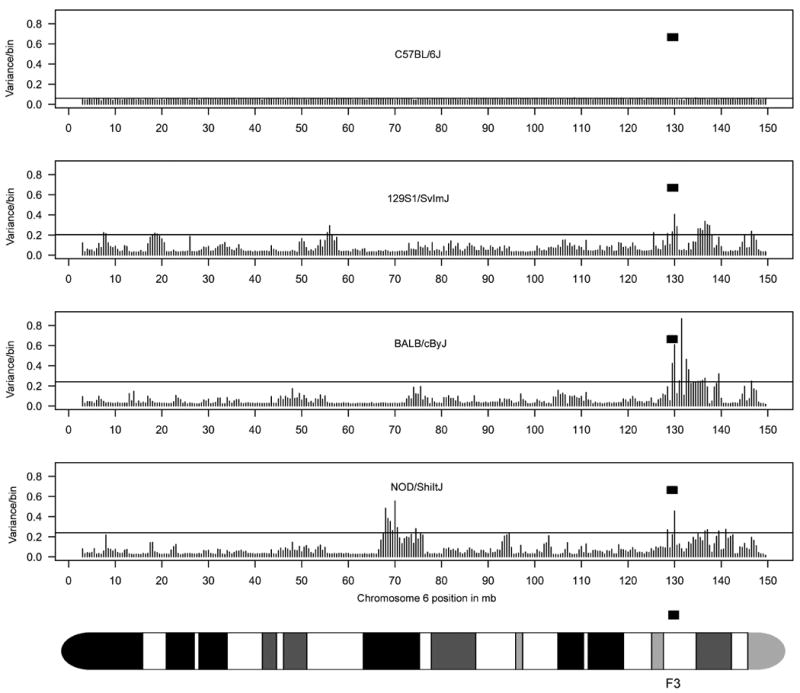
For the inbred mouse strains indicated, the variance of the log2 ratios of the signal intensities (see Supplemental Figure S1) for probes within each bin (500,000 bp/bin) was captured (see Methods for calculation). At 500,000 bp/bin, there is a total of 294 bins for chromosome 6. Each bar represents a bin along the chromosome. The bin size had minimal effect in capturing variability (Supplemental Figure S2). The horizontal line in each graph is at 2 standard deviations (sd). The NKC genomic region is highlighted by the filled rectangle. The NKC is positioned within the F3 band as indicated by the ideogram of chromosome 6.
When we extended this variability analysis for all 20 inbred mouse strains initially examined, high variability was consistently seen across all strains (except BTBR T+ft/J) as compared to C57BL/6 in the distal part of chromosome 6, encompassing the NKC (Figure 2). (See Supplemental Figure S3 for log2 ratios of probe signal intensities). Although there were other areas of variability on chromosome 6, such as the un-annotated region at ∼41-41.5 Mb position, the vomeronasal pheromone receptor family (∼56-56.5 Mb)20, the Igκ variable region (∼70-70.5 Mb)21, and the bitter taste receptor gene family (∼131.5-132 Mb)22, they were not consistently seen across all strains.
Figure 2. A comparison of the variability along chromosome 6 for 22 inbred mouse strains.
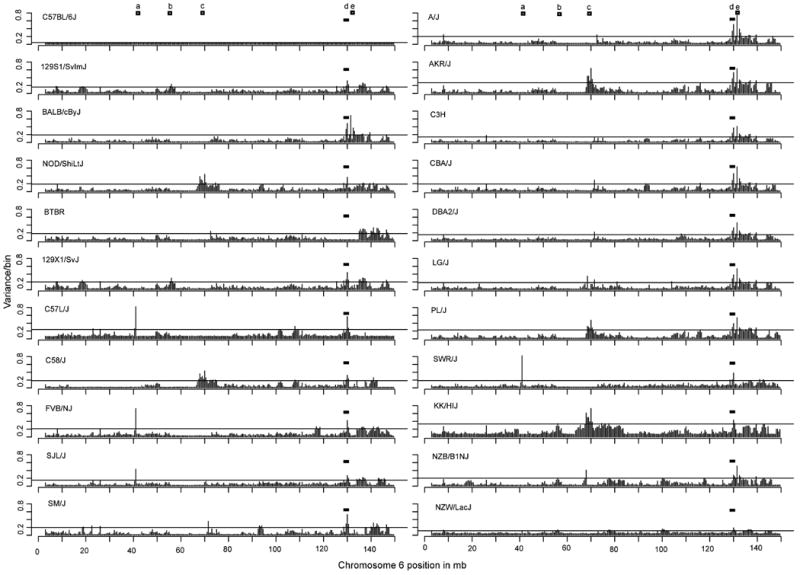
Shown is the variance within each 500,000 bp/bin. The regions of significant variability (greater than 2 sd) along the chromosome (other than the NKC) are represented by an open square. The NKC region (d) at ∼120.5 to 130 Mb is highlighted by the filled rectangle. Region a is at ∼41-41.5 Mb, region b is at ∼56-56.5 Mb, region c is at ∼70-70.5 Mb, and region e is at ∼131.5-132 Mb positions.
It should be noted that even in closely related strains as determined by pedigree, such as C57L/J and C58/J23, there was also variability in the NKC region as compared to each other and the C57BL/6 reference strain (Figure 2). Interestingly, BTBR T+tf/J was the only strain that showed no variability with C57BL/6 in the NKC region, which is surprising in that this strain is unknown to have a pedigree relationship with C57BL/624; 25. Thus, the NKC region consistently displays variability among the initial mouse strains analyzed, except for BTBR, as compared to C57BL/6 and the rest of chromosome 6.
Subregions of the NKC display differences in variability
In conventional analysis of the NKC, the NKC can be divided into two polymorphic regions, the centromeric Nkrp1 region and the telomeric Ly49 cluster6. The Nkrp1 region is thought to show less polymorphism than the Ly49 region4; 6. However, limited cDNA analysis suggests allelic polymorphisms of some expressed genes13-16 and detailed genome sequence information of different mouse strains is generally not available. By contrast, the Ly49 region displays considerable heterogeneity in both gene number and allelic polymorphisms of shared genes13-16. To determine if these differences were visible by aCGH, we analyzed the data at higher resolution (125,000 bp/bin) (Figure 3). When we focused on the region between 126 Mb and 135 Mb positions on chromosome 6, variability in the NKC was readily separable from variability in the taste receptors at position 130.5-132.5 Mb. Within the NKC, the variability within the Ly49 multigene family region was clearly evident among the prototype C57BL/6, BALB/c, 129 and NOD strains and indeed, among all 20 strains initially analyzed. In contrast, the region encompassing the Nkrp1 region was somewhat less variable than the Ly49 region among the prototype strains, and among all 20 strains initially analyzed. Thus, there are subregion differences in the NKC with respect to variability as detected by aCGH.
Figure 3. A comparison of the variability for 22 inbred mouse strains within a 9 million bp region encompassing the NKC.
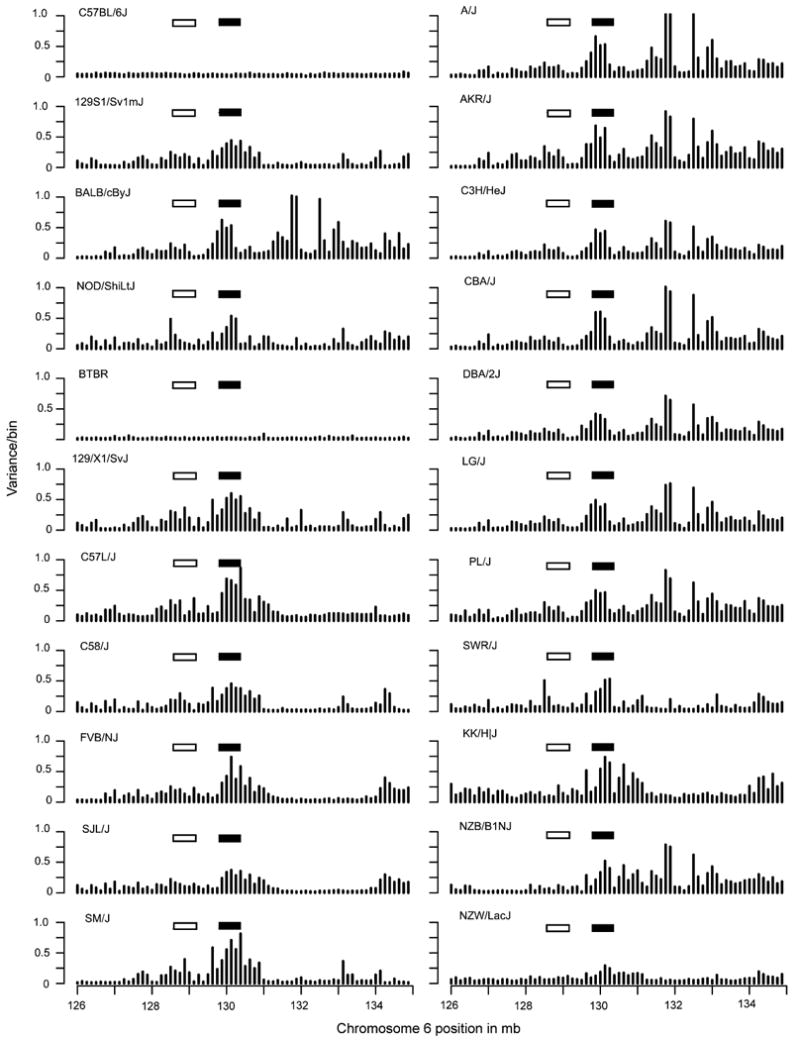
As captured by the variance within the 125,000 bp/bin index, the Ly49 region is highlighted by the filled rectangle and the Nkrp1 region is highlighted by the open rectangle.
Strain clustering of NKC variability by aCGH analysis corresponds to RFLP groupings
We also noticed that some of the mouse strains appeared to have similar patterns of variability that could be grouped. For example, it appeared that the mouse strains could be grouped in terms of their pattern of variability in the Ly49 region when the data were viewed at even higher resolution (15,625 bp/bin)(Figure S4). Grouping of strains by variability patterns was more evident upon examination of the summed absolute values within each bin (Figure 4) in which we factored in the differences in probe density within each bin (Figure S5). For example, by visual inspection, the 129S1 pattern looked similar to C58/J and different from BALB/c, etc. (Figure 4). For an unbiased quantitative approach, we performed unsupervised, non-hierarchical clustering (Table 1A) of the first set of inbred strains using the log2 signal ratios of each probe within the Ly49 (129.8 Mb to 130.3 Mb) genomic region (Figure S6). In this way, the aCGH data could be clustered into groups of inbred mouse strains based on their silhouette width value (Table 1A). Interestingly, this grouping is identical to the Ly49 groups previously generated by RFLP analysis on a subset of the strains examined here by aCGH12 Similarly, we performed a clustering analysis of the Nkrp1 region (128.6 Mb to 129.2 Mb). Even though this region appears to be less variable than the Ly49 regions (Figure 3), we were again able to obtain groups of mouse strains that were identical to the groups based on prior RFLP analysis of a subset of strains (Table 1B)2.
Figure 4. A comparison of the variability for 22 inbred mouse strains in the NKC as captured by the sum of absolute values of signal ratios within the 15,625 bp/bin index.
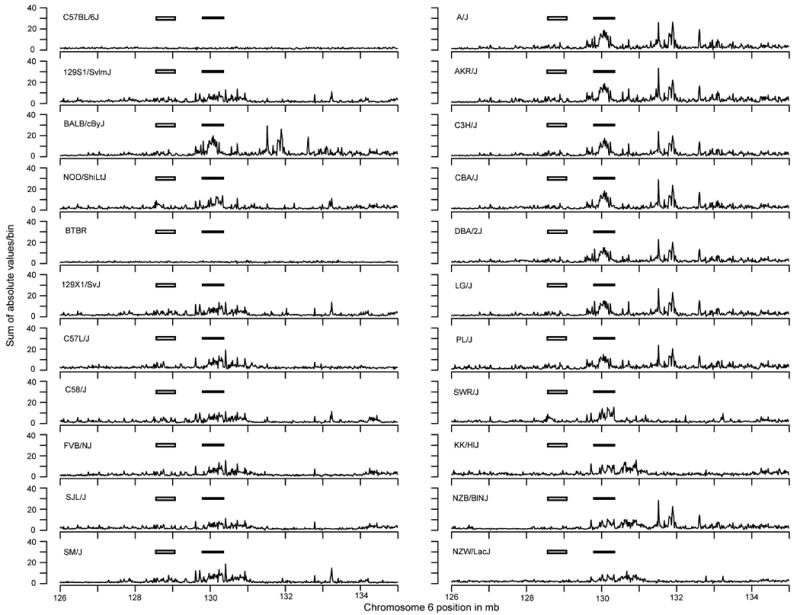
The Ly49 region is highlighted by the filled rectangle and the Nkrp1 region is highlighted by the open rectangle.
Table 1. Unsupervised, non-hierarchical clustering of the log2 ratios of the signal intensities of probes within the Ly49 and Nkrp1 genomic regions.
The Ly49 genomic region is from 129,796,847 bp to 130,348,221 bp positions. The Nkrp1 region is from 128,574,846 bp to 129,152,133 bp positions. The strains where both aCGH and RFLP data were previously available are underlined. The prospectively collected aCGH data are designated by *. RFLP analysis was prospectively performed for all other strains.
| A. Non-hierarchical clustering of a-CGH log2 ratios within Ly49 genomic region | ||||
| 1 | 2 | 3 | 4 | 5 |
| C57BL/6J | SJL/J | BALB/cByJ | SWR/J | NZB/B1NJ |
| BTBR T+tf/J | 129S1/SvImJ | A/J | NOD/ShiLt/J | KK/H1J |
| 129X1/SvJ | AKR/J | NZW/LacJ* | ||
| C57L/J | C3H/HeJ | |||
| C58/J | DBA/2J | |||
| FVB/NJ | LG/J | |||
| SM/J | PL/J | |||
| CBA/J* | ||||
| B. Non-hierarchical clustering of a-CGH log2 ratios within Nkrp1 genomic region | |||
| 1 | 2 | 3 | 4 |
| C57BL/6J | 129S1/SvImJ | C57L/J | SWR/J |
| NZB/B1NJ | A/J | C58/J | NOD/ShiLt/J |
| BTBR T+tf/J | AKR/J | SJL/J | |
| KK/H1J | C3H/HeJ | FVB/NJ | |
| NZW/LacJ* | BALB/cByJ | ||
| DBA/2J | |||
| 129X1/SvJ | |||
| LG/J | |||
| PL/J | |||
| SM/J | |||
| CBA/J* | |||
To further validate the non-hierarchical clustering analysis, we performed unsupervised, agglomerative hierarchical clustering of the aCGH data to generate a cluster dendrogram based on the probes within the Ly49 and the Nkrp1 gene families, respectively (Figure S7). Both clustering approaches yielded the same groups (Table 1A and 1B). More importantly, both clustering approaches yielded the same groups of mouse strains for the Ly49 and the Nkrp1 regions, respectively, as the RFLP results.
To validate the groupings by clustering algorithms of aCGH data and corresponding RFLP analysis, we prospectively collected aCGH data from two strains of mice (CBA/J and NZW/LacJ). These strains were not among the original 20 strains analyzed by aCGH but represented strains whose Ly49 and Nkrp1 RFLP patterns had been determined previously2; 12. By aCGH analysis, the CBA/J and NZW/LacJ strains clustered into the same aCGH group as determined by their Ly49 and Nkrp1 RFLP patterns (Figures 2-5, Table 1A and 1B).
Figure 5. RFLP variants correspond to aCGH grouped strains.
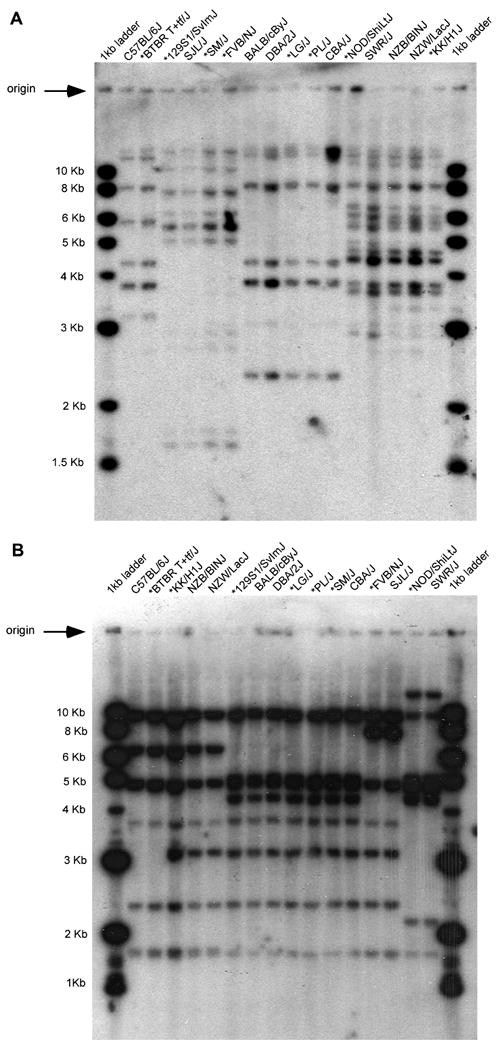
(A) Southern blot of HindIII digest of mouse DNA probed with a 659 bp PCR product of Ly49a. Five groups are indicated by their RFLP patterns. (B) Southern blot of EcoRI digest of mouse DNA probed with a 816 bp PCR product of Nkrpla. Five groups are indicated by their RFLP patterns. Strains not previously analyzed by Southern blots are indicated by an *.
In a reciprocal manner, Southern blots were prospectively performed on DNA from the BTBR, 129S1/J, FVB, LG/J, PL/J, NOD/J, SJL/J, SM/J, CBA/J, and KK/J inbred strains in order to obtain previously unavailable RFLP data with Ly49 and Nkrp1 cDNA probes (Figure 5). These strains demonstrated the RFLP variants predicted from the aCGH data (Table 1A and 1B). Taken together, these data indicate the high correlation between RFLP and aCGH analysis in detection of genome structural variation.
aCGH analysis predicts monoclonal antibody reactivity
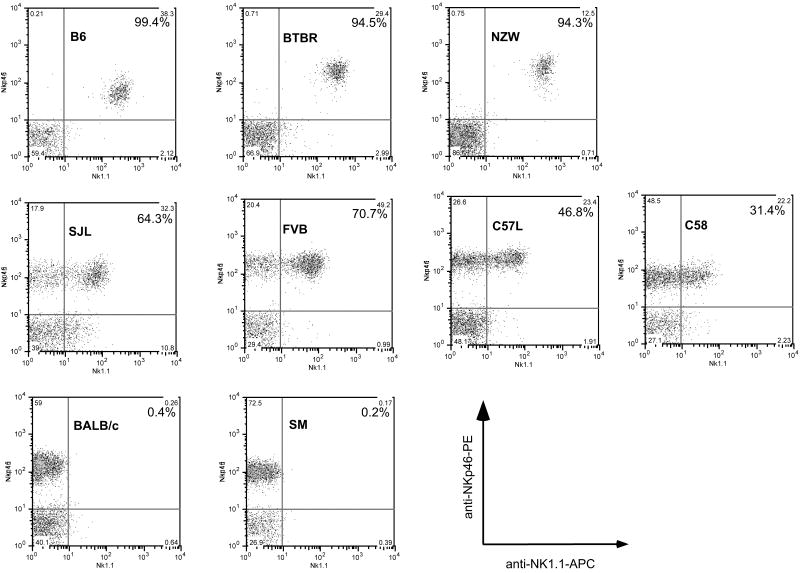
Corresponding to genetic differences based on RFLP and aCGH analyses, the NKC also displays allelic polymorphisms of genes that should be shared between mice in the same RFLP and aCGH groups. While it would be a daunting task to clone each of the NKC genes from all 22 strains of mice for detailed sequence comparison of the corresponding aCGH probes, we took advantage of a known monoclonal antibody (mAb) epitope to provide some insight into this issue. In particular, mAb PK136 binds an allotypic determinant on Nkrp1c that is expressed on all NK cells in C57BL/6. By contrast, mAb PK136 either is non-reactive in BALB/c mice or binds an allotypic determinant on Nkrp1b that is characteristically expressed on a subset of NK cells in SJL mice9; 10; 26. In the aCGH grouping, the BTBR and NZW strains are more closely related to C57BL/6 mice than to SJL mice which in turn are more closely related to the FVB, C58, and C57L strains (Figure S7). These data suggested that BTBR and NZW mice may possess a mAb PK136-reactive molecule expressed on all NK cells whereas FVB, C58, and C57L mice should express the PK136 epitope on a subset of NK cells. Indeed, these predictions were confirmed when mAb PK136 was used to stain NK cells from these strains of mice (Figure 6). Moreover, NK cells from SM mice are non-reactive with mAb PK136, consistent with their inclusion in the same aCGH group as BALB/c mice. In addition, previous studies indicate that mAb PK136 does not stain cells in other mice included in the BALB/c aCGH group (A, AKR, CBA, C3H, DBA/2, and 129 strains)27. Thus, the aCGH data predicted mAb reactivity with allotypic determinants on NKC-encoded molecules.
Figure 6. Flow cytometry profile of NK cells with allo-specific monoclonal antibody PK136.
CD3- and CD19- splenocytes were assessed for expression of NKp46 (expressed by all NK cells in all mouse strains, y-axis) and the NK1.1 epitope detected by monoclonal antibody PK136 (x-axis). Phycoerythrin (PE)-conjugated anti-NKp46, allophycocyanin (APC)-anti-NK1.1, fluorescein isothiocyanate (FITC)-conjugated anti-CD3, and FITC-anti-CD19 were used to stain the indicated mouse strains. The percentage of NK1.1+ cells among NKp46+ CD3− CD19− cells is shown in each dot plot.
Discussion
The NKC is known to display genetic variability by conventional approaches but prior analyses had been limited to a portion of the NKC in a small number of mouse strains. Particularly for the Ly49 genes that encode polymorphic receptors for the genetically diverse MHC class I molecules, it had been challenging to characterize the extent of this polymorphism by conventional approaches. Herein we used aCGH to efficiently analyze 22 (20 initially plus CBA/J and NZW/LacJ) inbred mouse strains and determined that the NKC is among the most consistently variable regions on chromosome 6 as compared to C57BL/6. We were also able to cluster mouse strains into the same groups previously identified by RFLP analysis of both Ly49 and Nkrp1 genes. Furthermore, we prospectively used aCGH data to predict RFLP variants and vice-versa. Moreover, the aCGH data provided insight into monoclonal antibody reactivity. While these findings demonstrate the power of aCGH for identifying and determining genome variability, and clustering of individuals with similar genomic structural variations, these data more importantly highlight the structural variation of the NKC region, one of the most variable regions in the entire mouse genome17.
Our data suggest that the different aCGH groups represent different haplotypes for the Ly49 and Nkrp1 gene clusters, respectively. While Ly49 genome sequence data were previously available for only 4 mouse strains, they fall into different Ly49 groupings that correspond to 4 of the 5 known groups based on RFLP variants13-16. Each of these strains display different haplotypes in terms of gene content. The clustering of the aCGH signal intensities and the related RFLP group suggest at least another Ly49 haplotype that includes the NZB, NZW, and KK inbred mouse strains. Similarly, we determined that there were 4 aCGH groups for the Nkrp1 gene cluster that correspond to identical RFLP groups. While there is little genomic sequence information on the Nkrp1 cluster in strains from each aCGH or RFLP group, the available data support allotypic differences, as evidenced by monoclonal antibody reactivity (described in greater detail below), between mice in the different groups. Thus, we detected major structural genome variants for the Ly49 and Nkrp1 clusters, respectively, that are indicative of haplotypes.
In contrast to aCGH in which physical relationships between genes in a test sample are inferred from a reference sequence, RFLP analysis results from a direct assessment of physically linked DNA elements by Southern blot. However, it is relatively insensitive because restriction enzymes survey dispersed, short DNA sequences, and it cannot detect diversity in genetic elements that do not hybridize to the probe utilized. In other cases, RFLP analysis may be too sensitive if the probe cross-hybridizes with different genes having related sequences, as in the case of gene families. Thus, it is striking that our clustering analysis of the aCGH data provided the same groupings of mice as the RFLP analysis, indicating that the aCGH data can provide complementary information, even though aCGH is an indirect measure of the physical relationships of genes.
More than just relating aCGH data to RFLP analysis, here we were able to use the aCGH data to quantitatively and efficiently assess NKC relationships between a large number of inbred mouse strains. Interestingly, the aCGH groupings for the NKC predict related haplotypes that are unrelated to strain relationships based on pedigree charts or even whole genome SNP analysis23; 28. For example, FVB and 129 mouse strains belong to separate groups (Swiss mice and Castle's mice, respectively) based on genealogy and SNP analysis but they are grouped together for Ly49 polymorphisms based on aCGH data. Another noteworthy strain is BTBR, which appears to have an NKC closely related, if not identical, to C57BL/6 by aCGH data. Although the genealogy of BTBR is unknown23, BTBR is more closely related to 129 strain mice than C57BL/6 mice by whole genome SNP analysis and had been chosen for resequencing efforts to maximize phenotypic diversity29. Thus, the aCGH groupings for the NKC do not strictly conform to established pedigree and genetic relationships of mouse strains.
Previous studies, however, have suggested that the classical inbred mouse strains are mosaics with genetic elements from different Mus domesticus and M. musculus origins30. Yet, even high density SNP analyses29 do not clearly discriminate the NKC relationships between these strains (http://mouse.perlegen.com) in the manner that we have done here with aCGH analyses. These differences may be due to ascertainment bias inherent with many informative SNPs31 or the high percentage of repetitive elements in the NKC [reviewed in6]. Despite these challenges, our aCGH data provide a means to group mice on the basis of their NKC relationships.
The Ly49 and Nkrp1 aCGH groups are similar but not identical. Perhaps this is not unexpected since these gene families are physically and therefore genetically linked. Moreover, previous studies have indicated that recombination events in this region are relatively rare but those studies were confined to only certain strain pairs32; 33. It is interesting to note that one of the Nkrp1 groups (group 1) is represented by two groups for Ly49 (groups 1 and 5). On the other hand, Nkrp1 group 2 is “recombined” into two groups for Ly49 groups 2 and 3, which in turn are represented by a different assortment of strains in Nkrp1 groups 2 and 3, suggesting recombination occurred in the NKC in these strains. Furthermore, while it is currently impossible to determine the recombination frequency from the aCGH data, the current data seem to indicate that the Nkrp1 and Ly49 families have been maintained en bloc in these inbred strains. While it remains possible that there has been recombination within the Nkrp1 and Ly49 gene families themselves, such analysis will probably require higher resolution such as detailed sequence information.
Prior studies on the NKC, and the Ly49 cluster in particular, have suggested that the NKC arose by individual gene duplication and potentially by duplication of genomic segments containing blocks of genes6; 13; 14. Moreover, such duplications appear to have occurred in the context of several genes which constitute “framework” Ly49 genes, as revealed by analysis of the BALB/c cluster since it has the least number of Ly49 genes15. Overall, the aCGH data support the putative duplication events in the Ly49 gene cluster.
Investigators have postulated that, following duplication of individual Ly49 genes or gene blocks, there was continued evolution with sequence divergence in individual Ly49 genes or gene conversion, giving rise to allelic polymorphisms13. Indeed, allelic polymorphisms of individual NKC genes have been previously noted by conventional analysis of a smaller number of strains and there is evidence for even more complex polymorphic relationships. For example, NK cells in the C57L strain express a Ly49 gene that resembles a chimeric C57BL/6 Ly49 gene34. Although C57L and C57BL/6 are in different Ly49 groups by aCGH/RFLP analysis, the C57L gene product contains an external domain that is 98% identical to the C57BL/6-derived Ly49D activation receptor (and only 91% identical to Ly49A), and stains with an anti-Ly49D-specific monoclonal antibody. However, the C57L molecule contains a transmembrane and cytoplasmic domain that is identical to Ly49A, a C57BL/6-derived inhibitory receptor, with only 56% identity to Ly49D in this region. Interestingly, the C57L molecule functions as an inhibitory receptor even though it recognizes the same ligand as Ly49D from C57BL/6, highlighting the complexity of allelic polymorphisms in the Ly49 gene cluster.
Whereas aCGH data are typically interpreted with respect to gene content as reflected by CNV, log2 signal ratios from probes within genomic regions that encompass multigene families with allelic polymorphisms are unlikely to have sufficient precision to discriminate between subtle differences in absolute copy number. In the data presented here, allelic polymorphisms of Ly49 genes probably affected the aCGH results analyzed here. For example, C57BL/6 mice have 15 identifiable Ly49 genes13. BALB/c mice have only 8 Ly49 genes, whereas 129 and NOD mice have an expansion in Ly49 genes with 19 and 22 genes, respectively14; 16; 35. When compared to the reference C57BL/6 strain, the 129 and NOD strains have increased numbers of Ly49 genes (15 versus 19 and 22, respectively) but these gene number differences are not even by a factor of 2. Yet, the log2 signal intensity data for these strains demonstrated a more robust difference, which showed up to a 16-fold loss of signal in BALB/c versus C57BL/6 and more than 4-fold gain in signal in NOD versus C57BL/6. Therefore, we suspect that allelic polymorphisms of individual genes also contributed to differences in hybridization signals.
Finally, the aCGH analysis provided the basis for classifying mAb allo-reactivity. Consistent with aCGH grouping, we found that all NK cells from BTBR and NZW mice reacted with mAb PK136, similar to B6 mice which express the NK1.1 epitope on Nkrp1c. In contrast, mAb PK136 reacted with a subset of NK cells in C58, C57L, and FVB mice which are grouped with SJL mice that express the NK1.1 epitope on Nkrp1b. Moreover, mAb pK136 did not react with many strains27, including SM as shown here, that are grouped with BALB/c by aCGH analysis. While additional cDNA cloning will be required to precisely ascertain the molecules reactive with mAb PK136 in the different strains, the aCGH data suggest that the relevant epitope will be on molecules very closely related to Nkrp1c or Nkrp1b, expressed on all NK cells or a subset of NK cells, respectively, in the indicated mouse strains. Regardless, the aCGH groups are also closely aligned with expressed proteins with differences in mAb allo-reactivity.
In summary, despite the complexities inherent in aCGH analysis of the polymorphic NKC, our current study provides structural genomic information on inbred mouse strains that were verified by RFLP analysis and antibody reactivity with allotypic determinants. Thus, beyond providing information about the NKC, our data provide insight into the use of aCGH for analysis of other polymorphic gene clusters in the genome.
Materials and methods
Sample preparation and aCGH hybridization
All animal studies were performed according to institutional guidelines for animal experimentation. DNA from the following 20 inbred mouse strains were analyzed by aCGH as previously described36 and data are available in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/index.cgi) under accession number GSE10656: C57BL/6J, 129S1/SvlmJ, BALB/cByJ, NOD/ShiLtJ, 129X1/SvJ, SJL/J, AKR/J, C58/J, BTBR, SM/J, A/J, FVB/NJ, C3H/J, NZB/B1NJ, PL/J, DBA2/J, LG/J, SWR/J, KK/HlJ, and C57L/J. In a second set of samples, CBA/J, and NZW/LacJ were analyzed using the same methodology and oligonucleotide arrays. In brief, DNA was extracted from the spleens and kidneys of healthy, male mice 8 to 10 wks of age obtained from The Jackson Laboratory (Bar Harbor, ME, USA). The aCGH studies were performed using tiling-path CGH long oligonucleotide arrays for whole-genome analysis in the mouse (mm8, NCBI Build 36.1), designed and manufactured by Roche NimbleGen (Madison, WI, USA). The array consisted of 2.1 million (M) unique probes (45-75 bp) with a median probe spacing of 1 kb. (Here we used the term “unique probe” to indicate probes that identified only one C57BL/6 genomic sequence by BLAST analysis. It is possible that some of the probes bound to more than one genomic segment in other strains, as might be expected from genes which are duplicated or display allelic polymorphisms.) Unamplified genomic DNA was labeled with Cy3 (test strains) or Cy5 (reference strain, C57BL/6J). Labeling, hybridization, washing and array imaging were performed by NimbleGen Systems in a two-color format as previously described37. The mad.1dr values generated from the image files using the Nimblescan software were well below the recommended value for high quality (<0.23). Data for the second set of strains will be available at GEO (http://www.ncbi.nlm.nih.gov/geo/index.cgi) (accession no. GSE20090).
Data Statistical Analysis
All mouse genome coordinates are based on the reference C57BL/6 strain using NCBI Build 36.1 (mm8). Signal intensities and genomic coordinates for all probes were compiled, and inherent differences between Cy3 and Cy5 dyes were normalized using normalize.qspline method from the Bio-conductor software package38. As a convenient way to capture the variability along chromosome 6, the log2 signal ratios were organized into bins (a defined chromosome length in terms of bp). The entire chromosome was divided into consecutive bins, beginning with the position of the first probe. We used the variance of each bin to represent the variability along the chromosome (both gains and losses of hybridization signals in a given genomic region). The size of the bin (length of genomic segment) appeared to have minimal effect on displaying the regions with the highest variability on chromosome 6 (Supplemental Figure S2).
To calculate the variance of the signal ratios within each bin, we used the following equation:
| where | N is number of probes in each bin |
| xi is the expression value in probe i, and | |
| x̄ is the mean of the probe values in each bin |
This calculation takes into account the variability of the number of probes in each bin. Python's ‘NumPy’ library was used to calculate the variance39. The standard deviation of the variability (as reflected by the variances) along chromosome 6 was calculated using R40.
Clustering Methodology
A) Visual Inspection
To capture the magnitude of the variability at the 15,625 bp/bin resolution, the summation of absolute values of the signal ratios within each bin was calculated. To account for the variability in probe density within each bin (Supplemental Figure S5), each sum was multiplied by the ratio of the average number of probes per bin (for the entire length of chromosome 6), and the actual number of probes in the corresponding bin (i.e.- each bin has its own normalization factor). This normalization allows a chromosome-wide comparison of the variability from which a visual clustering pattern emerged.
B) Unsupervised Clustering Algorithms
We first selected the genomic region of interest to cluster based on the probe positions. The Ly49 gene family region ranged from 129,796,847 bp position to 130,348,221 bp position (chromosome 6). This 551,375 bp genomic region included 383 probes. The Nkrp1 gene family region ranged from 128,574,846 bp position to 129,152,133 bp position (chromosome 6). This 577,288 bp genomic region included 449 probes. We performed two types of unsupervised clustering analysis of the log2 ratios of signal intensities of each probe (normalized). One is the non-hierarchical clustering using the partitioning around medoids (PAM) function as implemented in R's ‘cluster’ package40. The assigned number of medoids (n), ranged between 3 and 9, which encompassed the number of RFLP variants identified for the Ly49 and Nkrp1 haplotypes, respectively. The silhouette function in R was used to determine the appropriate number of clusters (based on the largest average silhouette width value). The other is the agglomerative (bottom-up) hierarchical clustering based on the Euclidean distance41. The average linkage method was used in clustering the strains as implemented in the hclust function in the ‘cluster’ package of R40.
RFLP Analysis
Splenic DNA was obtained from the Mouse DNA Resource at The Jackson Laboratory (Bar Harbor, Maine). A total of 5 μg of DNA was digested with HindIII or EcoRI, electrophoresed on 0.8% agarose gels, and transferred to nylon membranes as previously described42. The Ly49a probe was the 640 bp PstI fragment of the pA1.3 plasmid containing the Ly49a cDNA (accession # M25812)42. In some cases, a 659-bp probe was generated from the Ly49a cDNA (accession # M77753) using the following PCR primers: 5′-TTCTAAACCACCACAATAACTGCAG-3′ (forward) and 5′-GACTAAGTCCAATGGTCAAAACACT-3′ (reverse). The Nkrp1a probe was a 810 bp EcoRI/HindIII fragment of the Nkrp1a cDNA in a Bluescript plasmid2. In some cases, a 816 bp probe was generated from the Nkrp1a cDNA using the following PCR primers: 5′-CTGCAGGAATTCGGCACGAG (forward) and 5′-GCTTCATGTTGTAGCACTTCAGTTT-3′ (reverse). The probes were labeled with 32P using the Amersham Rediprime II DNA labeling system (GE Healthcare, Piscataway, NJ).
Flow Cytometry and Analysis
Mice used in the FACS analysis were obtained from The Jackson Laboratory (Bar Harbor, M E, USA). Anti-CD3 (clone 145-2C11), anti-CD19 (clone 1D3), anti-NK1.1 (clone PK136), and anti-NKp46/CD335 (clone 29A1.4) were purchased from BD Biosciences (San Jose, CA). Staining was performed by standard methods7. All flow cytometry data were collected using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Supplementary Material
Acknowledgments
The authors thank Megan Cooper and Beatrice Plougastel for careful reading of the manuscript.
Funding. Work in the Yokoyama laboratory is supported by grants from the National Institute of Allergy and Infectious Diseases and the Howard Hughes Medical Institute. The Graubert laboratory is supported by the National Cancer Institute (P01 CA101937).
Footnotes
Competing interests: The authors have declared that no competing interests exist.
Supplemental Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene).
References
- 1.Yokoyama WM. Chapter 16. Natural killer cells. In: Paul WE, editor. Fundamental Immunology. 6th. Lippincott-Raven; New York: 2008. pp. 483–517. [Google Scholar]
- 2.Yokoyama WM, Ryan JC, Hunter JJ, Smith HR, Stark M, Seaman WE. cDNA cloning of mouse NKR-P1 and genetic linkage with Ly-49. Identification of a natural killer cell gene complex on mouse chromosome 6. Journal of Immunology. 1991;147(9):3229–3236. [PubMed] [Google Scholar]
- 3.Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1(2):129–139. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4(8):801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 5.Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci U S A. 2004;101(10):3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlyle JR, Mesci A, Fine JH, Chen P, Belanger S, Tai LH, et al. Evolution of the Ly49 and Nkrp1 recognition systems. Semin Immunol. 2008;20(6):321–330. doi: 10.1016/j.smim.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358(6381):66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 8.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3(4):304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 9.Kung SK, Su RC, Shannon J, Miller RG. The NKR-P1B gene product is an inhibitory receptor on SJL/J NK cells. J Immunol. 1999;162(10):5876–5887. [PubMed] [Google Scholar]
- 10.Carlyle JR, Martin A, Mehra A, Attisano L, Tsui FW, Zuniga-Pflucker JC. Mouse NKR-P1B, a novel NK1.1 antigen with inhibitory function. J Immunol. 1999;162(10):5917–5923. [PubMed] [Google Scholar]
- 11.Carlyle JR, Mesci A, Ljutic B, Belanger S, Tai LH, Rousselle E, et al. Molecular and genetic basis for strain-dependent NK1.1 alloreactivity of mouse NK cells. J Immunol. 2006;176(12):7511–7524. doi: 10.4049/jimmunol.176.12.7511. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama WM, Kehn PJ, Cohen DI, Shevach EM. Chromosomal location of the Ly-49 (A1, YE1/48) multigene family. Genetic association with the NK 1.1 antigen. Journal of Immunology. 1990;145(7):2353–2358. [PubMed] [Google Scholar]
- 13.Wilhelm BT, Gagnier L, Mager DL. Sequence analysis of the ly49 cluster in C57BL/6 mice: a rapidly evolving multigene family in the immune system. Genomics. 2002;80(6):646–661. doi: 10.1006/geno.2002.7004. [DOI] [PubMed] [Google Scholar]
- 14.Makrigiannis AP, Patel D, Goulet ML, Dewar K, Anderson SK. Direct sequence comparison of two divergent class I MHC natural killer cell receptor haplotypes. Genes Immun. 2005;6(2):71–83. doi: 10.1038/sj.gene.6364154. [DOI] [PubMed] [Google Scholar]
- 15.Proteau MF, Rousselle E, Makrigiannis AP. Mapping of the BALB/c Ly49 cluster defines a minimal natural killer cell receptor gene repertoire. Genomics. 2004;84(4):669–677. doi: 10.1016/j.ygeno.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Belanger S, Tai LH, Anderson SK, Makrigiannis AP. Ly49 cluster sequence analysis in a mouse model of diabetes: an expanded repertoire of activating receptors in the NOD genome. Genes Immun. 2008;9(6):509–521. doi: 10.1038/gene.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3(1):e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, et al. Copy number variation: new insights in genome diversity. Genome Res. 2006;16(8):949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 19.Pinkel D, Albertson DG. Comparative genomic hybridization. Annu Rev Genomics Hum Genet. 2005;6:331–354. doi: 10.1146/annurev.genom.6.080604.162140. [DOI] [PubMed] [Google Scholar]
- 20.Grus WE, Zhang J. Rapid turnover and species-specificity of vomeronasal pheromone receptor genes in mice and rats. Gene. 2004;340(2):303–312. doi: 10.1016/j.gene.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 21.Brekke KM, Garrard WT. Assembly and analysis of the mouse immunoglobulin kappa gene sequence. Immunogenetics. 2004;56(7):490–505. doi: 10.1007/s00251-004-0659-0. [DOI] [PubMed] [Google Scholar]
- 22.Bachmanov AA, Beauchamp GK. Taste receptor genes. Annual Review of Nutrition. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, et al. Genealogies of mouse inbred strains. Nat Genet. 2000;24(1):23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 24.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press; 1994. p. 487. [Google Scholar]
- 25.Willison KR, Lyon MF. A UK-centric history of studies on the mouse t-complex. Int J Dev Biol. 2000;44(1):57–63. [PubMed] [Google Scholar]
- 26.Ryan JC, Turck J, Niemi EC, Yokoyama WM, Seaman WE. Molecular cloning of the NK1.1 antigen, a member of the NKR-P1 family of natural killer cell activation molecules. Journal of Immunology. 1992;149(5):1631–1635. [PubMed] [Google Scholar]
- 27.Koo GC, Peppard JR. Establishment of monoclonal anti-Nk-1.1 antibody. Hybridoma. 1984;3:301–303. doi: 10.1089/hyb.1984.3.301. [DOI] [PubMed] [Google Scholar]
- 28.Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14(9):1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448(7157):1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 30.Wade CM, Kulbokas EJ, 3rd, Kirby AW, Zody MC, Mullikin JC, Lander ES, et al. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420(6915):574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 31.Boursot P, Belkhir K. Mouse SNPs for evolutionary biology: beware of ascertainment biases. Genome Res. 2006;16(10):1191–1192. doi: 10.1101/gr.5541806. [DOI] [PubMed] [Google Scholar]
- 32.Depatie C, Muise E, Lepage P, Gros P, Vidal SM. High-resolution linkage map in the proximity of the host resistance locus CMV1. Genomics. 1997;39(2):154–163. doi: 10.1006/geno.1996.4498. [DOI] [PubMed] [Google Scholar]
- 33.Forbes CA, Brown MG, Cho R, Shellam GR, Yokoyama WM, Scalzo AA. The Cmv1 host resistance locus is closely linked to the Ly49 multigene family within the natural killer cell gene complex on mouse chromosome 6. Genomics. 1997;41(3):406–413. doi: 10.1006/geno.1997.4667. [DOI] [PubMed] [Google Scholar]
- 34.Mehta IK, Smith HRC, Wang J, Margulies DH, Yokoyama WM. A “chimeric” C57L-derived Ly49 inhibitory receptor resembling the Ly49D activation receptor. Cell Immunol. 2001;209(1):29–41. doi: 10.1006/cimm.2001.1786. [DOI] [PubMed] [Google Scholar]
- 35.Anderson SK, Dewar K, Goulet ML, Leveque G, Makrigiannis AP. Complete elucidation of a minimal class I MHC natural killer cell receptor haplotype. Genes Immun. 2005;6(6):481–492. doi: 10.1038/sj.gene.6364232. [DOI] [PubMed] [Google Scholar]
- 36.Cahan P, Li Y, Izumi M, Graubert TA. The impact of copy number variation on local gene expression in mouse hematopoietic stem and progenitor cells. Nat Genet. 2009;41(4):430–437. doi: 10.1038/ng.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, Nair P, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44(3):305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 38.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hetland M. Beginning Python: From Novice to Professional. Apress; Berkeley: 2005. Extending Python; pp. 357–372. [Google Scholar]
- 40.Verzani J. Using R for introductory statistics. Chapman and Hall/CRC Press; Boca Raton: 2005. p. 432. [Google Scholar]
- 41.Pevsner J. Bioinformatics and functional genomics. Wiley-Liss; Hoboken: 2003. Gene expression: Microarray data analysis; pp. 189–221. [Google Scholar]
- 42.Yokoyama WM, Jacobs LB, Kanagawa O, Shevach EM, Cohen DI. A murine T lymphocyte antigen belongs to a supergene family of type II integral membrane proteins. Journal of Immunology. 1989;143(4):1379–1386. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.