Abstract
The convergence of phage-displayed peptide libraries and recombinant viral vectors launched a promising new direction in targeted viral gene therapeutics, but the translation of targeting peptides to functional cancer therapeutic agents has been challenging. Here we report progress in developing a successful strategy to optimize targeted viral infection through adenovirus-displayed semi-random peptide libraries. A phage-derived peptide targeting the Prostate Specific Membrane Antigen (PSMA) was genetically incorporated into the adenoviral capsid Fiber protein and flanked by random peptide cassettes. The resulting adenovirus library was biopanned against PSMA-expressing cells and tumors to identify a PSMA-retargeted adenovirus. While the initial peptide alone was unable to target viral infection, the selected virus preferentially infects PSMA-expressing cells through the targeting peptide and infects LNCaP tumors after intravenous injection. Our results indicate that virus-displayed semi-random peptide libraries can be used to optimize targeting infection. This approach represents a novel principle for developing targeted agents in a variety of disease models.
Keywords: PSMA, Peptide Library, Genetically Modified Adenovirus, Cancer Gene Therapy, Prostate Cancer
Introduction
Cancer fighting adenoviral vectors are now clinically available for intra-tumoral injection; however, the translation of this approach to the systemic treatment of metastatic tumors requires solutions to multiple obstacles including broad viral tropism, liver sequestration, hepatic toxicity, low-level Coxsackie and Adenovirus Receptor (CAR) expression in some tumor types, and host immunity (1, 2). Some of these problems have been overcome by genetically targeting the therapeutic effect through tissue-specific promoters or oncolytic mechanisms. For example, the first tissue-specific lytic adenovirus was developed for the treatment of prostate cancer (PCa) by limiting viral replication to cells containing activated androgen receptor (AR) (3). These prostate-selective lytic vectors have been evaluated in clinical trials both as direct tumor injected agents as well as systemically administered virus for castration resistant metastatic PCa (4). While the therapeutic effect was limited to prostatic cells, the efficacy as a systemic therapy was hampered by viral sequestration and clearance. This reflects the loss of over 90% of intravenously (IV) administered adenovirus through liver sequestration (5). Next generation cancer gene therapy vectors seek to enhance efficacy through capsid modifications designed to limit viral clearance and/or improve tumor infection through alternative receptors.
Phage-displayed peptide libraries offer a powerful means to develop ligands for most any target molecule. The resulting targeting peptides are small and could theoretically be incorporated into viral capsid proteins to improve specificity or expand tropism to recalcitrant tumors. However, peptide-retargeted adenoviral infection rarely succeeds, most likely due to the negative effect of the peptide on fiber folding and capsid assembly, or the effect of capsid folding on the structural integrity of the targeting peptides (6). Significant efforts have been made to overcome these obstacles, by developing adenoviral capsid-displayed-peptide libraries (7, 8) or by utilizing larger whole-domain structures (1, 2). A major limitation of adenovirus-displayed peptide libraries is diversity, which is limited by difficulties associated with the large viral genome, poor transfection efficiencies for viral packaging, and library amalgamation during viral amplification and production. Nonetheless a lower diversity library is achievable (7, 8), which could theoretically be applied to improve targeting by an existing peptide.
PSMA is an attractive alternate receptor for targeted infection. It is an internalizing cell surface carboxypeptidase highly expressed on the surface of PCa cells (9). Because of this, PSMA is one of the most highly sought after targets for prostate cancer imaging and therapeutics today (10). We and others have developed PSMA-binding peptides through phage-display screens (7, 11). These peptides have been successfully applied by independent groups to target other therapeutic agents (12). However, when we translated these PSMA-targeting peptides into the Fiber capsid protein for retargeted adenoviral infection, PSMA-specificity was lost. Therefore, we sought to rescue PSMA-peptide targeting through an adenovirus-displayed semi-random peptide library approach.
Materials and Methods
Cell lines
293cre57 cells were provided by Stephen Langer (University of Colorado, Boulder, CO). Cre recombinase activity was authenticated by Cre Stop light assay. PC-3-PSMA cells were provided by Warren Heston (Cleveland Clinic, Cleveland, OH) and authenticated for PSMA expression by western blot and ELISA. DPL-S11 cells are generated at our lab (14). PC-3, 293, and LNCaP cells were obtained from ATCC and were not further authenticated.
Construction of the Adenovirus-displayed peptide Library
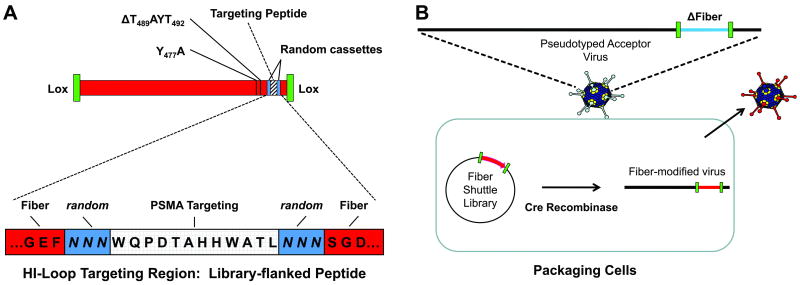
Oligonucleotides encoding PSMA-binding peptide (11), flanked by three random amino acid cassettes were synthesized and subcloned into the HI Loop of CAR-ablated Fiber in RPuc-FBR8 (ΔT489AYT492, Y477A) (Fig. 1A) at library scale. The resulting fiber-peptide shuttle library was recombined into the Fiber region of pseudotyped Adenovirus serotype 5 (Ad5), Ad5Track-Luc-Fex or Ad5-PSE-PBN-E1A-Fex (MOI=1), by Cre-recombinase-mediated cassette exchange in 6× 106 293cre57 cells and purified at 72hr as previously described (7) and summarized in Figure 1B.
Figure 1. Schematic illustration of semi-random Fiber-peptide library.
A, Fiber library. A preselected PSMA-targeting peptide, WQPDTAHHWATL, was cloned into the HI-loop of fiber knob and flanked by two random trimer peptide cassettes providing a potential diversity of 64 million different peptide orientations. The Fiber gene was further modified by ΔT489AYT492 and Y477A to ablate interaction with the natural receptor, CAR. The entire Fiber gene region is flanked by non-compatible unidirectional lox sites (green rectangle) for site directed cassette exchange. B, Generation of Adenoviral library. A pseudotyped acceptor virus, lacking any fiber coding region, was used to infect 293cre57 cells transfected with fiber-shuttle library cassettes. Cre-recombinase site-specifically transferred the modified fiber gene into the infected viral genome, resulting in amplification and packaging of the re-targeted virus.
In vitro library screening
Target cells were infected (MOI=1) for 2hr (round 1) or 1hr (subsequent rounds), washed, and incubated 3-4 days. Counter-selections were performed for 1hr in rounds 2-3, prior to positive selection (Table 1). The floxed Fiber-coding region was PCR rescued into the RPuc-Rescue shuttle vector (7) and 10-30 individual clones for each round were sequenced. The remaining library was amplified in large scale and applied as the shuttle-vector for the next round.
Table 1. Screening of Adenovirus-displayed peptide library in a PSMA-expressing cancer cell and tumor model.
| Screening Round | I | II | III | IV |
|---|---|---|---|---|
| Screening Type | In vitro | In vitro | In vitro | In vivo |
| E1 Gene Cassette | GFP | GFP | GFP | PSE-PBN-E1A |
| Counter Selection | N/A | 293 | PC-3 | Systemic Injection |
| Positive Selection | 293-PSMA | LNCaP | LNCaP | LNCaP Tumor |
In vivo selection
Subcutaneous LNCaP tumors in male athymic nu/nu mice (Charles River Laboratories) were grown to ∼0.5 cm3. 106 IU of Ad5-PSE-PBN-PSMA library was injected via tail vein. After three weeks, the tumor, kidney, lung, spleen and liver were collected, total DNA extracted, and the Fiber region PCR-amplified and subcloned as described above. Thirty clones were randomly sequenced.
Luciferase assays
Cells were infected (MOI =1) in 96 multiwell plates for 1hr, washed, fed complete media, and assayed for luciferase activity at 48hr (Promega, Madison, WI). Competition studies applied serially diluted peptide 30 minutes prior to infection.
Bioluminescence imaging
LNCaP tumor bearing mice were anesthetized with isoflurane three days after viral infection and D-luciferin was injected intraperitoneally. Bioluminescence images were observed with an in vivo imaging system (IVIS; Xenogen) and analyzed using Live Image software (Xenogen).
Virus purification
Recombinant adenovirus were purified by commercial adenovirus purification kit (AdenopureTM, Puresyn, PA) or iodixanol discontinuous density gradient ultracentrifugation and size exclusion column chromatography (13). Virus titer was determined by Adeno-XTM Rapid Titer Kit (BD Biosciences) or GFP using DPL-S11 cells which express an artificial anti-Fiber single-chain antibody receptor (14).
Results and Discussion
To generate an adenovirus-displayed semi-random peptide library we first subcloned a phage-derived PSMA-targeting peptide (WQPDTAHHWATL) (11), flanked by three random amino acid cassettes, into the HI-loop of an Ad5 fiber-shuttle vector (Fig. 1A). The host fiber gene was de-targeted from CAR-mediated infection through FG-loop T489AYT492 deletion and DE-loop Y477A mutation (15). The fiber-region of the shuttle-library was uni-directionally transferred into the native Ad5 fiber region by cre-recombinase-mediated-cassette-exchange in 6×106 packaging cells (Fig. 1B). Previous studies indicate that a library diversity of at least 105 can be achieved by this method (7). The resulting adenovirus library was harvested within a single life-cycle to prevent the generation of hybrid capsids, which could occur after virus spread and co-infection. In theory, this adenovirus-library approach can produce a high complexity of potential conformations for fiber protein folding, peptide folding, and orientation for peptide-target interaction.
The resulting adenovirus library was biopanned against PSMA-positive cell lines and tumors to isolate those adenoviruses which preferentially utilize PSMA as an alternative receptor (Table 1). Specifically, the initial adenovirus library was incubated with PSMA-expressing cells for one hour at a density of one infectious unit (IU) per cell; the cells were then washed and incubated for three days. Fiber gene cassettes from successfully infectious round one virus were rescued by PCR amplification and subcloning, via cre-recombinase-mediated-cassette-exchange, to generate a second round fiber-shuttle library, and finally a second round adenovirus displayed-peptide library. Various cell lines were used for counter- and positive-selections to minimize amplification of virus which infected via PSMA-independent mechanisms. Ten to twenty fiber-shuttle clones from each selection round were sequenced to ensure diversity and to identify potential candidates. By the third round, six unrelated candidate peptides were represented by multiple clones, indicating library maturation (supplemental Table S1). To identify the most successful targeted virus, we generated the fourth round library as a conditionally replicating virus library (Ad5PSE-PBN-E1A-PSMA-Lib), which utilizes an androgen receptor responsive cassette to drive E1A gene expression and viral replication. This replication-competent library was applied to a final and stringent in vivo selection, by intravenous injection, and allowed three weeks for viral infection, amplification, and spread within the LNCaP tumor. The tumor was harvested and peptide cassettes from the infectious viruses were rescued by PCR amplification and Cre-recombinase subcloning. Thirty randomly picked clones were sequenced and, surprisingly, a single peptide sequence (MetAEWQPDTAHHWATLPDP, named Saupw-1) was identified in all 30 clones. Diversity of the input fourth round library was re-confirmed by sequencing, suggesting that the in vivo selection was extremely stringent and resulted in a single successful targeting peptide.
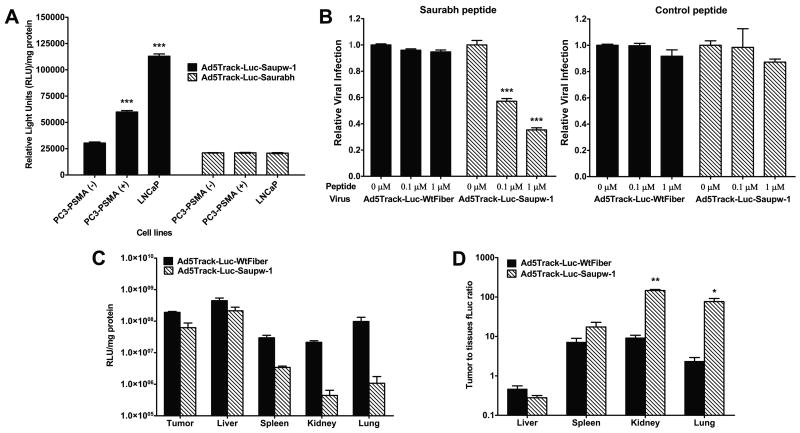
Reporter adenovirus displaying the in vivo selected peptide, Saupw-1, or the original non-flanked peptide (WQPDTAHHWATL), were evaluated for PSMA-mediated infection on three cell lines expressing varying levels of PSMA (Supplementary Fig. S1). Infection rate of the selected virus, Ad5Track-Luc-Saupw-1, directly correlated with PSMA-expression levels in these cells, while the virus-displayed parental peptide, Ad5Track-Luc-Saurabh, showed poor infection efficiency in all three lines (Fig. 2A). Infection of adenovirus with wild-type fiber, Ad5Track-Luc-WTFiber, correlated with CAR expression and failed to show any PSMA selectivity (Supplementary Fig. S1). These results indicate that the infection of Ad5Track-Luc-Saupw-1 is CAR independent and mediated by the peptide. To confirm this, competition experiments were performed with PSMA-targeting or control peptides. AdTrack-Luc-Saupw-1 was dose-dependently inhibited by the PSMA-targeting peptide (Fig. 2B), but not the control peptide. On the other hand, PSMA-targeting and control peptides had no effect on infection by wild-type fiber. These results support PSMA-targeted infection mediated through the fiber-displayed peptide. Similar inhibition studies with a peptide, containing the flanking regions, did not show an increased ability to inhibit viral infection (data not shown), indicating that the selected flanking peptides enhanced folding and display rather than peptide binding affinity. Interestingly, over 75% of the flanking sequences identified in each round contained at least one proline residue which suggests that peptides in the HI loop may be constrained or interfere with proper fiber folding and assembly. This was not unique to the PSMA-targeting peptide, but was also seen in other library selections.
Figure 2. PSMA mediated adenoviral infection in cell and tumor models.
A, PSMA-targeted infection. LNCaP, PC-3-PSMA (+) and PC-3-PSMA (-) cells were infected with Ad5Track-Luc-Saupw-1 (selected virus) or Ad5Track-Luc-Saurabh (parental virus), MOI =1. Virus infection was quantified by firefly luciferase activity (Relative Light Units, RLU, per mg protein). Columns, averages (n=3); Bars, SE. B, Infection is mediated by PSMA-binding peptide. LNCaP cells were pre-incubated with serially diluted PSMA-binding peptide or an unrelated control peptide for 30 min, followed by infection with Ad5Track-Luc-Saupw-1 or Ad5Track-Luc-WtFiber. Infection was quantified by luciferase activity, relative to non-inhibited wild-type fiber. Bars represent mean±SE infection of three replicates. C, Adenoviral infection following systemic injection. Mice bearing LNCaP tumors were intravenously injected with Ad5Track-Luc-Saupw-1 or Ad5Track-Luc-WtFiber (1 × 109 IU). After four days, viral infection was quantified by ex vivo luciferase activity of tissue lysates. Columns, averages (n=3); Bars, SE. D, Relative infection. Tumor to tissue luciferase ratios for each virus four days after virus administration. Statistical evaluation: *, P<0.05, **, P<0.01, ***, P < 0.001 (Student's t-test).
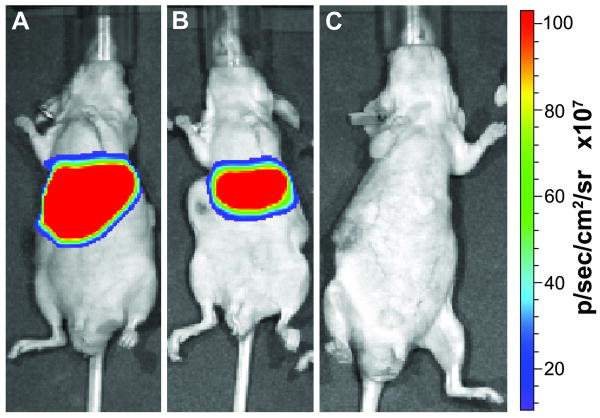
We finally sought to evaluate the ability of the selected virus to infect tumors in vivo after intravenous injection. Adenovirus biodistribution is complex and affected by many factors. In murine tissues, adenoviral biodistribution is independent of CAR binding. A series of pioneering papers recently revealed that hepatic infection is mediated by interactions between the viral hexon protein and blood factor X (16-18). Indeed, hexon gene modification or blockade can reduce hepatic infection and viral induced hepatic toxicity (5, 16). However, the majority of injected adenovirus still remains in the liver through non-infectious sequestration (19). Interestingly, while CAR is not the dominant receptor for hepatic infection, it remains important in tumor models. Tumor models which express low levels of CAR are poorly infected by Ad5 vectors after systemic injection; however, infection can be rescued by the use of alternate serotype Fiber genes. Similarly, we found that the selected PSMA-targeting peptide was capable of rescuing CAR-independent infection of LNCaP tumors (Fig. 2C). Following intravenous administration of Ad5Track-Luc-Saupw-1 (109 IU), LNCaP tumors were infected with similar efficiency when compared to adenovirus with wild-type fiber (Fig. 2C). These results reflect efficient retargeting by the PSMA-binding peptide. As expected, both virus presented significant liver transduction (Fig. 2C), as confirmed by in vivo luciferase imaging (Fig. 3). Transgene expression in tumor xenografts could not be observed from bioluminescence imaging with either Wild Type or PSMA-targeted Fiber genes, which is generally expected following systemic administration of replication-incompetent adenovirus reporters. On the other hand, Ad5Track-Luc-Saupw-1 infected kidney and lung with significantly lower efficiency. These results are consistent with previous studies utilizing Y477A detargeting mutations (20). Therefore, the combination of CAR detargeting and PSMA retargeting has affected the systemic infection pattern of the virus in some tissues while still retaining efficient tumor cell infection (Fig. 2D).
Figure 3. Luminescent imaging of viral infection and biodistribution.
Subcutaneous LNCaP tumors were prepared in nude mice. When the tumors reached approximately 0.8 cm3, 109 IU/per mouse of A, Ad5Track-Luc-WtFiber, B, Ad5Track-Luc-Saupw-1, or C, PBS were administered by tail-vein injection into mice, respectively. Three days after virus injection, mice were anesthetized and imaged.
In summary our study has demonstrated that adenovirus-displayed semi-random peptide libraries, in the form of flanking cassettes, can be applied to rescue the targeting ability of phage-derived peptides which were previously ineffective in such a complex environment. The result is a genetically modified adenovirus which successfully infects prostatic cells and tumors through PSMA. This approach can be applied to develop targeted agents for prostate cancer, or other cancers, not only in adenovirus but also in other experimental therapeutic platforms.
Supplementary Material
Acknowledgments
We thank Stephen Langer for providing 293cre57 cells, Victor Van Beusechem for the S11 Fiber pseudoreceptor, John Isaacs and Don Vindivich for technical assistance, and Warren Heston for the PC-3-PSMA cells. This study is funded by National Institutes of Health (1R01CA121153-01A2 to R.R.) and the Robert & Donna Tompkins Foundation.
References
- 1.Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8:573–87. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry MA, Hofherr SE, Chen CY, Senac JS, Hillestad ML, Shashkova EV. Systemic delivery of therapeutic viruses. Curr Opin Mol Ther. 2009;11:411–20. [PubMed] [Google Scholar]
- 3.Rodriguez R, Schuur ER, Lim HY, Henderson GA, Simons JW, Henderson DR. Prostate attenuated replication competent adenovirus (ARCA) CN706: a selective cytotoxic for prostate-specific antigen-positive prostate cancer cells. Cancer Res. 1997;57:2559–63. [PubMed] [Google Scholar]
- 4.Lupold SE, Rodriguez R. Adenoviral vectors for prostate cancer gene therapy. Cancer Therapy. 2005;3:267–84. [Google Scholar]
- 5.Shashkova EV, May SM, Doronin K, Barry MA. Expanded anticancer therapeutic window of hexon-modified oncolytic adenovirus. Mol Ther. 2009;17:2121–30. doi: 10.1038/mt.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh D, Barry MA. Selection of muscle-binding peptides from context-specific peptide-presenting phage libraries for adenoviral vector targeting. J Virol. 2005;79:13667–72. doi: 10.1128/JVI.79.21.13667-13672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupold SE, Kudrolli TA, Chowdhury WH, Wu P, Rodriguez R. A novel method for generating and screening peptides and libraries displayed on adenovirus fiber. Nucleic Acids Res. 2007;35:e138. doi: 10.1093/nar/gkm914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miura Y, Yoshida K, Nishimoto T, et al. Direct selection of targeted adenovirus vectors by random peptide display on the fiber knob. Gene Ther. 2007;14:1448–60. doi: 10.1038/sj.gt.3303007. [DOI] [PubMed] [Google Scholar]
- 9.Elgamal AA, Holmes EH, Su SL, et al. Prostate-specific membrane antigen (PSMA): current benefits and future value. Semin Surg Oncol. 2000;18:10–6. doi: 10.1002/(sici)1098-2388(200001/02)18:1<10::aid-ssu3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Foss CA, Byun Y, et al. Radiohalogenated prostate-specific membrane antigen (PSMA)-based ureas as imaging agents for prostate cancer. J Med Chem. 2008;51:7933–43. doi: 10.1021/jm801055h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aggarwal S, Singh P, Topaloglu O, Isaacs JT, Denmeade SR. A dimeric peptide that binds selectively to prostate-specific membrane antigen and inhibits its enzymatic activity. Cancer Res. 2006;66:9171–7. doi: 10.1158/0008-5472.CAN-06-1520. [DOI] [PubMed] [Google Scholar]
- 12.Rege K, Patel SJ, Megeed Z, Yarmush ML. Amphipathic peptide-based fusion peptides and immunoconjugates for the targeted ablation of prostate cancer cells. Cancer Res. 2007;67:6368–75. doi: 10.1158/0008-5472.CAN-06-3658. [DOI] [PubMed] [Google Scholar]
- 13.Peng HH, Wu S, Davis JJ, et al. A rapid and efficient method for purification of recombinant adenovirus with arginine-glycine-aspartic acid-modified fibers. Anal Biochem. 2006;354:140–7. doi: 10.1016/j.ab.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoti N, Chowdhury W, Mustafa S, et al. Armoring CRAds with p21/Waf-1 shRNAs - The next generation of oncolytic adenoviruses. Cancer Gene Therapy. 2010:1–13. doi: 10.1038/cgt.2010.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roelvink PW, Mi Lee G, Einfeld DA, Kovesdi I, Wickham TJ. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science (New York, NY. 1999;286:1568–71. doi: 10.1126/science.286.5444.1568. [DOI] [PubMed] [Google Scholar]
- 16.Waddington SN, McVey JH, Bhella D, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Kalyuzhniy O, Di Paolo NC, Silvestry M, et al. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5483–8. doi: 10.1073/pnas.0711757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vigant F, Descamps D, Jullienne B, et al. Substitution of hexon hypervariable region 5 of adenovirus serotype 5 abrogates blood factor binding and limits gene transfer to liver. Mol Ther. 2008;16:1474–80. doi: 10.1038/mt.2008.132. [DOI] [PubMed] [Google Scholar]
- 19.Di Paolo NC, van Rooijen N, Shayakhmetov DM. Redundant and synergistic mechanisms control the sequestration of blood-born adenovirus in the liver. Mol Ther. 2009;17:675–84. doi: 10.1038/mt.2008.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayo-Puxan N, Cascallo M, Gros A, Huch M, Fillat C, Alemany R. Role of the putative heparan sulfate glycosaminoglycan-binding site of the adenovirus type 5 fiber shaft on liver detargeting and knob-mediated retargeting. J Gen Virol. 2006;87:2487–95. doi: 10.1099/vir.0.81889-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.