Abstract
Background
Mitochondrial DNA (mtDNA) influences metabolic diseases and perhaps antiretroviral therapy (ART) complications. We explored associations between European mtDNA haplogroups and metabolic changes among A5142 participants.
Methods
757 ART-naïve subjects were randomized to one of three class-sparing ART regimens including efavirenz and/or lopinavir/ritonavir with or without nucleoside reverse transcriptase inhibitors (NRTIs). Non-randomized NRTIs included stavudine, tenofovir, or zidovudine, each with lamivudine. Fasting lipid profiles and whole-body dual-energy X-ray absorptiometry (DEXA) were performed. Nine European mtDNA haplogroups were determined for 231 self-identified non-Hispanic white subjects. Metabolic changes from baseline to 96 weeks were analyzed by haplogroup.
Results
Median age was 39 years, 9% were female, and 37%, 32%, and 30% were randomized to NRTI-containing regimens with either efavirenz or lopinavir/ritonavir, and an NRTI-sparing regimen respectively. Among NRTI-containing regimens, 51% included zidovudine, 28% tenofovir, and 21% stavudine. Compared with other haplogroups, mtDNA haplogroup I (N=10) had higher baseline non-HDL cholesterol (160 mg/dL [interquartile range 137–171] vs. 120 mg/dL [104–136]; p=0.005), a decrease in non-HDL cholesterol over 96 weeks (−14% [−20-+6] vs. +25% [+8-+51]; p<0.001), tended to have more baseline extremity fat, and had more extremity fat loss by DEXA (−13% [−31-+12] vs. +9% [−13-+26]; p=0.08) and lipoatrophy (50% vs. 20%; p=0.04). Haplogroup W (N=5; all randomized to NRTI-sparing regimens) had the greatest increase in extremity fat (+35.5% [+26.8 - +54.9]; P=0.02).
Conclusions
Lipids and extremity fat were associated with European mtDNA haplogroups in this HIV-infected population. These preliminary results suggest that mitochondrial genomics may influence metabolic parameters before and during ART.
Keywords: Mitochondrial genome, DNA, Mitochondrial, Antiretroviral Therapy, Highly Active, Dyslipidemias, HIV-Associated Lipodystrophy Syndrome, Metabolic Diseases
Introduction
Mitochondrial DNA (mtDNA) is distinct from nuclear DNA, encodes thirteen subunits of the electron transport chain, and exhibits abundant genetic variation across >16,000 base pairs. Human mtDNA sequences have diverged over approximately the last 150,000 years due to natural selection and human migration, resulting in distinct patterns of single nucleotide polymorphisms (SNPs), called haplogroups.[1] Evidence for functional differences among mtDNA haplogroups has been demonstrated in studies of human longevity,[2] neurodegenerative disorders,[3] and metabolic disease and cardiovascular risk.[4–7]
Antiretroviral therapy (ART) improves HIV morbidity and mortality. Unfortunately ART also has treatment-limiting metabolic complications such as dyslipidemia, insulin resistance, abnormal fat accumulation, and peripheral fat loss (lipoatrophy). Inhibition of mtDNA replication by some nucleoside reverse transcriptase inhibitors (NRTIs) is believed to lead to a cascade of mitochondrial dysfunction with impaired oxidative phosphorylation, overproduction of free radicals, tissue injury, and ultimately symptomatic toxicity.[8, 9] There is wide variation in the clinical manifestations of NRTI toxicities. This phenotypic variation together with their putative mechanism(s) suggests a role for host mtDNA variation in determining susceptibility. Our group has examined the role of mtDNA variation in susceptibility to NRTI-associated peripheral neuropathy, identifying preliminary associations with mtDNA haplogroups in U.S. clinical trial participants of both European[10] and African[11] descent. We[12] and others[13, 14] have also explored European mtDNA haplogroups and ART-associated metabolic effects, primarily lipoatrophy.
AIDS Clinical Trials Group (ACTG) study A5142 was a trial designed to determine virologic efficacy and adverse metabolic effects of three class-sparing ART regimens.[15] The important metabolic effects which have been attributed to various ART classes, including lipoatrophy, were key secondary study outcomes and have been presented elsewhere.[16] Briefly, A5142 confirmed that lipoatrophy was greatest in participants treated with thymidine analogue NRTIs, and, more surprisingly, in those randomized to receive the non-NRTI (NNRTI), efavirenz. Lipid effects were greatest in the NRTI-sparing arm. We hypothesize that mtDNA variation as represented in haplogroups confers subtle differences in oxidative phosphorylation, mtDNA replication, and/or apoptotic regulation, and thus is a marker for relative susceptibility to ART complications. In the pharmacogenomic analyses reported herein we explore associations between European mtDNA haplogroups and metabolic changes during ART in non-Hispanic whites who participated in A5142.
Methods
Study subjects
This exploratory analysis included participants from ACTG study A5142 (NCT# 00050895), a phase III, multicenter, randomized, open-label trial that enrolled HIV-1-infected, ART-naïve volunteers in the United States with plasma HIV-1 RNA of at least 2000 copies/mL.[15] Participants used self-identified categories for race/ethnicity. Due to the relative ease in genotyping and defining European mtDNA haplogroups compared with other racial/ethnic groups, this initial analysis focused on those self-identified as “white, non-Hispanic.”
Study design
A5142 participants were randomized to one of three ART regimens: lopinavir/ritonavir plus efavirenz (lopinavir-efavirenz arm) or two NRTIs plus either lopinavir/ritonavir (lopinavir arm) or efavirenz (efavirenz arm). As described in detail elsewhere,[15, 16] investigators selected NRTIs (zidovudine, stavudine extended release [XR], or tenofovir) to be given with lamivudine if the subject was randomized to the NRTI-containing arms. Randomization was stratified by NRTI choice. Subjects were followed for 96 weeks after the last subject was enrolled. Changes in randomized regimen or NRTI were allowed for toxicity or intolerance and were considered endpoints in the efficacy analysis. Body composition was determined by whole-body dual-energy X-ray absorptiometry (DEXA) at entry, 48 weeks, and 96 weeks. Scan results were determined at a central reading site (Tufts University, Boston, MA, USA) by readers blinded to treatment assignments. Fasting serum lipids were measured at each site by commercial laboratories at entry, weeks 12, 24, and every 24 weeks.
The present study was a retrospective analysis of non-Hispanic white participants with DNA available under ACTG protocol A5128[17]. Both retrospective cohort (for changes in lipids and extremity fat) and case-control (for lipoatrophy) study designs were utilized. A5142 and A5128 protocols were approved by institutional review boards at each study site, and participants provided written informed consent. The Vanderbilt Committee for the Protection of Human Subjects and the ACTG approved the use of DNA.
DNA sequencing and mitochondrial haplogroup determination
DNA was isolated from whole blood using PUREGENE® (Gentra Systems Inc., Minneaplois, MN). Genotyping was performed with the ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems Inc., Foster City, CA) using the 5’ nuclease allelic discrimination Taqman assay. Based on the work of Torroni, et al.[18], we characterized single nucleotide polymorphisms at positions 1719, 4580, 7028, 8251, 9055, 10398, 12308, 13368, 13708, and 16391. Probes and primers were based on previously reported genotyping[3] and have been presented in detail elsewhere.[12] Raw genotypic data were analyzed using ABI Sequence Detection System version 2.0 software and genotype calls were confirmed by manual inspection of the plots.
Statistical analysis
Simple proportions are used to describe demographic and genetic data. Medians and interquartile ranges (IQR) are presented for continuous data. Fisher’s exact or Pearson Chi-squared tests and Wilcoxon rank-sum (Mann-Whitney U) test were used for comparisons of categorical and continuous covariates, respectively. Univariate and multivariate logistic regression were used to determine exact odds ratios (OR) and 95% confidence intervals (CI) for haplogroup associations with lipoatrophy. In the multivariate model, ART exposure was included as intent-to-treat based on randomization. In the primary metabolic study, results did not differ substantially in intent-to-treat versus as-treated analyses.[16] Analyses were not corrected for multiple comparisons in this exploratory study. Stata SE version 10 (Stata Corp., College Station, TX, USA) was used for statistical analyses.
Results
Study population demographics and baseline HIV disease parameters
A total of 245 self-identified white, non-Hispanic A5142 participants (32% of all study participants; 89% of white, non-Hispanic participants) had DNA available for analysis. Of these, 231 (94%) were classified into one of nine major European mtDNA haplogroups and included in subsequent analyses. Fourteen participants (6%) were excluded due to non-European mtDNA haplogroup classification (N=11) or genotyping no-calls at haplogroup-defining base positions (N=3). The 231 included 20 (9%) females, and had a median (IQR) baseline age of 39 (17–73) years, CD4+ T cell count of 248 (83–357) cells/mm3, and log10 plasma HIV-1 RNA of 4.9 (4.5–5.5) copies/mL.
Mitochondrial DNA haplogroups
European mtDNA haplogroup distributions were similar to those reported in other U.S. populations. The majority (48%) belonged to haplogroup H, followed in frequency by haplogroups U (15%), T (11%), and J (7%). The remainder comprised haplogroups I, K (5% each), W, X (3% each), and V (<1%).
Baseline metabolic characteristics
Of the 231 participants evaluable for mtDNA haplogroups, 218 (94%) and 220 (95%) had baseline DEXA and fasting lipid data available, respectively. Baseline metabolic parameters and prescribed NRTIs are shown for the overall group and by randomized treatment arms in Table 1, and are similar to those in the primary study population.[16] Median baseline body mass index (BMI) was 25 kg/m2, extremity and trunk fat mass by DEXA were 7.1 and 9.2 kg, respectively, and non-HDL cholesterol was 121 mg/dL. None of the participant demographics, baseline HIV disease or metabolic parameters differed significantly by treatment arm. Among the study participants receiving NRTIs, 50% received zidovudine, 29% tenofovir, and 21% stavudine. There were no significant differences in NRTIs selected by randomized treatment arm.
Table 1.
Baseline metabolic parameters and selected NRTI, by randomized treatment arm.
| Randomized treatment arma |
||||
|---|---|---|---|---|
| Total | Efavirenz | Lopinavir | Lopinavir-efavirenz | |
| N (for DEXA) | 218 | 79 | 74 | 65 |
| N (for cholesterol) | 220 | 84 | 71 | 65 |
| Body mass index (kg/m2) | 25 (23–28) | 25 (22–27) | 24 (23–29) | 26 (23–29) |
| Extremity fat (kg) | 7.1 (5.1–10.3) | 7.1 (4.6–10.1) | 6.7 (5.0–9.7) | 7.7 (5.7–11.5) |
| Trunk fat (kg) | 9.2 (6.1–12.6) | 8.5 (5.4–11.9) | 9.5 (6.6–12.5) | 9.5 (6.2–13.4) |
| Total cholesterol (mg/dL) | 154 (137–173) | 151 (136–173) | 158 (133–177) | 152 (139–172) |
| HDL cholesterol (mg/dL) | 33 (27–39) | 34 (26–40) | 32 (27–39) | 33 (28–39) |
| Non-HDL cholesterol (mg/dL) | 121 (104–137) | 120 (103–138) | 123 (103–142) | 122 (105–135) |
| LDL cholesterol (mg/dL) | 92 (76–110) | 91 (79–108) | 89 (75–114) | 94 (77–110) |
| Triglycerides (mg/dL) | 127 (88–185) | 136 (84–203) | 131 (92–173) | 114 (91–165) |
| Lipid lowering therapyb | 6 (3) | 3 (4) | 1 (1) | 2 (3) |
| NRTI selectedc | ||||
| Stavudine XR | 32 (21) | 18 (23) | 14 (19) | - |
| Tenofovir | 44 (29) | 22 (28) | 22 (30) | - |
| Zidovudine | 77 (50) | 39 (49) | 38 (51) | - |
Data shown are median (interquartile range) or N (%) except where noted
All comparisons across randomized treatment arms p>0.15 by Kruskal-Wallis test
Percentages based on total with baseline cholesterol data
Percentages based on total from NRTI-containing arms with baseline DEXA (overall total n=153)
NRTI=nucleoside reverse transcriptase inhibitor; DEXA=dual energy x-ray absorptiometry
Baseline metabolic parameters by mtDNA haplogroups are shown in Table 2 (haplogroup V is not shown due to only 2 individuals in this group). The only significant differences from the overall group were among persons belonging to haplogroup I who had significantly greater median total (195 vs. 152 mg/dL), non-HDL (160 vs. 120 mg/dL), and LDL (127 vs. 91 mg/dL) cholesterol (P≤0.01 for each), and tended to have greater baseline extremity (9.9 vs. 7.1 kg; P=0.09) and trunk fat (12.3 vs. 8.8 kg; P=0.05) than those belonging to non-I haplogroups. Median baseline fasting triglycerides were elevated among persons belonging to haplogroup I (169 vs. 126 mg/dL among other haplogroups), but this difference was not statistically significant (P=0.4). There were no statistically significant differences across haplogroups with respect to age, sex, baseline HIV-1 RNA level or CD4+ T cells (data not shown), baseline BMI, HDL cholesterol, or, among those randomized to NRTI-containing treatment arms, NRTIs selected (Table 2).
Table 2.
Baseline metabolic parameters, by European mitochondrial haplogroup.
| European mtDNA haplogroup |
||||||||
|---|---|---|---|---|---|---|---|---|
| H | I | J | K | T | U | W | X | |
| N (for DEXA) | 103 | 10 | 19 | 12 | 25 | 33 | 7 | 7 |
| N (for cholesterol) | 106 | 10 | 20 | 10 | 25 | 33 | 7 | 7 |
| Body mass index (kg/m2) |
24 (22–27) | 26 (25–28) | 26 (23–29) | 27 (22–29) | 24 (23.6–28) | 25 (23–32) | 27 (20–30) | 27 (22–32) |
| Extremity fat (kg) | 7.2 (5.2–9.4) | 9.9 (7.0–13.0) | 6.4 (5.2–10.1) | 7.8 (5.4–11.5) | 6.4 (4.7–9.9) | 7.5 (4.9–13.9) | 6.4 (2.9–9.8) | 7.9 (3.3–18.6) |
| Trunk fat (kg) | 9.0 (6.1–12.3) | 12.3 (11.9–15.4) | 8.6 (6.2–11.9) | 10.0 (6.8–14.6) | 8.2 (5.5–11.3) | 8.6 (6.5–16.4) | 9.2 (3.0–12.0) | 11.2 (4.6–19.0) |
| Total cholesterol (mg/dL) |
154 (133–171) | 195 (173–201) | 152 (139–179) | 142 (131–165) | 155 (141–163) | 149 (139–172) | 147 (123–159) | 154 (147–190) |
| HDL cholesterol (mg/dL) |
32 (28–39) | 33 (31–36) | 31 (25–34) | 32 (26–40) | 37 (31–48) | 33 (27–40) | 27 (25–30) | 30 (25–35) |
| Non-HDL cholesterol (mg/dL) |
120 (104–136) | 160 (137–171) | 124 (105–144) | 113 (104–125) | 111 (103–127) | 121 (105–135) | 117 (98–132) | 128 (122–145) |
| LDL cholesterol (mg/dL) |
94 (73–110) | 127 (106–136) | 94 (72–106) | 88 (70–111) | 87 (77–95) | 88 (79–105) | 93 (62–110) | 102 (88–123) |
| Triglycerides (mg/dL) |
122 (90–178) | 169 (104–184) | 136 (96–196) | 146 (125–235) | 120 (78–155) | 130 (66–240) | 118 (103–158) | 112 (99–161) |
| Lipid lowering therapya |
2 (2) | 0 | 2 (10) | 1 (10) | 0 | 1 (3) | 0 | 0 |
| Randomized treatment armb |
||||||||
| Efavirenz | 36 (35) | 5 (50) | 7 (37) | 2 (17) | 8 (32) | 16 (48) | 1 (14) | 3 (43) |
| Lopinavir | 36 (35) | 4 (40) | 7 (37) | 5 (42) | 8 (32) | 10 (30) | 1 (14) | 3 (43) |
| Lopinavir- efavirenz |
31 (30) | 1 (10) | 5 (26) | 5 (42) | 9 (36) | 7 (21) | 5 (71) | 1 (14) |
| NRTI selectedb | ||||||||
| Stavudine XR | 14 (19) | 2 (22) | 2 (14) | 3 (43) | 3 (19) | 7 (27) | 0 | 1 (17) |
| Zidovudine | 35 (49) | 5 (56) | 8 (57) | 3 (43) | 9 (56) | 12 (46) | 1 | 3 (50) |
| Tenofovir | 23 (32) | 2 (22) | 4 (29) | 1 (14) | 4 (25) | 7 (27) | 1 | 2 (33) |
Data shown are median (interquartile range) or N (%) except where noted
Percentages based on total with baseline cholesterol data
Percentages based on total arms with baseline DEXA (overall total n=153)
NRTI=nucleoside reverse transcriptase inhibitor; DEXA=dual energy x-ray absortiometry
DEXA changes and lipoatrophy
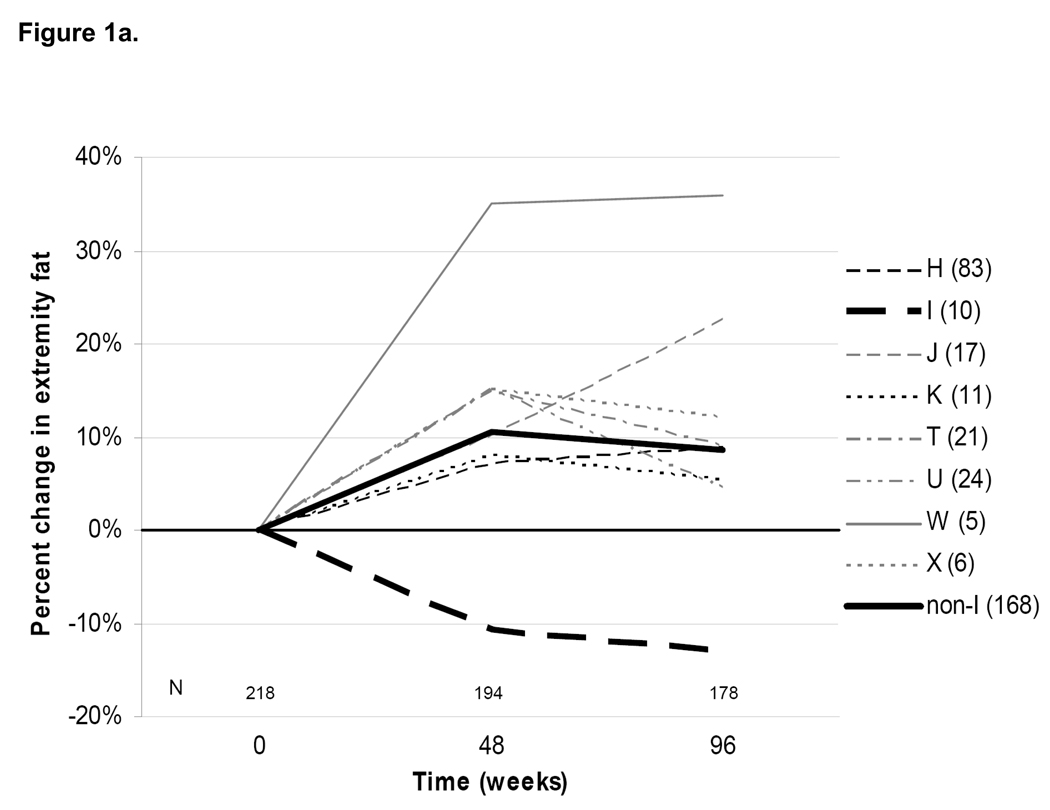
Of 218 persons with baseline DEXA data, 178 (82%) had paired 96-week data. Overall, there was a median (IQR) percent increase in extremity fat of +7.5% (−15.4 - +23.7), corresponding to a median absolute increase of +0.6 kg (−1.6 - +1.7) from baseline to 96 weeks (Table 3). This change was consistent with that seen in the primary study population.[16] Median week 48 and 96 changes in DEXA extremity fat stratified by mtDNA haplogroups are shown in Figure 1a. The only statistically significant difference in percent extremity fat change was in subjects belonging to haplogroup W (N=5; median [IQR] change +35.5% [+26.8 - +54.9]; P=0.02). Only haplogroup I demonstrated a trend toward an overall percent decrease in extremity fat (N=10; median change −13.2% [−31.2 - +11.6]; P=0.08).
Table 3.
Baseline and week 96 DEXA results, by European mitochondrial haplogroup
| European Haplogroup |
Total N (%)a |
Baseline extremity fat (kg) |
Week 96 extremity fat (kg) N=178b |
Absolute change (kg), week 96 |
%change, week 96 |
p-valuec |
|---|---|---|---|---|---|---|
| Total | 218 | 7.1 (5.1–10.3) | 7.6 (5.2–11.3) | +0.6 (−1.1- +1.7) | +8 (−15- +24) | - |
| H | 103 (47) | 7.2 (5.2–9.4) | 7.8 (5.4–10.9) | +0.8 (−1.1- +1.6) | +9 (−16- +24) | 0.8 |
| I | 10 (5) | 9.9 (7.0–12.9) | 7.8 (6.3–9.7) | −1.2 (−3.5- +0.8) | −13 (−31- +12) | 0.08 |
| J | 19 (9) | 6.4 (5.2–10.1) | 6.7 (5.2–12.0) | +0.6 (−0.6- +2.5) | +14 (−10- +32) | 0.5 |
| K | 12 (6) | 7.8 (5.4–11.5) | 9.8 (3.7–12.2) | +0.6 (−1.1- +2.0) | +5 (−18- +29) | 0.9 |
| T | 25 (11) | 6.4 (4.7–9.9) | 5.9 (4.4–9.4) | +0.5 (−1.5- +1.5) | +9 (−15- +26) | 0.9 |
| U | 33 (15) | 7.5 (4.9–13.9) | 7.1 (6.3–12.0) | +0.2 (−0.9- +1.2) | +4 (−12- +14) | 0.4 |
| V | 2 (<1) | - | - | - | - | - |
| W | 7 (3) | 6.4 (2.9–9.8) | 10.5 (9.9–17.5) | +3.5 (+0.8- +4.6) | +36 (+27- +55) | 0.2 |
| X | 7 (3) | 7.9 (3.3–18.6) | 7.0 (3.5–13.2) | +1.9 (−2.1- +3.8) | +12 (−26- +38) | 0.8 |
Data are median (interquartile range) except where noted.
Sample size with DNA, haplogroup, and baseline DEXA data available.
Sample size with DNA, haplogroup, and 96-week data available.
Wilcoxon rank-sum test for 96 week % change vs. all other haplogroups.
Figure 1. Median percent changes in (a) extremity fat by DEXA and (b) fasting non-HDL cholesterol from baseline to 48 and 96 weeks after randomization, by mtDNA haplogroup.
Dark horizontal line indicates no change. Individual lines represent different haplogroups. Thick lines denote changes within haplogroup I (dashed lines) and combined non-I haplogroups (solid lines). For Figure 1a: P=0.08 for 96-week difference in median percent changes in extremity fat between I and non-I groups; P=0.02 for haplogroup W versus combined non-W haplogroups. Baseline and 96-week DEXA data are also shown in Table 3. For Figure 1b: P<0.001 for 96-week difference in median percent changes in non-HDL cholesterol between I and non-I groups. Sample sizes with available data at each time point are shown above the horizontal axis; individual haplogroup sample sizes with data available at 96 weeks are shown in the haplogroup legend. Note: the total non-I haplogroup sample sizes shown include a single subject belonging to haplogroup V.
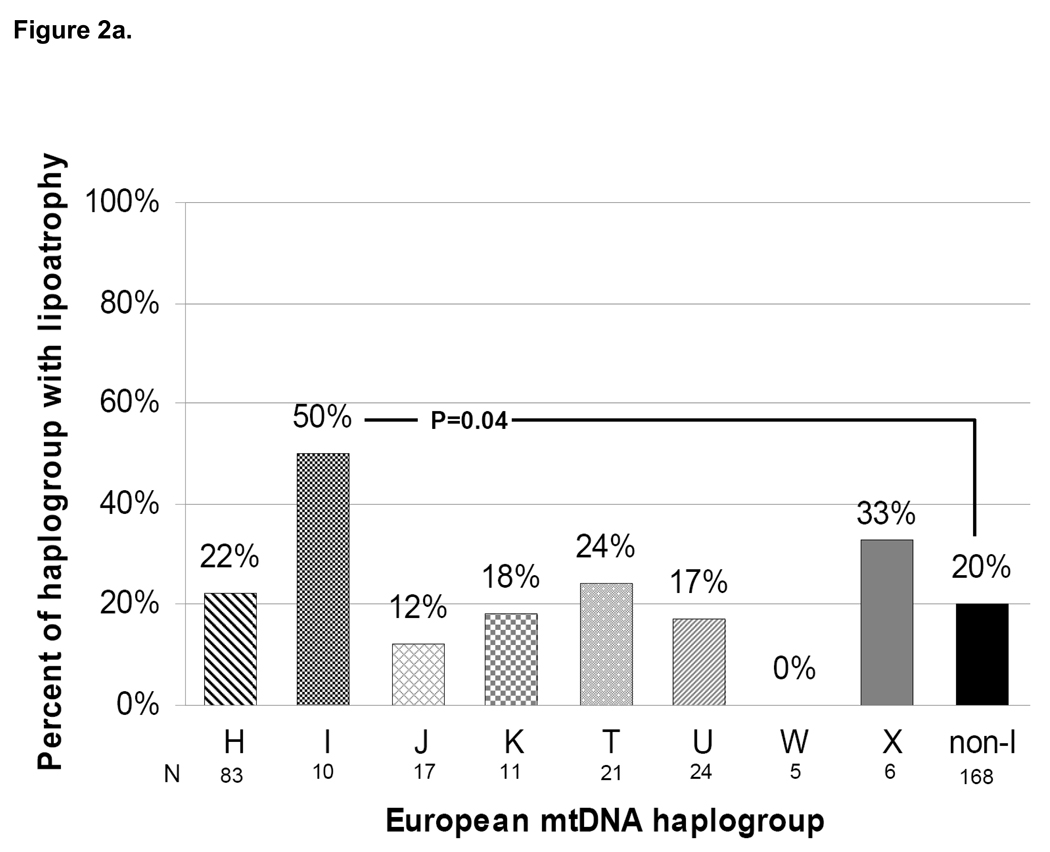
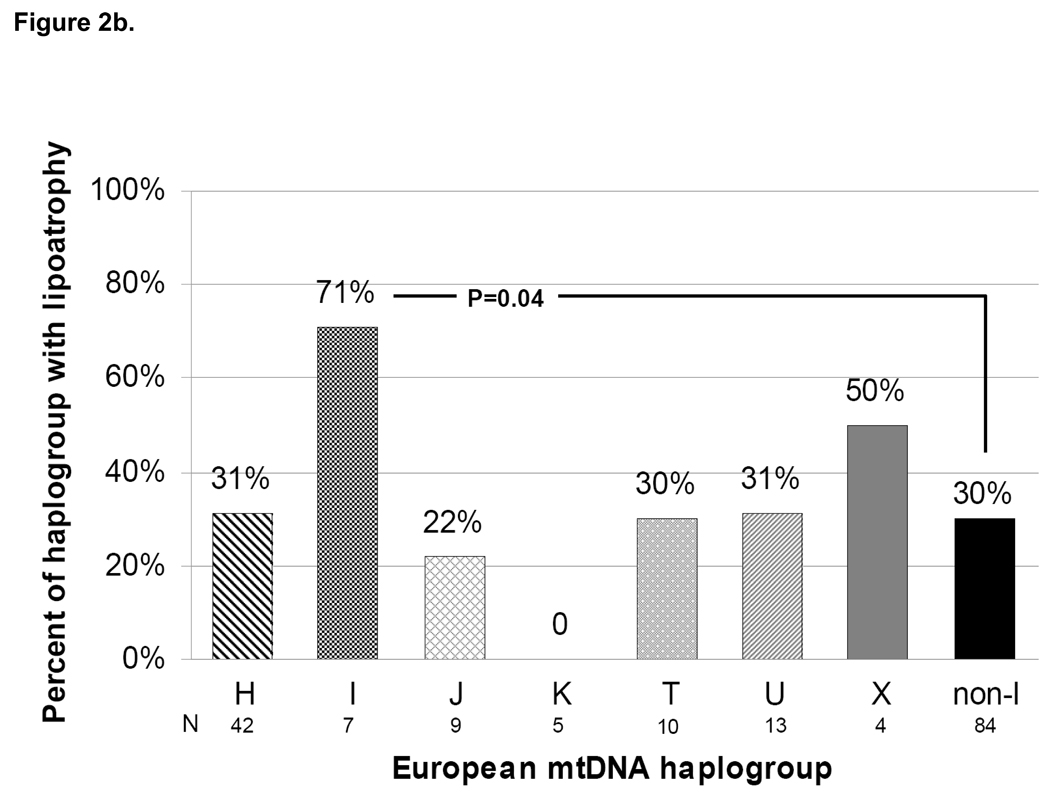
As in the primary metabolic analysis, lipoatrophy was defined as ≥20% decrease in extremity fat from baseline at 96 weeks. Overall, 39 of 178 persons (22%) with 96 week DEXA data met this definition. This proportion was 32% of persons randomized to the efavirenz arm; 21% of those randomized to the lopinavir arm; and 11% of those randomized to the NRTI-sparing lopinavir-efavirenz arm (P=0.02 for three-arm comparison). Not surprisingly, 96-week lipoatrophy rates were 50%, 26%, and 9% for persons receiving stavudine, zidovudine, and tenofovir, respectively (P=0.002) and were similar to those in the primary study population.[16] When compared across mtDNA haplogroups, persons belonging to haplogroup I were more likely to have met the definition of lipoatrophy than those belonging to non-I haplogroups (50% vs. 20%; P=0.04; Figure 2a). No other haplogroup differed significantly with respect to lipoatrophy. In a subgroup analysis limited to persons receiving the thymidine analogue NRTIs zidovudine or stavudine (N=91; 30 [33%] developed lipoatrophy), haplogroup I remained significantly over-represented compared with non-I haplogroups (71% vs. 30%; P=0.04; Figure 2b).
Figure 2. Proportion of subjects with lipoatrophy defined as ≥20% loss of extremity fat by DEXA at 96 weeks (a) overall, and (b) among those who received thymidine analogue NRTIs zidovudine or stavudine, by mtDNA haplogroup.
Individual haplogroup sample sizes with DEXA data available at 96 weeks are shown beneath the horizontal axis labels. Total N=178 for (a), N=91 for (b). Note: the total non-I haplogroup sample sizes shown include a single subject belonging to haplogroup V.
Logistic regression was used to determine ORs of baseline demographic, immunologic, and ART-related factors for lipoatrophy, and to adjust for possible confounding (Table 4). In unadjusted models, randomization to the NRTI-containing efavirenz arm was associated with lipoatrophy (OR=2.4 [95% CI 1.2–5.4]; P=0.02), and randomization to the NRTI-sparing arm was protective (0.3 [0.1–0.9]; P=0.03), consistent with the primary metabolic analyses.[16] Among the selected NRTIs, stavudine use was associated with development of lipoatrophy (4.8 [2.0–11.6]; P<0.001), and tenofovir use tended to be protective (0.3 [0.1–1.1]; P=0.06). Of mtDNA haplogroups, only haplogroup I was significantly associated with lipoatrophy (3.9 [1.1–14.4]; P=0.04). In a model adjusted for randomization to the NRTI-sparing regimen, baseline extremity fat mass, and mtDNA haplogroup I, NRTI-sparing treatment remained protective of lipoatrophy (0.3 [0.1–0.9]; P=0.03), and the effect size of haplogroup I decreased and the P-value no longer demonstrated statistical significance (3.0 [0.8–11.4]; P=0.11). In separate models adjusting for individual thymidine analogue NRTI use, stavudine remained significantly associated with lipoatrophy (5.2 [2.1–12.9]; P<0.001), zidovudine was not (1.4 [0.7–3.0]; P=0.3), and although haplogroup I retained similar OR (3.7 [0.9–14.6]; P=0.06 and 3.5 [0.9–12.9]; P=0.07, respectively; Table 4), these associations were no longer statistically significant. Baseline extremity fat was not associated with lipoatrophy in unadjusted or adjusted analyses.
Table 4.
Unadjusted and adjusted odds ratios (OR) for selected predictors of lipoatrophya
| Model 1- NRTI-sparing vs. other regimens |
Model 2- Stavudine XR vs. other regimens |
Model 3- Zidovudine vs. other regimens |
||||||
|---|---|---|---|---|---|---|---|---|
| Covariateb | Unadjusted OR (95% CI) |
P-value | Adjusted OR (95% CI) |
P-value | Adjusted OR (95% CI) |
P-value | Adjusted OR (95% CI) |
P-value |
| Randomized study arm | ||||||||
| Efavirenz | 2.4 (1.2–5.4) | 0.02 | ||||||
| Lopinavir | 0.9 (0.4–1.9) | 0.8 | ||||||
| Lopinavir-efavirenz | 0.3 (0.1–0.9) | 0.03 | 0.3 (0.1–0.9) | 0.03 | ||||
| Selected NRTI | ||||||||
| Stavudine XR | 4.8 (2.0–11.6) | <0.001 | 5.2 (2.1–12.9) | <0.001 | ||||
| Zidovudine | 1.5 (0.7–3.0) | 0.3 | 1.4 (0.7–3.0) | 0.3 | ||||
| Tenofovir | 0.3 (0.1–1.1) | 0.06 | ||||||
| Baseline extremity fat (per kg increase) |
1.1 (0.98–1.2) | 0.13 | 1.1 (0.98–1.2) | 0.13 | 1.1 (0.98–1.2) | 0.13 | 1.1 (0.97–1.2) | 0.18 |
| mtDNA haplogroup I (vs. all others) |
3.9 (1.1–14.4) | 0.04 | 3.0 (0.8–11.4) | 0.11 | 3.7 (0.9–14.6) | 0.06 | 3.5 (0.9–12.9) | 0.07 |
Lipoatrophy defined as ≥20% extremity fat loss at 96 weeks.
Baseline age, CD4 T cells, HIV RNA, and sex were not significant predictors of lipoatrophy (data not shown) and were not included in adjusted models.
NRTI=nucleoside reverse transcriptase inhibitor; CI=confidence interval.
Fasting serum lipid changes
Of 220 persons with any baseline fasting lipid data, 174 (79%) had paired 96 week data. These sample sizes were 219 and 171 (78%), respectively for non-HDL cholesterol. Overall, median percent changes from baseline to 96 weeks for fasting total, LDL, non-HDL, HDL cholesterol, and triglycerides of +22.5%, +19.5%, +21.8%, +27.6%, and +50.4%, respectively. Changes from baseline in the primary lipid outcome of non-HDL cholesterol stratified by mtDNA haplogroups are shown in Figure 1b. Only haplogroup I demonstrating a percent decrease in non-HDL cholesterol at 96 weeks (median −14.0% [IQR −19.9 - +6.2] vs. +25.5% [+8.1 - +51.1] among combined non-I haplogroups; P<0.001). This change corresponded to a median decrease from baseline of 20 mg/dL to an absolute 96 week value of 132 mg/dL for haplogroup I compared to a median increase from baseline of 30 mg/dL to 152 mg/dL for non-I haplogroups. Both comparisons were statistically significant (P=0.001 for absolute change and 0.05 for absolute 96 week values) despite the higher baseline non-HDL cholesterol among persons belonging to haplogroup I. In addition, persons belonging to haplogroup I had significantly less median percent increases in total (+3% vs. +23%; P=0.005) and LDL (−5% vs. +21%; P=0.01) cholesterol, and triglycerides (−4% vs. +55%; P=0.04) over 96 weeks than those belonging to other haplogroups.
Discussion
Metabolic complications of ART have become prominent treatment-limiting adverse effects that increase the complexity of management of HIV infection and likely contribute to excess morbidity and mortality due to cardiovascular disease. Accurately predicting persons at greatest risk of these complications is not possible at present, and though several studies have reported genetic predictors of ART-associated metabolic complications,[19–24] none have been well-validated or incorporated into prospective clinical trials. As the HIV-infected, ART-treated population ages, metabolic complications of ART are likely to become more prominent. Better strategies to prevent these complications are needed, including expanded studies of the role of pharmacogenomics in these drug toxicities.
The present study is the largest to explore associations between mtDNA haplogroups and changes in metabolic parameters among HIV-infected persons starting ART. We found differences in baseline serum lipids among persons belonging to mtDNA haplogroup I, with these persons having higher total, LDL-, and combined non-HDL-cholesterol than persons from other haplogroups. Differences in baseline median body fat measurements by DEXA were also substantial (>2.5 kg more extremity fat and >3 kg more trunk fat), but marginally statistically significant. Persons belonging to haplogroup W demonstrated extremity fat gain and no cases of lipoatrophy. These observations should be interpreted cautiously, however, given that only five individuals had 96 week DEXA data available for analyses, and five were randomized to NRTI-sparing treatment. To our knowledge, these metabolic parameters have not been reported based on mtDNA haplogroups in the general population, and reasons for these pre-ART differences are unknown. There are data demonstrating associations between a specific mtDNA SNP and lipid parameters in Asians.[4, 5] Based on our hypothesis that mtDNA variation represented by haplogroups confers differences in oxidative phosphorylation, mtDNA replication, and/or apoptotic regulation, it is plausible that these differences may affect baseline lipid and fat metabolism prior to initiating ART.
Persons belonging to haplogroup I had more baseline fat by DEXA, but in contrast to the overall group and all other haplogroups, lost extremity fat over 96 weeks of ART. A significantly greater proportion of haplogroup I lost at least 20% of extremity fat during the study, meeting the pre-specified definition of lipoatrophy. However, due at least in part to excess baseline extremity fat, the absolute extremity fat mass in I and non-I haplogroups did not differ at 96 weeks. In multivariate models, baseline extremity fat was not associated with lipoatrophy, and did not fully attenuate the association between haplogroup I and lipoatrophy, nor did adjustment for use of thymidine analogue NRTIs. However (and perhaps not surprisingly), adjustment for randomization to NRTI-sparing treatment attenuated the association between haplogroup I and lipoatrophy, suggesting that the absence of NRTI-containing ART limited the influence of mtDNA variation on this outcome. Also of note was that despite baseline differences in serum lipids, persons belonging to haplogroup I still demonstrated significant differences in non-HDL cholesterol change from baseline. This led to a statistically and clinically significant difference in absolute week 96 values, with persons from haplogroup I having a median non-HDL cholesterol of 132 mg/dL, 20 mg/dL lower than non-I haplogroups.
These results did not confirm previous studies that had explored mtDNA haplogroups and metabolic changes in ART-treated persons of European descent. In ACTG study A5005s, the metabolic substudy of ACTG 384, individuals belonging to haplogroup J had increases in extremity fat when compared to other haplogroups (+26% vs. −8%; P=0.07).[12] In the present study the increases in fat for haplogroup J did not reach statistical significance. There were only two persons with paired DEXA data available belonging to haplogroup I in the previous study. A cross-sectional study of more than 340 Italian HIV-infected patients referred to a metabolic clinic did not identify any associations between European mtDNA haplogroups and several metabolic outcomes.[14] Only four subjects (1.2%) in this population belonged to haplogroup I, and they were excluded from analyses. More recently, data from more than 400 self-reported white participants in the Multicenter AIDS Cohort Study (MACS) underwent mtDNA haplogrouping and were analyzed for changes in peripheral lipoatrophy across three body regions (buttocks, legs, arms) determined by standardized physical exam assessment.[13] Persons belonging to haplogroup H had greater lipoatrophy severity in the arms and legs than persons from other haplogoups. The investigators noted that haplogroup J was associated with less severe lipoatrophy in all three regions, but these associations were not statistically significant. Haplogroup I was only analyzed as part of a combined IWX group, which was not statistically associated with lipoatrophy.
Our study has several limitations which should be noted. Although this is the largest published sample of HIV-infected subjects with mtDNA haplogroups and longitudinal metabolic data from a pre-ART baseline, the sample size for many haplogroups is small, especially for subgroups of subjects randomized to and/or exposed to various ART. This limited our power to detect associations with some haplogroups, and could explain the borderline statistical significance seen with extremity fat changes and lipoatrophy. The lack of replication of the possible association of haplogroup J with less extremity fat loss[12] in the study presented here (and in the MACS analysis[13]) could be due to different ART with higher rates of lipoatrophy (all subjects included in the ACTG 384 study were randomized to include a thymidine analogue NRTI, and half received stavudine and didanosine in combination), different phenotypes (longitudinal DEXA from pre-ART in a clinical trial versus cross-sectional clinical lipoatrophy assessments during ART [13]), and/or a combination of these factors. Aside from the extremity fat gain seen with haplogroup W, persons belonging to haplogroup J (N=17 with 96 week DEXA data) had the most extremity fat gain of any haplogroup (+14% [IQR −10%-+32%]). Although this difference was not statistically significant, results from three independent populations have demonstrated either a gain in limb fat by DEXA or less severe lipoatrophy among persons belonging to haplogroup J. The lack of association between haplogroup H and lipoatrophy that was reported previously[13] may also be due to the issues noted above. Given the exploratory nature of these analyses, we did not correct for multiple comparisons, thus some associations based on the traditional p-value threshold may be spurious. It should be noted, however, that 96 week percent changes in both total and non-HDL cholesterol among haplogroup I all remain statistically significant with simple Bonferroni correction (P<0.006). Lastly, our analyses did not include nuclear gene variants that have been associated with lipodystrophy[21–23] and dyslipidemia[19, 20, 24] in ART-treated populations, or assessments of facial lipoatrophy, and data on other non-genetic factors (e.g. diet and exercise) that may influence lipids and body fat were not available.
There is little biological evidence to explain associations observed between haplogroup I and metabolic phenotypes at baseline or during ART. Haplogroup I is relatively uncommon in persons of European descent (~5%), and has not previously been associated with human diseases. This haplogroup is characterized by synonymous SNPs at mtDNA positions: 1719G-A located in the ribosomal RNA gene; 8251G-A in the cytochrome C oxidase subunit II gene; and 16391G-A in the D-loop/control region. Shared SNPs with haplogroups W and X at positions 8251 and 1719, respectively, have provided rationale for combining these relatively infrequent haplogroups into an IWX clade for analyses. This may in part explain why analyses using similar phenotypes have not reported associations with haplogroup I.[13] Haplogroup I is also related to haplogroups J and K in the sharing of a non-synonymous SNP at position 10398A-G. This change leads to a Thr-Ala amino acid substitution in the NADH dehydrogenase subunit 3 gene and has been associated with neurodegenerative diseases and cancer.[3, 25] Mechanisms by which mtDNA variation may influence baseline and/or ART-associated dyslipidemia are not known, but recent literature highlights the role of mitochondrial function in common metabolic and cardiovascular diseases,[26–28] with studies examining the specific influence of mtDNA variation on risk of metabolic and cardiovascular disease in Asian[4–7, 29, 30] and non-Asian[31–37] populations. Obviously, findings from these studies in HIV-negative populations do not directly address the influence of mtDNA variation on acquired metabolic effects of ART.
These results demonstrate preliminary associations between a European mtDNA haplogroup and metabolic parameters in an HIV-infected population, both prior to and during ART exposure as part of a clinical trial. Many genetic and non-genetic factors likely influence risk for metabolic changes in this population; mtDNA variation may be one of these. Continued study is needed to replicate these associations and determine mechanisms by which mtDNA variation may be associated with different metabolic effects. More extensive mtDNA genotyping in large, well-characterized populations is needed to identify additional functional variants (i.e. SNPs that change amino acid sequence and/or protein function) that may underlie this and other reported associations, and to expand these analyses to other racial/ethnic groups with different mtDNA lineages.
Acknowledgements
The authors acknowledge Greg DiRenzo, Ph.D., and Lynne Peeples, M.S. (Statistical Data Analysis Center, Harvard University, Boston, MA) for assistance with clinical datasets, and Cara Sutcliffe and Ping Mayo (DNA Resources Core, Center for Human Genetics Research, Vanderbilt University, Nashville, TN) for assistance with mtDNA genotyping.
The authors wish to thank the study volunteers from A5142 and A5128 who contributed data and DNA for this analysis. Other members of the A5142 study team included: Barbara Brizz and Joelle Touw (SSS), protocol specialists; Pat Cain (Stanford University Medical Center), protocol field representative; Marlene Cooper and Mary Dobson (Frontier Science and Technology Research Foundation), laboratory data coordinators; Michael Dorosh (University of Colorado Health Sciences Center), community representative; Pualani Kondo (University of Hawaii), protocol laboratory technologist; David Rusin (Frontier Science and Technology Research Foundation), data manager; Kathleen Squires (Thomas Jefferson University), Co-Investigator; Paul Tran (Division of AIDS, National Institute of Allergy and Infectious Disease), protocol pharmacist. Additional high enrolling investigators: Mitchell Goldman (Indiana University); Hector Bolivar (University of Miami). Additional pharmaceutical collaborators include Mick Hitchcock and Michael Wulfsohn of Gilead Sciences; and Scott C. Brun and Richard A. Rode of Abbott Laboratories.
Funding support:
A5142 (NCT#00050895) was supported by grants AI 068636 (AIDS Clinical Trials Group Central Grant), AI 068634, AI 069471, AI 27661, AI 069439, AI 25859, AI 069477, AI 069513, AI 069452, AI 27673, AI 069470, AI 069474, AI 069411, AI 069423, AI 069494, AI 069484, AI 069472, AI 38858, AI 069501, AI 32783, AI 069450, AI 32782, AI 069465, AI 069424, AI 38858, AI 069447, AI 069495, AI 069502, AI 069556, AI 069432, AI 46370, AI 069532, AI 46381, AI 46376, AI 34853, AI 069434, AI 060354, AI 064086, AI 36214, AI 069419, AI 069418, AI 50410, AI 45008, RR 00075, RR 00032, RR 00044, RR 00046, RR 02635, RR 00051, RR 00052, RR 00096, RR 00047, RR 00039, and DA 12121 from the National Institute of Allergy and Infectious Disease, National Institutes of Health. The collaborating pharmaceutical companies provided lopinavir–ritonavir (Abbott), efavirenz and stavudine XR (Bristol-Myers Squibb), and tenofovir DF (Gilead).
The ACTG Human DNA Repository is also supported by NIH grant RR024975.
Other support included NIH grants AI 64086, AI 36214, AI 69432, AI 60484.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
These data were presented in part at the 11th International Workshop on Adverse Drug Reactions and Co-morbidities in HIV, Philadelphia, PA, U.S.A., October 28, 2009 [Abstract ADRLH-55].
Potential conflicts of interest:
Dr. Hulgan reports having received research support from Merck.
Dr. Haubrich reports having received speaking honoraria or consultant fees from Abbott, Boehringer Ingelheim, Bristol-Myers Squibb, Gilead Sciences, Merck, Schering and Roche and has received research support from Abbott, GlaxoSmithKline, Pfizer and Tibotec.
Dr. Riddler reports having received lecture or consultation fees from Bristol-Myers Squibb and grant support from Schering-Plough and Hoffman-LaRoche.
Dr. Tebas reports having received consultation fees from Glaxo, Merck, Pfizer and Tibotec.
Dr. Ritchie reports having received consultation fees from Boehringer-Ingelheim.
Dr. McComsey reports having received speaking honoraria or consultant fees from Bristol-Myers Squibb, Gilead Sciences, GlaxoSmithKline and Abbott and has received research support from Bristol-Myers Squibb, GlaxoSmithKline, Merck, Gilead, and Abbott.
Dr. Haas has received research support from Bavarian Nordic, Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead Sciences, Merck, and Tibotec, and has served on Scientific Advisory Boards for Boehringer-Ingelheim and Tibotec.
Dr. Canter reports no conflicts.
Author Contributions:
The initial concept for the study was conceived by Drs. Hulgan, Haubrich, Riddler, Tebas, McComsey, Haas, and Canter. Drs. Hulgan, Canter, and Ritchie were responsible for data coordination and analyses. Drs. Hulgan and Canter wrote the first draft of the manuscript with critical review by all authors. All authors participated in the development of the study protocol and analysis plan, reviewed the study data/analysis reports, and approved the final manuscript.
A5142 Participating Sites and Site Personnel:
Karen Coleman, RN (Northwestern University), Beverly Sha, MD (Rush University), and Oluwatoyin Adeyemi, MD (Cook County CORECenter) - Northwestern University; W. Keith Henry, MD and Winston Calvert, MD - University of Minnesota; Michael Morgan, FNP and Brenda Jackson, RN - Vanderbilt University; Mitchell Goldman, MD and Janet Hernandez, RN- Indiana University; Margaret A. Fischl, MD - University of Miami School of Medicine; Carl J. Fichtenbaum, MD and Jenifer Baer, RN - University of Cincinnati; Suzette Byars and Mae Stewart- University of Alabama; Hannah Edmondson-Melancon, RN, MPH and Connie A. Funk, RN, MPH - University of Southern California; Jolene Noel Connor, RN, BSN and Madeline Torres, RN, BSN - Columbia Collaborative HIV/AIDS Clinical Trials Unit; William E. Maher, MD and Laura Laughlin, RN - The Ohio State University; Mary Adams, RN and Christine Hurley, RN - University of Rochester; Clara Zelasky, PA-C and David Wohl, MD - University of North Carolina-Chapel Hill; Deborah McMahon, MD and Barbara Rutecki, MSN, MPH, CRNP - University of Pittsburgh ; Princy Kumar, MD and Ioulia Vvedenskaya - Georgetown University; Gary M. Cox, MD and Deitra Wade, RN - Duke University Medical Center; Paul Sax, MD and Jon Gothing, RN - Harvard/BMC ACTU; A. A. Amod, FCPath, Durban International Clinical Trials Unit; Benigno Rodriguez, MD, MSc and Barbara Philpotts, BSN, RN - Case Western Reserve University; Harvey Friedman, MD and Aleshia Thomas, RN - University of Pennsylvania, Philadelphia ; Beverly Putnam, MSN and Cathi Basler, MSN - Colorado ACTU; William A. O’Brien, MD, MS and Gerianne Casey, RN - UTMB-Galveston; Ilene Wiggins, RN and Gerianne Casey, RN - Johns Hopkins University; Margrit Carlson, MD and Eric Daar, MD - University of California, Los Angeles; Abby Olusanya, NP and Melissa Schreiber, PA - University of California, Davis Medical Center; Charles Davis, MD and Becky Boyce, RN - University of Maryland, Inst. of Human Virology; Ge-Youl Kim, RN, BSN and Kimberly Gray RN, MSN- Washington University in St. Louis; Joann Volinski, RN - University of California, San Francisco; Jane Norris, PA-C and Sandra Valle PA-C- Stanford University; Julie Hoffman, RN and Susan Cahill, RN - University of California, San Diego; Donald Garmon, NP and Donna Mildvan, MD - Beth Israel Medical Center: Janet Forcht, RN and Charles Gonzalez, MD - New York University/NYC HHC at Bellevue Hospital Center; Karen Tashima, MD and Deborah Perez, RN - The Miriam Hospital; Philip Keiser, MD and Tianna Petersen, MS - UT Southwestern Medical Center at Dallas; Nancy Hanks, RN and Scott Souza, PharmD - University of Hawaii at Manoa and Queen’s Medical Center; Ann C. Collier, MD and Sheryl Storey, PA-C - University of Washington, Seattle; Valery Hughes, FNP and Todd Stroberg, RN - Cornell University; Gregory Smith RN and Ighovera Ofotokun MD - Emory University.
References
- 1.Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci U S A. 1994;91:8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. Epub 2002 Oct 2017. [DOI] [PubMed] [Google Scholar]
- 3.van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. doi: 10.1086/373937. Epub 2003 Feb 2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kokaze A, Ishikawa M, Matsunaga N, Yoshida M, Sekine Y, Teruya K, et al. Association of the mitochondrial DNA 5178 A/C polymorphism with serum lipid levels in the Japanese population. Hum Genet. 2001;109:521–525. doi: 10.1007/s004390100602. [DOI] [PubMed] [Google Scholar]
- 5.Lal S, Madhavan M, Heng CK. The association of mitochondrial DNA 5178 C > a polymorphism with plasma lipid levels among three ethnic groups. Ann Hum Genet. 2005;69:639–644. doi: 10.1111/j.1529-8817.2005.00192.x. [DOI] [PubMed] [Google Scholar]
- 6.Mukae S, Aoki S, Itoh S, Sato R, Nishio K, Iwata T, Katagiri T. Mitochondrial 5178A/C genotype is associated with acute myocardial infarction. Circ J. 2003;67:16–20. doi: 10.1253/circj.67.16. [DOI] [PubMed] [Google Scholar]
- 7.Park KS, Chan JC, Chuang LM, Suzuki S, Araki E, Nanjo K, et al. A mitochondrial DNA variant at position 16189 is associated with type 2 diabetes mellitus in Asians. Diabetologia. 2008;51:602–608. doi: 10.1007/s00125-008-0933-z. [DOI] [PubMed] [Google Scholar]
- 8.Lewis W, Kohler JJ, Hosseini SH, Haase CP, Copeland WC, Bienstock RJ, et al. Antiretroviral nucleosides, deoxynucleotide carrier and mitochondrial DNA: evidence supporting the DNA pol gamma hypothesis. AIDS. 2006;20:675–684. doi: 10.1097/01.aids.0000216367.23325.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis W, Copeland WC, Day BJ. Mitochondrial dna depletion, oxidative stress, and mutation: mechanisms of dysfunction from nucleoside reverse transcriptase inhibitors. Lab Invest. 2001;81:777–790. doi: 10.1038/labinvest.3780288. [DOI] [PubMed] [Google Scholar]
- 10.Hulgan T, Haas DW, Haines JL, Ritchie MD, Robbins GK, Shafer RW, et al. Mitochondrial haplogroups and peripheral neuropathy during antiretroviral therapy: an adult AIDS clinical trials group study. AIDS. 2005;19:1341–1349. doi: 10.1097/01.aids.0000180786.02930.a1. [DOI] [PubMed] [Google Scholar]
- 11.Canter JA, Robbins GK, Selph D, Clifford DB, Kallianpur AR, Shafer R, et al. African Mitochondrial DNA Subhaplogroups and Peripheral Neuropathy during Antiretroviral Therapy. J Infect Dis. 2010 doi: 10.1086/652419. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulgan T, Tebas P, Canter JA, Mulligan K, Haas DW, Dube M, et al. Hemochromatosis Gene Polymorphisms, Mitochondrial Haplogroups, and Peripheral Lipoatrophy during Antiretroviral Therapy. J Infect Dis. 2008;197:858–866. doi: 10.1086/528697. [DOI] [PubMed] [Google Scholar]
- 13.Hendrickson SL, Kingsley LA, Ruiz-Pesini E, Poole JC, Jacobson LP, Palella FJ, et al. Mitochondrial DNA Haplogroups Influence Lipoatrophy After Highly Active Antiretroviral Therapy. J Acquir Immune Defic Syndr. 2009;51:111–116. doi: 10.1097/QAI.0b013e3181a324d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nasi M, Guaraldi G, Orlando G, Durante C, Pinti M, Nemes E, et al. Mitochondrial DNA Haplogroups and Highly Active Antiretroviral Therapy-Related Lipodystrophy. Clin Infect Dis. 2008;47:962–968. doi: 10.1086/591706. [DOI] [PubMed] [Google Scholar]
- 15.Riddler SA, Haubrich R, DiRienzo AG, Peeples L, Powderly WG, Klingman KL, et al. Class-sparing regimens for initial treatment of HIV-1 infection. N Engl J Med. 2008;358:2095–2106. doi: 10.1056/NEJMoa074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haubrich RH, Riddler SA, DiRienzo AG, Komarow L, Powderly WG, Klingman K, et al. Metabolic outcomes in a randomized trial of nucleoside, nonnucleoside and protease inhibitor-sparing regimens for initial HIV treatment. AIDS. 2009;23:1109–1118. doi: 10.1097/QAD.0b013e32832b4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas DW, Wilkinson GR, Kuritzkes DR, Richman DD, Nicotera J, Mahon LF, et al. A multi-investigator/institutional DNA bank for AIDS-related human genetic studies: AACTG Protocol A5128. HIV Clin Trials. 2003;4:287–300. doi: 10.1310/MUQC-QXBC-8118-BPM5. [DOI] [PubMed] [Google Scholar]
- 18.Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, et al. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnedo M, Taffe P, Sahli R, Furrer H, Hirschel B, Elzi L, et al. Contribution of 20 single nucleotide polymorphisms of 13 genes to dyslipidemia associated with antiretroviral therapy. Pharmacogenet Genomics. 2007;17:755–764. doi: 10.1097/FPC.0b013e32814db8b7. [DOI] [PubMed] [Google Scholar]
- 20.Foulkes AS, Wohl DA, Frank I, Puleo E, Restine S, Wolfe ML, et al. Associations among race/ethnicity, ApoC-III genotypes, and lipids in HIV-1-infected individuals on antiretroviral therapy. PLoS Med. 2006;3:e52. doi: 10.1371/journal.pmed.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maher B, Alfirevic A, Vilar FJ, Wilkins EG, Park BK, Pirmohamed M. TNF-alpha promoter region gene polymorphisms in HIV-positive patients with lipodystrophy. AIDS. 2002;16:2013–2018. doi: 10.1097/00002030-200210180-00005. [DOI] [PubMed] [Google Scholar]
- 22.Nolan D, Moore C, Castley A, Sayer D, Mamotte C, John M, et al. Tumour necrosis factor-alpha gene −238G/A promoter polymorphism associated with a more rapid onset of lipodystrophy. AIDS. 2003;17:121–123. doi: 10.1097/00002030-200301030-00017. [DOI] [PubMed] [Google Scholar]
- 23.Ranade K, Geese WJ, Noor M, Flint O, Tebas P, Mulligan K, et al. Genetic analysis implicates resistin in HIV lipodystrophy. AIDS. 2008;22:1561–1568. doi: 10.1097/QAD.0b013e32830a9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarr PE, Taffe P, Bleiber G, Furrer H, Rotger M, Martinez R, et al. Modeling the influence of APOC3, APOE, and TNF polymorphisms on the risk of antiretroviral therapy-associated lipid disorders. J Infect Dis. 2005;191:1419–1426. doi: 10.1086/429295. [DOI] [PubMed] [Google Scholar]
- 25.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 26.Maassen JA, t Hart LM, Ouwens DM. Lessons that can be learned from patients with diabetogenic mutations in mitochondrial DNA: implications for common type 2 diabetes. Curr Opin Clin Nutr Metab Care. 2007;10:693–697. doi: 10.1097/MCO.0b013e3282f0b774. [DOI] [PubMed] [Google Scholar]
- 27.Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res. 2008;102:401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madamanchi NR, Runge MS. Mitochondrial dysfunction in atherosclerosis. Circ Res. 2007;100:460–473. doi: 10.1161/01.RES.0000258450.44413.96. [DOI] [PubMed] [Google Scholar]
- 29.Takagi K, Yamada Y, Gong JS, Sone T, Yokota M, Tanaka M. Association of a 5178C->A (Leu237Met) polymorphism in the mitochondrial DNA with a low prevalence of myocardial infarction in Japanese individuals. Atherosclerosis. 2004;175:281–286. doi: 10.1016/j.atherosclerosis.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Okura T, Koda M, Ando F, Niino N, Tanaka M, Shimokata H. Association of the mitochondrial DNA 15497G/A polymorphism with obesity in a middle-aged and elderly Japanese population. Hum Genet. 2003;113:432–436. doi: 10.1007/s00439-003-0983-8. [DOI] [PubMed] [Google Scholar]
- 31.Poulton J, Brown MS, Cooper A, Marchington DR, Phillips DI. A common mitochondrial DNA variant is associated with insulin resistance in adult life. Diabetologia. 1998;41:54–58. doi: 10.1007/s001250050866. [DOI] [PubMed] [Google Scholar]
- 32.Poulton J, Luan J, Macaulay V, Hennings S, Mitchell J, Wareham NJ. Type 2 diabetes is associated with a common mitochondrial variant: evidence from a population-based case-control study. Hum Mol Genet. 2002;11:1581–1583. doi: 10.1093/hmg/11.13.1581. [DOI] [PubMed] [Google Scholar]
- 33.Crispim D, Canani LH, Gross JL, Tschiedel B, Souto KE, Roisenberg I. The European-specific mitochondrial cluster J/T could confer an increased risk of insulin-resistance and type 2 diabetes: an analysis of the m.4216T > C and m.4917A > G variants. Ann Hum Genet. 2006;70:488–495. doi: 10.1111/j.1469-1809.2005.00249.x. [DOI] [PubMed] [Google Scholar]
- 34.Crispim D, Fagundes NJ, Canani LH, Gross JL, Tschiedel B, Roisenberg I. Role of the mitochondrial m.16189T>C variant in type 2 diabetes mellitus in southern Brazil. Diabetes Res Clin Pract. 2006;74:204–206. doi: 10.1016/j.diabres.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Chinnery PF, Mowbray C, Patel SK, Elson JL, Sampson M, Hitman GA, et al. Mitochondrial DNA haplogroups and type 2 diabetes: a study of 897 cases and 1010 controls. J Med Genet. 2007;44:e80. doi: 10.1136/jmg.2007.048876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, Hirschhorn JN, et al. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am J Hum Genet. 2006;79:54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benn M, Schwartz M, Nordestgaard BG, Tybjaerg-Hansen A. Mitochondrial haplogroups: ischemic cardiovascular disease, other diseases, mortality, and longevity in the general population. Circulation. 2008;117:2492–2501. doi: 10.1161/CIRCULATIONAHA.107.756809. [DOI] [PubMed] [Google Scholar]