Abstract
Protein-protein interactions are important to understanding cell functions; however our theoretical understanding is limited. There is a general discontinuity between the well-accepted physical and chemical forces that drive protein-protein interactions and the large collections of identified protein-protein interactions in various databases. Minimotifs are short functional peptide sequences that provide a basis to bridge this gap in knowledge. However, there is no systematic way to study minimotifs in the context of protein-protein interactions or vice versa. Here we have engineered a set of algorithms that can be used to identify minimotifs in known protein-protein interactions and implemented this for use by scientists in Minimotif Miner. By globally testing these algorithms on verified data and on 100 individual proteins as test cases, we demonstrate the utility of these new computation tools. This tool also can be used to reduce false positive predictions in the discovery of novel minimotifs. The statistical significance of these algorithms is demonstrated by an ROC analysis (p = 0.001).
Keywords: Minimotif Miner, HomoloGene, BLAST, Grb2, SLiM, short linear motifs
Introduction
Protein-protein interactions (PPIs) are central to understanding how cell functions are integrated. At this point scientists have deciphered a well-developed theory that explains the physical and chemical forces that underlie the non-covalent chemistry of PPIs. Electrostatic, dipole-dipole, van der Waals forces are well quantified into accepted force fields such as AMBER that are used for protein structure determination by NMR, modeling of macromolecules, refinement of X-ray structures, macromolecular dynamics, and drug design1–3. A more recent knowledge base of PPIs has emerged from the growing number of PPI databases built upon data largely from high-throughput techniques such as yeast 2-hybrid screens and Tandem affinity purification/Mass-spectrometry analysis, as well as manual curation of the scientific literature. Collectively BIND, DIP, BioGrid, IntAct, MIPS, MINT, HPRD, the YPD and others contain several hundred thousand PPIs4–8. Despite these efforts, we have yet to develop a comprehensive enumeration of PPIs.
One of the major efforts addressing the development of a theory that reliably predicts new PPIs is that of minimotifs (also called short linear motifs or SLiMs), which bind to protein domains and provide a key connection of the physical and chemical forces with the large PPI data sets. Minimotifs are contiguous peptide elements in proteins, generally less than 15 residues in length that have a defined function. One class of functions includes binding minimotifs such as those that engage SH2, SH3, PDZ, and a number of other modular protein domains9. These minimotifs are of known molecular function and are distinct from de novo prediction of motifs by several approaches including MEME, Gibbs Sampler, PRATT, TEIRESIAS, D-MOTIF and other algorithms10–14.
While some PPIs involve extensive surface contact, PPIs driven by minimotifs are generally simpler, with a reduced surface of contact. Analysis of minimotif-driven PPIs simplifies the problem of theoretically predicting new PPIs by limiting the residues that need to be considered. Generally minimotifs are identified by studying sequences of a collection of instances that are known to interact with a protein or by analysis of the permutational space of each position in a peptide sequence that can bind to a domain by phage display, screening random peptide libraries, SPOT peptide arrays, or site-directed mutagenesis. Most often interpretation of this data reduces the series of instances down to a consensus minimotif that accounts for degeneracy at each position. Alternatively, degeneracy and variation can be quantitatively represented in a position specific-scoring matrix (PSSM), which has the advantage that it captures the probability of the collection of instances for each position.
Consensus sequences and PSSMs have some predictive value but have limitations. While high-throughput experimentation has helped us to begin to understand some of the specificity determinants of many minimotifs, sequence alone is not an accurate predictor of novel minimotif instances and does not, by itself, account for the higher degree of specificity observed in minimotif-driven PPIs.
There has been much effort to increase the specificity of and reduce false-positive minimotif predictions15–19,9. In the Minimotif Miner (MnM) application for predicting minimotifs, three approaches can be used: frequency analysis relies on the simple premise that minimotifs with more complex definitions are less likely to have false-positives15,16. Since minimotifs must be on the surface of a protein, a surface prediction algorithm can be used to minimize the prediction of buried minimotifs. Likewise, those minimotifs that are conserved in different species provide another measure to reduce false positives. Another major minimotif database, Eukaryotic linear motif server (ELM) utilizes different filters that are complementary to MnM17,18. A cell compartment filter identifies minimotifs in appropriate cellular compartments, while a globular domain filter can be used to restrict predictions to intrinsically disordered regions. ELM uses taxonomy in a different way than MnM to identify minimotifs in organisms with a conserved minimotif partner. ELM also has a surface filter and a secondary structure prediction filter.
In MnM a query protein (minimotif source protein) is entered and, in part, the sequence is analyzed for minimotifs that encode putative interactions with target proteins. Each query generally produces many target predictions; however, like other minimotif prediction programs, there is a relatively high number of false-positive predictions. To address this limitation we have now adapted a concept previously used to identify novel minimotifs19. Neduva and Russel examined sets of protein-protein interactions to identify proteins that interacted with a common target protein and shared a unique minimotif signature19.
Here, our goal was to engineer a new tool that would have two principle uses: 1) it would improve prediction of new minimotifs in MnM, by reducing the false-positives predictions. A filter would restrict the target predictions to those proteins where the minimotif source protein and the target protein are already known to interact. We implemented several strategies to modulate filter stringency. 2) it can be used to facilitate the study of PPI theory by identifying minimotifs between two proteins that are already known to interact, but the interface is not yet known. For example, in an example analysis of Discs, large homolog associated protein (DLGAP-1, NP_075235) Minimotif Miner predicts 123 potential binding motifs, however, only 2 are previously known to have a known protein-protein interaction. Thus, using PPIs reduced the number of minimotif predictions. In the second application above, an example analysis of DLGAP-1 shows that it contains PxxPxK and YxxP minimotifs. These minimotifs are known to bind to the SH3 and SH2 domains of Crk, respectively20. A protein-protein interaction of DLGAP1 with Crk was previously identified by an array screen of SH3 binding motifs that included Crk peptides21. Thus two mechanisms for this PPI are now suggested.
Materials and Methods
A. Protein-protein interaction filters
In order to refine minimotif predictions using known PPIs, we first needed sources of protein-protein interaction data. The minimal PPI data we need is two interacting proteins with accession numbers. We accepted the limitation that some protein-protein interaction databases may contain data with a low intrinsic false-positive rate, containing a few incorrect or artifactual interactions. We selected five databases primarily based on the public availability, amount of data, and reliability of the data. The Database of Interacting Proteins (DiP), Entrez Gene, Human Protein Resource Database (HPRD) release 8, Molecular Interaction database (MINT) release 2.5, VirusMINT and IntAct contain protein-proteins interactions annotated from the literature or derived from experimentation5,22,23. Statistics for these databases are provided in Table I.
Table I.
Sources of protein-protein interaction data
| Database | # interactions | # proteins | # species | Data source | Reference # | Date Downloaded |
|---|---|---|---|---|---|---|
| DiP | 57,683 | 20,728 | 274 | Literature curation | 37 | Aug., 2009 |
| Entrez Gene | 387,159 | 19,205 | unknown | Linkout databases | 38 | Sep., 2009 |
| HPRD | 38,806 | 27,081 | 1 | Yeast 2-hybrid, in vitro or in vivo experiments, Proteinpedia | 39 | Aug., 2009 |
| MINT | 83,321 | 29,774 | unknown | Literature curation | 40 | Jun., 2009 |
| Virus MINT | 1,854 | 468 | 99 | Literature curation | 23 | Nov., 209 |
| IntAct | 216394 | 63654 | unknown | Experiments | 22 | Jun., 2010 |
| Total | 785217 |
Total does not consider that different databases may use different accession numbers for the same protein.
Collectively, these databases have >785,000 total interactions. The total number of non-redundant interactions is likely more similar to the 322,579 unique PPIs consolidated in the Agile Protein Interaction DataAnalyzer8.
B. Annotation of minimotifs
All minimotifs in the Minimotif Miner database were curated from the literature and have supporting experimental evidence15,16. The data was refactored into a new data model24. The basic elements of this model have a ‘motif source’, which contains the Minimotif, a “target’, which engages the Minimotif, and an ‘activity’ which describes the function outcome of the minimotif source engaging its target. In order to use this information for the computational PPI filters, we examined the references for all 5313 minimotif definitions and assigned accession numbers to the majority of Minimotif sources and targets. In a few cases the accession number could not be assigned for one of two reasons: 1) an unambiguous assignment could not be made based on the information in the paper or in referenced material, or 2) if the minimotif source was identified in a phage display, or peptide based screen then there is no assignment of an accession number.
C. Protein-protein interaction filtering algorithms based on homology and similarity
We designed several Protein-Protein Interaction filtering algorithms that could be compared for their utility in refining minimotif definitions. The basic algorithm is as follows: For the purpose of describing these algorithms, let p be the putative minimotif, let S be its source protein, and let T be the target protein which interacts with the source protein that contains p as previously described in our model of minimotifs24. Each PPI pair will be assigned to (A, B) and (B, A) where A and B are proteins, thus there are two assignments to (S, T) for each PPI. Then each database is searched for an exact (S, T) match. This filter is designated the “PPI-filter”.
The source of minimotifs for this analysis will be the minimotif miner (MnM) database, which has over 3000 minimotifs for protein-protein interactions and is modeled with pairs of minimotif source and target proteins (S, T)15,16. To use MnM for this filter, we annotated both Source and Target accession numbers for all of the ~3000 PPI minimotifs that could be unambiguously assigned.
Several studies suggest that the minimotifs that drive PPIs are often conserved between species25–27. Therefore, we designed an extension of the above protein-protein interaction filter, designated the “HomoloGene-PPI” filter that uses the HomoloGene database to extend PPI predictions into other species. HomoloGene is a database, which clusters paralogues and orthologues into gene families28. For a given (A, B) pair in a PPI database, we can assign (Ai, Bi) and (Bi, Ai) to (Si, Ti), where i indicates a species. We examine HomoloGene clusters for Si and then match the species to Ti, which evaluates both proteins of a (A, B) pair. Using this approach, we can now expand the number of PPI interactions based on the assumption that the interactions are conserved in orthologues and paralogues across species and taxa.
While the HomoloGene-PPI filter assesses conservation in orthologues and paralogues, minimotifs may also be conserved in a broader range of homologues. For example, a PxxP minimotif is likely to bind to many of the 100’s of SH3 domains present in different proteins, even if the proteins are homologues in the same species. To develop this “Similarity-PPI” filter we used all the protein-protein interaction databases. Each protein in these databases was used to form a cluster using BLAST29. Each cluster will have proteins with sequence similarity. By varying the threshold cutoff value in the BLAST analysis we generated clusters with different stringency. We refer to these versions of PPI databases as Extended-PPI. A pair in the extended-PPI is of the form (A, B’) or (A’, B) where A and B are proteins and A’ and B’ are protein clusters. In the Similarity-PPI filter, for any given source target pair (S, T), all the entries in the database are examined to identify interacting protein-protein pairs (S′, T′) where S′ is similar to S and T′ is similar to T. A pseudocode for the detailed algorithm is given in the Appendix. To reduce false positive predictions of PPIs by this approach, we optionally enforced the additional constraint that S′ should contain p, the putative motif.
D. ROC curves
Relative Operating Characteristic (ROC, also named receiver operating characteristic) curves are commonly used to evaluate the sensitivity and specificity of algorithm performance and were used to evaluate the PPI filters.. We used the R project software suite to compare ROC curves of the PPI filter with the frequency score filter used on the MnM website30. In the R package the area under the curve (AUC) is calculated following Mason and Graham’s methods31. The ROC area can be interpreted as re-parameterized forms of the Mann-Whitney U-statistic32. The statistical significance (p-value) of the ROC area is calculated from the Mann-Whitney U-distribution. In this paper, AUC and p-values are calculated based on empirical curves. We have compared the empirical curves to a ROC curve fit with a binormal function, which assumes that the positive data follow a normal distribution. The robustness of binormal model shows high similarity with experimental curves, even when the data follow some other distribution33.
Results
Evaluation of Protein-Protein Interaction filters
We wanted to create a set of filters that reduce the number of false-positive minimotifs predicted by Minimotif Miner. Given a query protein, we plan to use PPI filters to restrict these predictions to those that are previously known to have a PPI. We also generated and tested less stringent versions of these filters (PPI-HomoloGene, and PPI-Similarity). The PPI, PPI-HomoloGene, and PPI-Similarity filtering algorithms were evaluated using the Minimotif Miner 2 (MnM2) database. MnM2 has 2941 PPI minimotifs where accession numbers for both the minimotif source protein (that contains the minimotif) and the target protein (that binds to this minimotif) are known. We used two metrics to evaluate the success of these filters.
To assess sensitivity of these filters, the percentage of protein pairs that were identified in both a PPI database and MnM database was computed. This percentage metric can be thought of as the sensitivity of the algorithm filter with higher percentage recovery representing higher sensitivity.
We also wanted to test the selectivity of each filter for true minimotifs. We randomly selected 20 proteins to be processed using MnM. Let A be one such query protein. A list of L of putative minimotifs identified by MnM was collected. Let r be an entry in L. Let the target protein for the r motif in A be T. We sent (A, T) to the PPI filter and checked if this pair will pass the filter (that is, we sent a minimotif-containing protein and an interaction partner predicted by MnM into the PPI algorithm). We did this for every element in L generated by MnM for the 20 randomly picked proteins. As a result, we computed the percentage of minimotifs (r’s) known to mediate binding with targets (T’s) contained in the 20 proteins (A’s). This is the percentage of interactions (A, T), which passed the filter. We refer to this percentage as the selectivity of the filter; lower numbers indicate more selectivity for valid minimotifs. We use the ratio of sensitivity/selectivity as a single metric called Discrimination Ratio (DR) that evaluates the success of the filter.
Since we did not know the redundancy and coverage of each PPI database, the PPI-filter was evaluated for each of the PPI databases (Table II). The results obtained for MINT, HPRD and Entrez Gene database showed that many PPIs were also present in MnM; whereas few were identified in a set of randomly synthesized PPI interacting pairs; this is reflected as higher sensitivity. Collectively, 58% of minimotifs had a PPI in at least 1 of the 5 PPI databases examined. We compared the results of the filter using MINT, HPRD, and Entrez Gene databases with a set of randomly synthesized PPI interacting pairs. Overall, integration of all five PPI databases yielded the highest overall Discrimination Ratio of 31 (higher scores indicate better performance), revealing the effectiveness of this filter and suggesting that when applied to novel queries this filter would help to reduce false positive predictions.
Table II.
Evaluation of the PPI filter
| 1 Sensitivity (%) | 2 Selectivity (%) | 3 Discrimination ratio | |
|---|---|---|---|
| DiP | 1.6 | 0.0 | x |
| Entrez Gene | 40.7 | 3.3 | 12.5 |
| HPRD | 31.0 | 2.7 | 11.6 |
| MINT | 41.2 | 1.5 | 27.7 |
| VirusMint | 0.2 | 0.0 | x |
| IntAct | 7.4 | 0.7 | 11.4 |
| 4At least one | 61.6 | 2.1 | 29.3 |
′ Sensitivity’ refers to the percentage of positive instances that are accepted by the filter.
′ Selectivity’ refers to the percentage of negative instances accepted by the filter.
′ Discrimination ratio’ is sensitivity/selectivity.
′At least one’ refers to a minimotif that was identified in at least one of the PPI databases listed in this table.
We next evaluated the HomoloGene-PPI filter to determine if extending the sets of PPIs to include orthologues and paralogues reduces the stringency of the PPI filter. The HomoloGene-PPI filter did not produce any significant increase in sensitivity when the MnM data set was analyzed, however, there was a modest reduction of selectivity reducing the Discrimination ratio score when compared to the PPI filter (Table III). The reason that the HomoloGene-PPI did not yield major improvements is likely because there are few minimotifs in the MnM data set where a minimotif in an interaction is defined for more than one species. Although this algorithm did not improve results in analysis of the MnM data set, we can envision situations where this filter is valuable as shown applying this filter to a test case in the next section.
Table III.
Evaluation of the HomoloGene-PPI filter
| 1 Sensitivity | 1 Selectivity | 1 Discrimination Ratio | |
|---|---|---|---|
| DIP | 1.9 | 0 | x |
| Entrez Gene | 44.7 | 3.5 | 12.7 |
| HPRD | 33.3 | 2.7 | 12.2 |
| MINT | 41.2 | 2.4 | 17 |
| VirusMint | 1.5 | 0 | x |
| IntAct | 8.3 | 1.2 | 7 |
| 2At least one | 63.7 | 5.1 | 12.5 |
Measurements in column are as defined in Table I.
′At least one’ refers to a minimotif that with a source and target that matched in at least one of the PPI databases listed in this table.
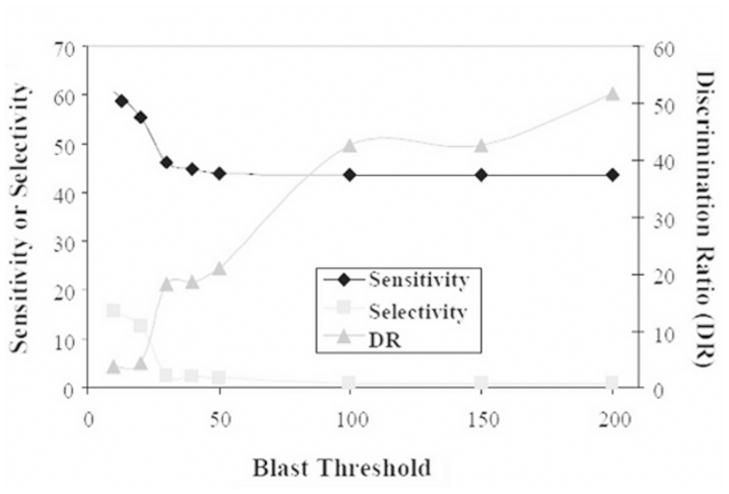
To further reduce stringency of the PPI filter, we created and tested the Similarity-PPI filter, which uses BLAST instead of HomoloGene clusters to identify proteins with sequence similarity. The BLAST threshold used to identify similar proteins was varied and the performance of this filter was assessed as for the other filters (Figure 1). The Discrimination ratio (sensitivity/selectivity) correlated positively with increasing BLAST scores, indicating that the more stringent the sequence similarity, the better the filter performance. The discrimination ratio was better than that of the HomoloGene-PPI filter when BLAST thresholds above 20 were used.
Figure 1. Evaluation of the BLAST threshold used to create extended-MINT in the Similarity-PPI filter.
The Similarity-PPI filter was applied to various datasets and sensitivity, specificity and the discrimination ratio were measured and plotted. Different datasets were created by using BLAST to identify proteins with sequence similarity; the BLAST threshold was varied.
Better selectivity scores were obtained when using high BLAST thresholds with the Similarity-PPI filter. With BLAST thresholds of 10 and 20 we observed increases in sensitivity as well as less stringent selectivity. However, the gains in selectivity were not sufficient to improve the overall DR. This was expected; when a wider breadth of homologues is selected with lower BLAST scores selectivity should become worse. The potential use of this filter is that it is capable of generating broader (albeit less stringent) predictions of minimotifs than either of the PPI and HomoloGene-PPI filters.
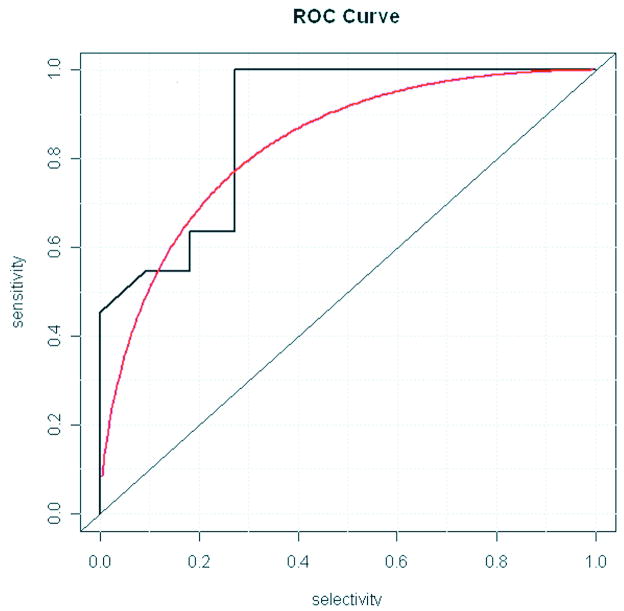
An ROC curve was used to assess the Similarity-PPI filter; the underlying variable parameter was the BLAST score. Note that for PPI-filter and HomoloGene-PPI filter there is no such parameter and hence ROC curves are not relevant. This ROC curve (Figure 2) clearly demonstrates the statistical significance of our algorithm. The area under the curve is 0.9 and the p-value is 0.001. Note that the p-value corresponds to the probability that a random predictor (i.e., filter algorithm) will produce the same results as our algorithm. We also wanted to compare the performance of our algorithm in relation to the existing frequency filter of MnM. The frequency score filter scores minimotifs based on the amino acids in the minimotif sequence definition and the frequency of these amino acids in the proteome15. The area under the ROC curve for the frequency filter is 0.7 and the p-value is 0.08 (Figure 3). Comparison of ROC curves for the two filters shows that the PPI filter has a better performance than the previously reported frequency score filter15. Table IV compares these two filters on many parameters of interest. In this table n.total is the total number of tests, n.events is the total number of tests on true data, n.noevents is the total number of tests on random (negative) data.
Figure 2. ROC curve for Similarity- PPI Filter.
ROC curves for the Similarity-PPI curve were generated with the R project software package. The empirical curve (black) and binormal curve (red) are shown. The binormal curve is calculated based on the assumption that the data is from a normal distribution. The area under the empirical curve is 0.9 (p = 0.001).
Figure 3. ROC curve for Frequency Score Filter.
The Frequency Score Filter previously implemented in MnM was analyzed and an ROC curve was plotted for the sensitivity and specificity of the filter by varying the frequency threshold. Curves are as in Fig. 2. The area under the empirical curve is 0.7 (p = 0.08).
Table IV.
Comparison between filters based on ROC curves
| PPI Filter | Frequency Score Filter | |
|---|---|---|
| 1area | 0.9 | 0.7 |
| 2n.total | 22 | 16 |
| 3n.events | 11 | 8 |
| 4n.noevents | 11 | 8 |
| p-value | 0.001 | 0.08 |
area is the area under the ROC curve.
n.total is the number of tests.
n.events is the total number of tests on true data.
n.noevents is the total number of tests on random (negative) data.
The other principle goal was to identify minimotifs that are present in one of two proteins known to interact. By using any of these filters on the MnM website users can enter a query protein and use MnM to find a minimotif that potentially drives the mechanism of the interaction.
Examination of Grb2 using PPIs
In order to further test the different PPI filters we examined Growth Factor Receptor Binding protein 2 (Grb2) as an example minimotif source query. Grb2 is an adaptor protein involved in receptor tyrosine kinase signaling that has many known protein interaction partners, thus provides a good test case. We analyzed Grb2 proteins from human, mouse, rat and fly and indicate the percentage of the minimotifs that pass the filter (Table V). With the PPI filter 50% of the predicted minimotifs for human and 19% of rat Grb2 had previously known interactions; fly and mouse Grb2 has not reported minimotifs with known PPIs in the MnM database. The PPI filter would provide benefit to users by allowing selection of known minimotifs and predicted minimotifs for many human and rat Grb2 PPIs.
Table V.
Percentages of minimotifs in Grb2 that pass different PPI filters
Grb2 proteins from different species were analyzed with the PPI and HomoloGene-PPI filters.
The percentage of minimotif predictions that pass the filter.
If two proteins are known to interact in human and rat, as in the case for Grb2, then it is reasonable to assume that they also interact in mouse and perhaps also in fly. When we analyzed the same Grb2 proteins with the HomolGene-filter, 15 and 11 minimotifs in mouse and fly Grb2, respectively, now passed the filter whereas none had passed the PPI filter (Table V). Although the HomoloGene-filter algorithm showed no distinct advantage when globally applied to MnM data, it is clear for the case of Grb2 that the filter worked as designed by extending minimotif predictions to proteins in other species.
We next examined the Similarity-PPI filter on the Grb2 proteins using different thresholds in BLAST to cluster proteins. Higher BLAST scores indicate more stringent protein similarity. Even when the highest BLAST threshold of 200 was used we observed significantly more minimotifs passing the Similarity-PPI filter (Table VI). This result is consistent with more homologues being present in each protein cluster. As expected for all 4 species examined lower BLAST thresholds produced more predictions and the profiles were consistent with the variation in sensitivity, selectivity and score observed with when the BLAST threshold was titrated as shown in Figure 1.
Table VI.
Percentages of minimotifs in Grb2 that pass the Similarity-PPI filter
| 1 Grb2 | 1 BLAST score threshold | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 20 | 30 | 40 | 50 | 100 | 150 | 200 | 500 | 1000 | |
| Human | 74 | 63 | 61 | 59 | 55 | 55 | 55 | 55 | 55 | 55 |
| Mouse | 61 | 43 | 30 | 30 | 28 | 20 | 20 | 20 | 20 | 20 |
| Rat | 64 | 48 | 37 | 37 | 35 | 31 | 31 | 31 | 31 | 31 |
| Fly | 61 | 50 | 28 | 28 | 25 | 19 | 19 | 19 | 19 | 19 |
Analysis as in Table V except that the Similarity PPI filter was used and BLAST threshold was varied to created different PPI datasets for the analysis.
Since the percentage of minimotifs was higher than expected for the Grb2 proteins, even at the highest BLAST threshold we implemented the additional constraint that Si should contain pi, the putative minimotif. Results from this algorithm shown in Table VII show that in all cases less minimotifs passed the filter than when there was no requirement for the presence of the minimotif in the Similarity PPI filter.
Table VII.
Percentages of minimotifs in Grb2 that pass the Similarity-PPI filter that contain the predicted minimotif
| 1 Grb2 | 1 BLAST score threshold | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 20 | 30 | 40 | 50 | 100 | 150 | 200 | 500 | 1000 | |
| Human | 55 | 44 | 42 | 36 | 36 | 36 | 36 | 36 | 36 | 36 |
| Mouse | 61 | 43 | 30 | 30 | 28 | 20 | 20 | 20 | 15 | 7 |
| Rat | 55 | 40 | 28 | 28 | 26 | 22 | 20 | 20 | 15 | 13 |
| Fly | 61 | 50 | 28 | 28 | 25 | 19 | 19 | 19 | 17 | 5 |
In Analysis as in Table VI except the additional constraint that Si should contain pi, the putative minimotif was implemented.
We wanted to develop a better feel for results we would obtain with a novel protein query. We randomly selected 100 proteins from the RefSeq database ((growth associated protein 43), CD2, DYRK1A, TRPM6, IRS-1, etc.) and analyzed them with Minimotif miner using the Similarity-PPI filters with different BLAST thresholds. When the homology filter was used with a promiscuous BLAST score threshold of 10, no predicted minimotifs were eliminated, whereas when stringent BLAST thresholds of 500 or 1000 were used, only 11% of the predictions were selected by the filter as having a previously known interaction in a related protein (Table VIII). Thus, a wide range of filtering was observed. The results are shown in the Appendix in a series of Tables.
Table VIII.
Analysis of 100 random query proteins with the Similarity-PPI filter in MnM
| 1 Blast score threshold | 1# not removed by filter | 1 Removed by filter (%) |
|---|---|---|
| 10 | 4658 | 28% |
| 20 | 2133 | 54% |
| 30 | 904 | 80% |
| 40 | 668 | 86% |
| 50 | 550 | 88% |
| 100 | 396 | 91% |
| 150 | 347 | 93% |
| 200 | 347 | 93% |
| 500 | 274 | 94% |
| 1000 | 274 | 94% |
Analysis as in Table VI except on 100 randomly selected proteins.
We examine the putative minimotifs that were retained by the Similarity-PPI filter by examining IRS-1 target predictions. Several previously known interactions with IRS-1 were retained by the filter (Grb2, PI3K, Insulin receptor and Epidermal Growth Factor Receptor). In addition, a number of proteins with a related signaling function were also retained (Integrin, PTPN11, Fyn, Crk, Casein Kinase 2, and JAK2). These results indicate that the filter selects known positives and also identifies reasonable candidates for novel minimotif driven protein-protein interactions.
Adapting the Minimotif Miner 2 user interface to include PPI algorithms
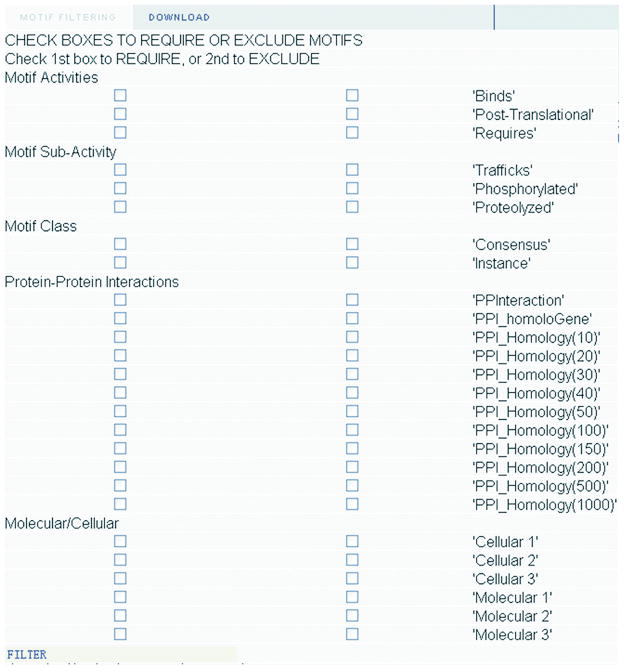
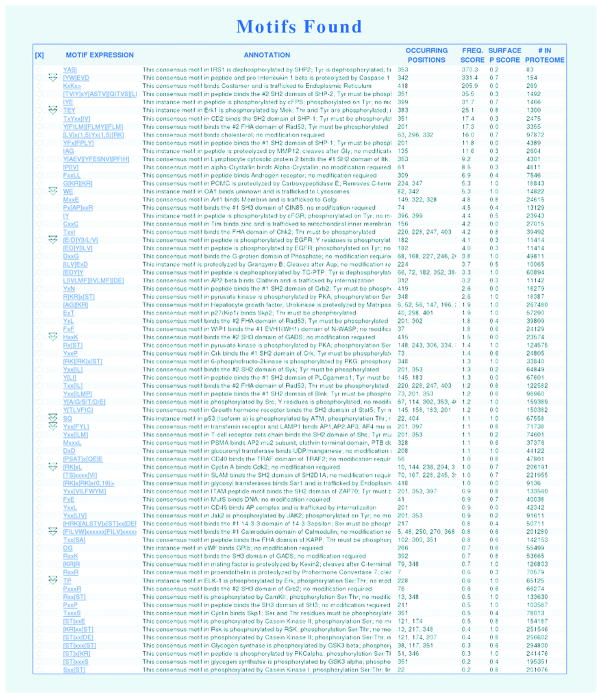
In order to provide an interface for scientists to use the PPI filters we added the PPI filter section to the Minimotif Miner 2.0 web application (Figure 4). This filter section contains PPI, PPI-HomoloGene, and PPI-Similarity filters; BLAST thresholds of 10, 20, 30, 40, 50, 100, 150, 200, 500 and 1000 are available for the PPI-Similarity filter. For each filter, options to use the filter, examine and select motifs retained or eliminated by the filter are available. The PPI filters can be used in combinations with other minimotif scoring metrics. Furthermore, in addition to searching for minimotifs with previously known PPIs, the user can use the exclude option to also identify minimotifs that do not have a previously known PPI. Figures 5 and 6 show screen shots of the MnM website with and without the PPI filter; 48 of 75 minimotif predictions were eliminated by the filter in this analysis of acyl-CoA dehydrogenase.
Figure 4. Image of filter selector and results modification added to the Minimotif Miner 2 web application.
The category filters in the Protein-Protein Interaction section was added for this paper.
Figure 5. Screenshots of Results table in MnM.
Image of restuls table from an MnM analysis of acyl-CoA dehydrogenase (NP_000007). MnM returns 75 minimotifs predictions.
Figure 6. Screenshots of MnM website with PPI filter applied.
Image of results table from an MnM analysis of specific acyl-CoA dehydrogenase (NP_000007) after applying the PPI filter. MnM returns 27 minimotif predictions after applying the PPI filter.
The minimotif results table has also been modified to make each score column sortable by clicking at the top of the column. This will better allow users to select the criteria they want to use to filter out false-positives or to focus their search. The MnM help section has been updated to help users with the new filter and reporting functions.
Discussion
In developing a theory of PPIs, scientists have made significant progress in understanding the physical forces that drive chemical interactions and also have generated large datasets of known PPIs. Minimotifs can drive protein-protein interactions, thus provide a simplified system in which we begin to develop a protein-protein interaction theory.
Currently, there are no broad based tools with which a scientist can systematically search for protein-protein interactions that have minimotifs, an important tool for investigating the role of minimotifs in PPI theory. For example, if two proteins are known to interact, then identifying a minimotif in one that mediates an interaction with the other protein in the protein complex helps users formulate a hypothetical mechanism of the interaction. Since minimotif prediction has a high false positive rate, this tool would have a second use in reducing false positives in the prediction of new minimotifs.
To address these problems, we have built several different PPI filters and implemented them in the Minimotif Miner 2 website16. In building the PPI filters we examined PPI data from existing PPI databases. When comparing the PPIs in the MnM database with known PPI databases that collectively have over 500,000 interactions we noticed that ~40% of the interactions in the MnM database were not yet observed in other PPI databases. Since the MnM annotation is a completely independent literature curation effort, this suggests, that even collectively the other existing PPI databases are not yet comprehensive for all PPIs reported in the literature.
In using the PPI filter, there was a strong preference for minimotifs being in protein with known PPI rather than randomly generated PPIs. In the example analysis of Grb2, 15 of the predictions with MnM that passed the PPI filter involved predictions that were not previously known to occur by the proposed mechanism. For example, several of the proteins that both interact with Grb2 and have a SH2 domain that binds to a consensus sequence that is consistent with a minimotif in Grb2 are Rasa (Y[ILV]x[FPYW]), Shp-1 (Y[IV]x[ILPV]) bLnk (Yxx[ILMP]) and Crk (YxxP). These proteins are predicted to bind at Tyr 209 near the end of the C-terminal SH3 domain of Grb2. Grb2 is also tyrosine phosphorylated on Tyr 209 (Human Protein Reference Database; HPRD), a requirement for these SH2 interactions34,35. Examination of the structure of Grb2 (1GRI) shows that these minimotifs are on the surface and available for binding. Considering these data we can generate several new hypotheses for the mechanisms by which these proteins interact with Grb2, which is not yet known.
PPIs are very often conserved between similar species, and are often conserved over a wider taxonomical range36. This is especially true in mammals where orthologous proteins in different species (which often have >90% amino acid identity) are inferred to have conserved interaction partners and minimotifs. The PPI-HomoloGene filter was designed to take advantage of PPI conservation, but did not show any improvement in discrimination ratio on the MnM data set when compared to the PPI filter; in fact it dropped significantly. We propose that this is due to that fact that most data in the MnM database does not have that same minimotif annotated for different species. This was supported by our analysis. The obvious need for this filter was evident in the analysis of Grb2. In Grb2, if one was studying a human protein 50% of the predictions pass this filter. This reflects that most work on Grb2 in the PPI databases was done in human and rat proteins. However, there were no proteins that passed this filter in mouse and fly orthologues. Since the domains and minimotifs are well conserved, we expect that any interaction observed in human and rat would likely also be observed in mouse, and likely fly orthologues. Thus, despite the poorer global performance of the PPI HomoloGene filter, it is clear that at least in isolated cases, it will be valuable to those studying PPIs and minimotifs.
Our initial implementation of the Similarity PPI filter showed that more minimotifs passed the filter than expected, even when high BLAST thresholds were used. This was not observed in the global analysis of the MnM data. Therefore, in order to adjust the stringency of this filter we provide users the option to use a wide range of BLAST thresholds, so that the number of resulting minimotifs can be adjusted on a case by case basis if desired. The related ROC curve and the p-value indicate that our algorithm is highly statistically significant. A comparison of the Similarity PPI filter with the previously reported frequency score filter indicates that the Similarity PPI filter is more significant statistically15.
In summary, the PPI, PPI-HomoloGene, and PPI-Similarity filters provide new computational tools to study the mechanisms of PPIs and to reduce false positive prediction in the discovery of novel minimotifs.
For further details please see http://137.99.1.76:8080/MNM/.
Supplementary Material
Acknowledgments
We would like to thank members of the Minimotif Miner team for critical input in the daily operations of our group. Grant Support: This work was supported by National Institutes of Health grant number GM079689 to M.R.S., an NSF Grant 0829916 to S.R., and by the Universities of Nevada Las Vegas and Connecticut.
APPENDIX
Pseudocode for HomoloGene-PPI filter
Input: Query Protein Q, a Set of Target Proteins T1, .. Tn.
Algorithm:
Get the homologene cluster for Q. Let it be {Q1, Q2..Qn}.
-
For each i = 1 to n do
Get the interactors of Qi from the database. This results in the set {Qpi1, Qpi2, .., Qpim}.
Now we have the interactors pair as set Q′ as
{(Q1, Qp11),(Q1, Qp12), …, (Q1, Qp1m)
(Q2, Qp21),(Q2, Qp22), …, (Q2, Qp2m), …, (Qn, Qpn1), (Qn, Qpn2), …, (Qn, Qpnm)}
Remove from Q′ all the pairs that is not of the same species, to get Q″.
-
For each i = 1 to n do
Get the homologene cluster of the target protein Ti. Let it be {Ti1, Ti2,.. T1m}.
For each j = 1 to m do
Check if there exists a pair in Q″ such that Qpxy = Tij.
If yes, (Q, Ti) passes the filter. Else it fails.
Pseudocode for Similarity-PPI filter
Input: Query Protein Q, a set of Target Proteins (T1, ..Tn), Threshold t, PPI Databases with interactors as {(I1, I1′), (I2, I2′).. {Ix, Ix′)}
Algorithm:
Blast Q against all the entries in the database. Get all the entries within the threshold. Let the resulting set be S{s1, s2, s3.., sn}. (S has, say example, {I3, I7′, I15, I19, I34′,…})
-
For each i = 1 to n in S do
Get the interacting pair of interactors from the database. Say if the interactors is I1, then get I1′. The resulting set of all the partners of interactors in S be S′. (Now S′ has entries {I3′, I7, I15′, I19′, I34,…})
-
For each i = 1 to n do
Blast the target Ti against S′.
If the score is within the threshold t, (Q, Ti) passes the filter. Else it fails.
References
- 1.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 2.Kennedy MA, McAteer K. Force field dependence of NMR-based, restrained molecular dynamics DNA structure calculations including an analysis of the influence of residual dipolar coupling restraints. Abstracts of Papers of the American Chemical Society. 2002;224:U482–U483. doi: 10.1080/07391102.2003.10506867. [DOI] [PubMed] [Google Scholar]
- 3.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. AMBER 10. University of California; San Francisco: 2008. [Google Scholar]
- 4.Lehne B, Schlitt T. Protein-protein interaction databases: keeping up with growing interactomes. Human genomics. 2009;3:291–7. doi: 10.1186/1479-7364-3-3-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathivanan S, Periaswamy B, Gandhi TK, Kandasamy K, Suresh S, Mohmood R, et al. An evaluation of human protein-protein interaction data in the public domain. BMC bioinformatics. 2006;7(Suppl 5):S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costanzo MC, et al. The Yeast Proteome Database (YPD) and Caenorhabditiselegans Proteome Database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivas JDL, Luis AD. Interactome data and databases: different types of protein interaction. Comp Funct Genomics. 2004;5(2):173–178. doi: 10.1002/cfg.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prieto C, Rivas JDL. APID: Agile Protein Interaction DataAnalyzer. Nucleic Acids Res. 2006;34:W298–W302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootton JC. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–14. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 12.Jonassen I. Efficient discovery of conserved patterns using a pattern graph. Comput Appl Biosci. 1997;13:509–522. doi: 10.1093/bioinformatics/13.5.509. [DOI] [PubMed] [Google Scholar]
- 13.Rigoutsos I, Floratos A. Combinatorial Pattern Discovery in Biological Sequences: the TEIRESIAS Algorithm. Bioinformatics. 1998;14(1):55–67. doi: 10.1093/bioinformatics/14.1.55. [DOI] [PubMed] [Google Scholar]
- 14.Tan SH, Willy H, Sung WK, Ng SK. A correlated motif approach for finding short linear motifs from protein interaction networks. BMC Bioinformatics. 2006;7:502. doi: 10.1186/1471-2105-7-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balla S, Thapar V, Luong T, Faghri T, Huang CH, Rajasekaran S, del Campo JJ, Shin JH, Mohler WA, Maciejewski MW, et al. Minimotif Miner, a tool for investigating protein function. Nat Methods. 2006;3:175–177. doi: 10.1038/nmeth856. [DOI] [PubMed] [Google Scholar]
- 16.Rajasekaran S, Balla S, Gradie P, Gryk MR, Kadaveru K, Kundeti V, Maciejewski MW, Mi T, Rubino N, Vyas J, Schiller MR. Minimotif miner 2nd release: A database and web system for motif search. Nucleic Acids Res. 2009;37(Suppl 1):D185–D190. doi: 10.1093/nar/gkn865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould CM, Diella F, Via A, Puntervoll P, Gemünd C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Via A, Gould CM, Gemünd C, Gibson TJ, Helmer-Citterich M. A structure filter for the Eukaryotic Linear Motif Resource. BMC Bioinformatics. 2009;10:351. doi: 10.1186/1471-2105-10-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neduva V, Russell RB. Linear motifs: evolutionary interaction switches. FEBS Lett. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72(5):767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 21.Wu C, Ma MH, Brown KR, Geisler M, Li L, Tzeng E, Jia CY, Jurisica I, Li SS. Systematic identification of SH3 domain-mediated human protein-protein interactions by peptide array target screening. Proteomics. 2007;7(11):1775–85. doi: 10.1002/pmic.200601006. [DOI] [PubMed] [Google Scholar]
- 22.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, Roechert B, van Eijk K, Hermjakob H. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–31. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatr-aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, Sacco F, Tinti M, Smolyar A, Castagnoli L, Vidal M, et al. VirusMINT: a viral protein interaction database. Nucleic Acids Res. 2009;37:D669–D673. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vyas J, Nowling RJ, Maciejewski MW, Rajasekaran S, Gryk MR, Schiller MR. A proposed syntax for minimotif semantics, version 1. BMC Genomics. 2009;10:360. doi: 10.1186/1471-2164-10-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren S, Uversky VN, Chen Z, Dunker AK, Obradovic Z. Short Linear Motifs recognized by SH2, SH3 and Ser/ThrKinase domains are conserved in disordered protein regions. BMC Genomics. 2008;9(Suppl 2):S26. doi: 10.1186/1471-2164-9-S2-S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chica C, Diella F, Gibson TJ. Evidence for the concerted evolution between short linear protein motifs and their flanking regions. PLoS One. 2009;4(7):e6052. doi: 10.1371/journal.pone.0006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards RJ, Davey NE, Shields DC. SLiMFinder: A probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins. PLoS One. 2007;2(10):e967. doi: 10.1371/journal.pone.0000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. A Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.R Development Core Team. R: A language and environment for statistical computing. Vol. 1. R Foundation for Statistical Computing; 2009. [Google Scholar]
- 31.Mason SJ, Graham NE. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: Statistical significance and interpretation. QJR Meteorol Soc. 2002;30(1982):291–303. [Google Scholar]
- 32.Bamber D. The area above the ordinal dominance graph and the area below the receiver-operating characteristic graph. Journal of Mathematical Psychology. 1975;12(4):387–415. [Google Scholar]
- 33.Hanley JA. The robustness of the ‘binormal’ assumptions used in fitting ROC curves. Medical Decision Making. 1988;8 (3):197–203. doi: 10.1177/0272989X8800800308. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Wu C, Huang H, Zhang K, Gan J, Li SS. Prediction of phosphotyrosine signaling networks using a scoring matrix-assisted ligand identification approach. Nucleic Acids Res. 2008;10:3263–73. doi: 10.1093/nar/gkn161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machida K, et al. High-throughput phosphotyrosine profiling using SH2 domains. Mol Cell. 2007;26(6):899–915. doi: 10.1016/j.molcel.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 36.Choi YS, Yang JS, Choi Y, Ryu SH, Kim S. Evolutionary conservation in multiple faces of protein interaction. Proteins: Structure, Function, and Bioinformatics. 2009;77(1):14–25. doi: 10.1002/prot.22410. [DOI] [PubMed] [Google Scholar]
- 37.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–51. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–9. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.