Abstract
Protein carbonylation is a major form of protein oxidation and is widely used as an indicator of oxidative stress. Carbonyl groups do not have distinguishing UV or visible, spectrophotometric absorbance/fluorescence characteristics and thus their detection and quantification can only be achieved using specific chemical probes. In this paper, we review the advantages and disadvantages of several chemical probes that have been and are still being used for protein carbonyl analysis. These probes include 2, 4-dinitrophenylhydazine (DNPH), tritiated sodium borohydride ([3H]NaBH4), biotin-containing probes, and fluorescence probes. As our discussions lean toward gel-based approaches, utilizations of these probes in 2D gel-based proteomic analysis of carbonylated proteins are illustrated where applicable. Analysis of carbonylated proteins by ELISA, immunofluorescent imaging, near infrared fluorescence detection, and gel-free proteomic approaches are also discussed where appropriate. Additionally, potential applications of blue native gel electrophoresis as a tool for first dimensional separation in 2D gel-based analysis of carbonylated proteins are discussed as well.
Keywords: Biotin, carbonylation, carbonyls, carbonylated, chemical probes, infrared fluorescence, oxidative stress, proteomics, tritiated sodium borohydride
1 Introduction
Oxidative stress is commonly viewed as a condition under which the generation of reactive oxygen species (ROS) within a cellular system exceeds the buffering capacity of endogenous antioxidant defenses [1], leading to oxidative damage involving lipids, DNA, and proteins [2]. Given the multitude of sources involved in the generation of ROS and the variety of enzymatic and non-enzymatic oxidant defenses, the condition of oxidative stress is most often an inference based upon the presence of an excess of oxidative damage to macromolecules. Among the numerous oxidation products, carbonylation of proteins may be the most widely used type of damage used to infer oxidative stress [3–5], in part based on the fact that carbonyl modifications can be produced by wide variety of ROS as well as by-products of lipid oxidation. However, specific protein carbonylations are thought to be of additional significance, beyond their use as a biomarker, because they can function as biological signals [6,7] or confer irreversible loss of protein function in connection with disease [4,5,8,9]. Generally, there are three types of amino acid oxidative modifications that can give rise to protein carbonyls: (1) direct attack by reactive oxygen species on certain amino acid side chains (Glu, Thr, Asp, Lys, Arg, and Pro) [10]; (2) modification of histidine, cysteine, and lysine residues by lipid peroxidation products such as malondialdehyde and 4-hydroxynonenal [11–13]; and (3) reaction with reducing sugars, forming advanced glycation end products adducts [14,15]. The existence of all three mechanisms of protein carbonylation have been well documented in aging and in age-related degenerative diseases [4,15].
2 Analysis of carbonylated proteins relies on the use of chemical probes
Because protein carbonyls have no distinguishing UV or visible spectrophotometric absorbance/fluorescence properties, they can not be directly determined. Instead, detection and quantification of protein carbonyls require the use of specific chemical probes that serve as handles for determination. In this review, we will discuss several probes that have been in use for the analysis of protein carbonyls, including 2,4-dinitrophenylhydrazine (DNPH) [16], tritiated sodium borohydride [17,18], biotin-containing probes [19,20], and fluorescence probes [21,22]. Except for tritiated sodium borohydride, a common feature of all probes is a hydrazine-like moiety that can react with carbonyl groups.
2.1 2, 4-Dinitrophenylhydrazine (DNPH)
2.1.1 Spectrophotometric measurements
DNPH was first introduced to the measurement of protein carbonyls by Levine et al. [16] and is still widely used. The unique feature of this probe is a peak absorbance around 360 nm that remains after its conjugation to proteins, allowing protein carbonyl content to be measured spectrophotometrically. The labeling process usually takes place under acidic conditions, whereby DNPH is dissolved in a 2N HCl solution. As an excess of DNPH is always added during the labeling, the samples usually undergo further processing involving precipitation of protein by TCA (10%, final concentration) and extensive washing with an organic solvent that is usually comprised of ethanol/ethyl acetate (1:1, v/v). An important caveat to be considered when DNPH is used for spectrophotometric determination of protein carbonyl content, is that proteins such as cytochrome c and hemoglobin have absorbance wavelengths similar to DNPH and may interfere with its measurement [23], leading to inaccurate estimation of protein carbonyls. If this is the case, other probes, such as tritiated sodium borohydride (section 2.2) [18], may be used.
2.1.2 Gel-based analysis
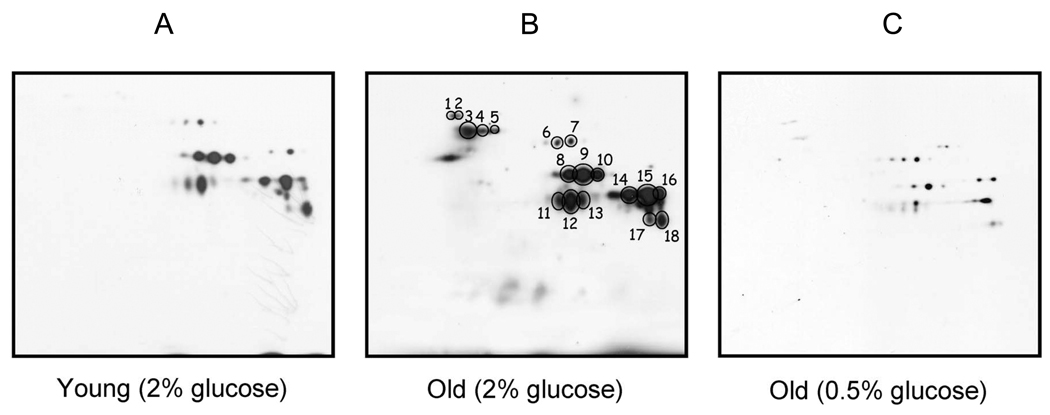
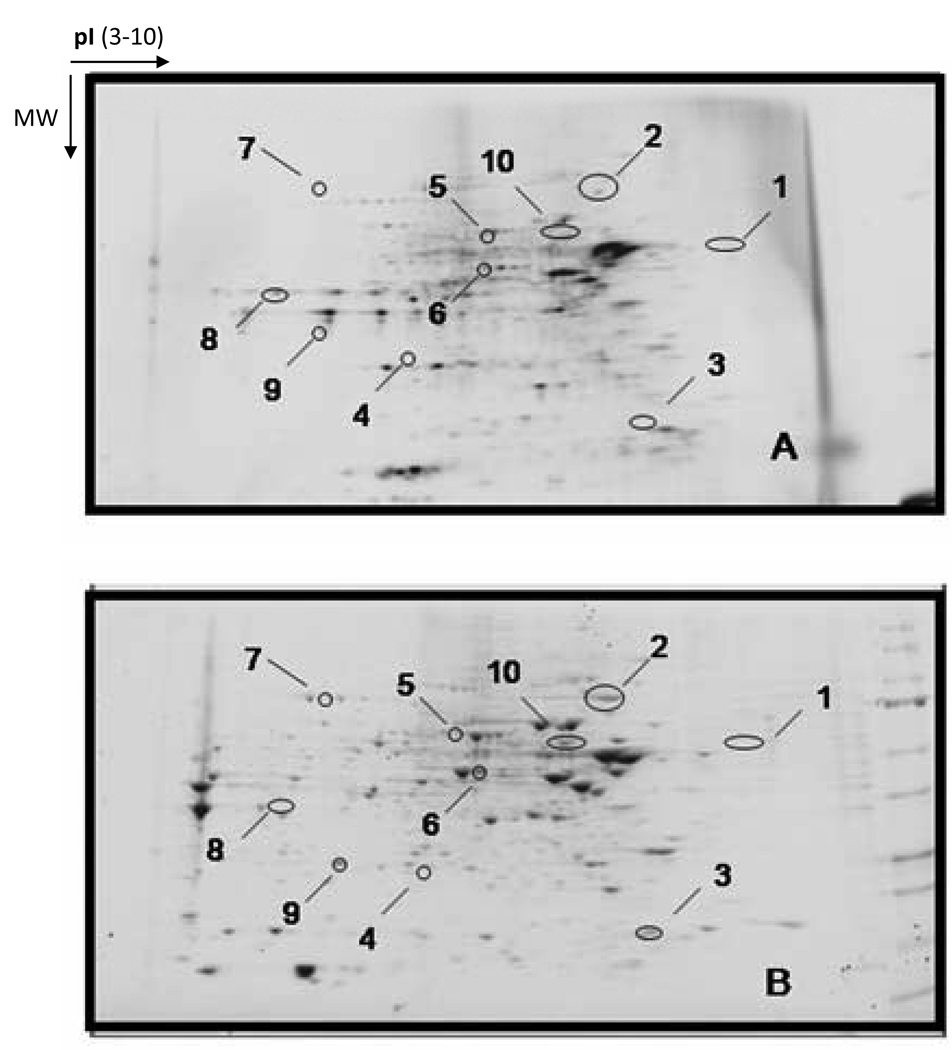
Protein samples treated with DNPH can be resolved by SDS-PAGE, and carbonylation associated with specific bands can be detected on Western blots using commercially available anti-DNPH antibodies [24,25]. Initial studies adopting this 1-D approach led to the unexpected observation that not all proteins in a given proteome were subject to equivalent oxidative attacks, supporting the current view that protein oxidation during aging and disease is a selective rather than a random process [26,27]. A multitude of more recent studies have successfully analyzed DNPH-treated samples using 2D IEF/SDS-PAGE, in a variety of experimental systems [28–31]. Fig. 1 shows a very good example of anti-DNP 2D immunoblot detection of carbonylated proteins during aging in yeast, as reported by Reverter-Branchat et al. [32]. In this study, many yeast proteins were found to exhibit an age-related increase in protein carbonyl content (Panels A and B) and the increase could be remarkably attenuated by caloric restriction (Fig. 1, panel C), a regimen that is known to increase cellular redox potential and decrease age-related oxidative stress [33–36].
Fig. 1.
Anti-DNP 2D immunoblot detection of carbonylated proteins during yeast replicative aging and the effects of caloric restriction. Numbered spots (Panel B) indicate identification of major targets that showed age-related increase in protein carbonylation. The increase in each target’s protein carbonyl content was markedly attenuated by caloric restriction that was achieved by growing yeast cells in the presence of 0.5% glucose (Panel C), as opposed to normal growth conditions whereby the concentration of glucose was 2%. Young: 2-generation-old; Old: 16–18-generation-old. (Figure reproduced with permission from American Society for Biochemistry and Molecular Biology, Ref. [32].)
2.1.3 Immunofluorescent imaging of carbonylated proteins
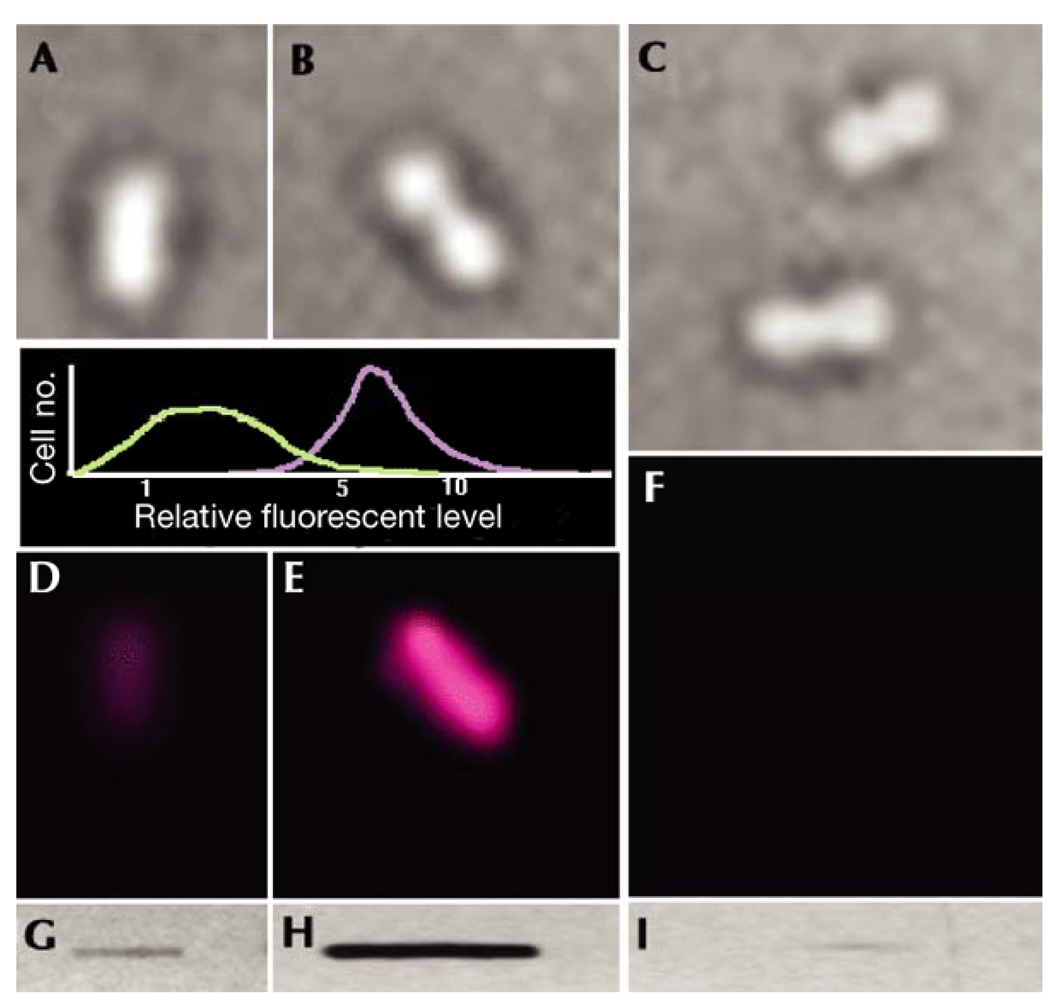
With the use of anti-DNP antibodies and fluorescent-secondary antibodies, protein carbonyls labeled with DNPH can also be visualized via in-situ immunofluorescent imaging [37–41]. This detection method can provide further insights into the mechanisms of cellular oxidative stress that occurs under physiological and pathophysiological conditions [42,43]. Fig. 2 shows an excellent example of in-situ immunofluorescent imaging of carbonylated proteins, as reported by Desnues et al [39]. In this study, level of protein carbonylation in whole E. coli cells was shown to be greatly elevated by H2O2 treatment (Fig. 2, E vs. D).
Fig. 2.
Immunofluorescent visualization of in situ carbonylated proteins in single Escherichia coli cells. Cells were grown in Luria–Bertani medium at 37 °C in the absence (A,D,G) or presence of 200 mM H2O2 (B,E,H). Transmission images of cells (A–C) and the fluorescence emission from fluorescently labeled antibodies (D–F) are shown. Cell samples from H2O2-stressed cells were treated with proteinase K (C,F,I). Slot-blot analysis of carbonyl levels in protein extracts (G–I) was carried out in parallel with carbonyl imaging to ensure that the carbonyl signal intensities agreed with the expected signal from the total-protein extracts. The inset graph shows the results of a statistical analysis of the carbonyl-intensity signal in 200 control (green line) and 200 H2O2-treated (purple line) cells. Protein carbonyls were derivatized with DNPH and probed with anti-DNP antibodies followed by visualization with TexasRed-conjugated secondary antibodies [38,41]. “Cell no.” = number of cells. (Figure reproduced with permission from Ref. [39])
2.1.4 ELISA measurement of protein carbonyls
Protein carbonyl content can also be measured using enzyme-linked immunosorbent assay (ELISA). The method was originally developed [44,45] by Dr. Christien Winterbourn’s group at Christchurch School of Medicine, New Zealand, and has since received wide applications. The assay involves the use of biotin-linked anti-DNP antibodies that not only bind to the coated DNP-derivatized proteins, but also facilitate detection with streptavidin [44,45]. The assay was later modified by Alamdari et al., in which proteins were first coated onto plates and then labeled with DNPH [46]. This modification makes it unnecessary to concentrate, otherwise, DNP-derivatized proteins for samples containing low amount of proteins [46]. The advantage of these ELISA methods is that the assay only requires microgram amounts of proteins and is capable of measuring many samples simultaneously. It should be noted that the obtained carbonyl values depend very much on the oxidized protein standard that is used for each assay. Additionally, it is known that ELISA results do not correlate well with those from DNPH measurements, but the reasons for this discrepancy have not been investigated. The biotin/hydrazide-streptavidin system, without the involvement of any antibodies, has also been adapted in the ELISA assay of protein carbonyls [47].
2.1.5 Potential problems associated with DNPH measurement using commercial kit
For immunochemical detection of carbonylated proteins separated by 1D or 2D gel electrophoresis, many investigators use commercial Western blot kit such as OxyBlot (Chemicon/Millipore, USA). However, two groups have recently pointed out problems associated with the use of these commercial kits [48,49]. These include overestimation of protein carbonyl content due to the recommended use of a high concentration of β-mercaptoethanol in the homogenizing buffer that presumably induces protein auto-oxidation [48], a diminished DNPH labeling efficiency upon storage of the kit due to a decrease in acidity of the DNPH solution [49], and instability of certain DNPH-derivatized protein samples upon storage either under reducing conditions at 4°C [48] or at low pH values [50]. Therefore, cautions should be exercised when the commercial kit is used. Additionally, for spectrophotometric quantification of protein carbonyl content in crude bacterial homogenates, nucleic acids should be removed with streptomycin sulfate (usually 1%, final concentration) to avoid overestimation of protein carbonyl content [48,51]. For spectrophotometric quantification of protein carbonyl content in eukaryotic cells, however, contamination from nucleic acids is usually negligible, but may need to be evaluated depending on the method of homogenization [48]. For gel-based analysis of protein carbonyls, no contamination from nucleic acids is expected as nucleic acids separate from proteins during gel electrophoresis. Relevantly, it has been recently reported that DNPH is also reactive toward protein sulfenic acids in the absence of reducing reagents such as β-mercaptoethanol [52]. However, whether this reaction leads to overestimation of protein carbonyl content has not been determined.
2.1.6 Unit expression of protein carbonyl content
Protein carbonyl content is usually expressed as nmol carbonyl/mg protein [16]. As such, both protein levels and carbonyl levels need to be determined. For crude homogenates, carbonyl content is determined spectrophotometrically at 360 nm by the quantities of DNPH incorporated, while protein amount is determined by protein assay. For gel-based analysis, both the carbonyl levels and the protein levels, with the latter often being reflected by Coomassie blue staining, can be densitometrically determined for given gel bands or spots. The ratio between the carbonyl signal intensity and the protein signal intensity would then give specific carbonyl content of the protein that is of interest. It should be noted that such a ratio is as expected unitless, but can be expressed as nmol carbonyl/mg protein provided that an oxidized protein standard is used [53–56].
2.2 Tritiated sodium borohydride ([3H]NaBH4)
Protein carbonyls can be reduced by sodium borohydride (NaBH4) to corresponding ethanol groups. When tritiated NaBH4 is used in the reduction, tritium is incorporated selectively into the carbonylated proteins [17] and, after TCA precipitation and extensive washing, the radioactivity can be measured using a scintillation counter. Additionally, the radio-labeled proteins can also be separated by SDS-PAGE and the protein band of interest can be excised [18]; the amount of tritium associated with the band can be released by incubating the gel band in a 30% hydrogen peroxide solution and the radioactivity can then be measured [18]. This probe offers a method of choice when target proteins have absorbance around 360 nm that would otherwise interfere with DNPH measurement [23].
2.3 Biotin probes
Biotin probes take advantage of the affinity binding between biotin and avidin (or streptavidin), among the strongest known non-covalent interactions [57]. Two such probes have been applied for the analysis of protein carbonyls, biotin-hydrazide [19] and N’-aminooxymethylcarbonylhydrazino-D-biotin (ARP) [58]. Proteins that are biotinylated using these probes can be affinity captured using avidin or streptavidin beads [59], or analyzed using 2D gel-based approaches. No secondary antibodies are involved when biotin-containing probes are used for Western blot detection of carbonyls, rendering the detection process more efficient and less subject to error.
2.3.1 Biotin-hydrazide
Dr. Fred Regnier’s group at Purdue University has used biotin-hydrazide quite extensively and in a variety of experimental systems [19,59–61]. During labeling with this probe, the initial reaction between a carbonyl group and the hydrazide forms a Schiff-base that is unstable and must be reduced to a stable hydrazine bond [19]. The additional reduction step necessitates further procedures involving either buffer exchanges via filtration or TCA precipitation/washing prior to further analysis of the biotinylated proteins. An excellent application of this probe is the profiling of carbonylated proteins in human plasma [59]. In this study, the use of biotin-hydrazide, in conjunction with affinity purification by avidin columns, allowed accurate estimation of total carbonylated proteins in plasma of healthy humans (0.2%), in addition to establishing identity of the proteins and their tissue sources. Additionally, in this and other studies, in conjunction with the use of mass spectrometry techniques, the biotin-hydrazide approach has been instrumental in identifying the sites and types of oxidation involved in carbonylation of specific proteins [59,62].
2.3.2 N’-aminooxymethylcarbonylhydrazino-D-biotin (ARP)
ARP was introduced to the analysis of carbonylated proteins by Dr. Claudia Maier’s group at Oregon State University [20,58,63,64]. The advantage of this probe is that removal of excess ARP is not needed if the biotinylated samples are to be subjected to gel-based analysis [20], because the bond formed between ARP and the carbonyl group is stable. However, when ARP-treated proteins are to be purified by affinity capture, further steps such as buffer exchange using centrifugal filters or TCA precipitation/washing are indeed required to completely remove excess ARP [58,63]. Using the ARP probe, Dr. Maier’s group identified 7 mitochondrial proteins that showed an age-dependent increase in protein carbonyls [20], including a confirmation of aconitase and adenine nucleotide translocase identified previously as targets of oxidative damage using other methods [26,27,65,66]. Similar to biotin-hydrazide [59], ARP was also successfully used in mass spectrometric identification of specific sites that had been modified [63].
2.3.3 Quantification of incorporated biotin
In addition to Western blot detection and affinity capture of carbonylated proteins or peptides, another application of the biotin-containing probes is the quantification of carbonyls in target proteins, based on incorporated biotin. This approach makes use of 4’-hydroxyazobenzene-2-carboxylic acid (HABA) and avidin that are pre-mixed at a proper ratio [67,68]. The HABA, when complexed with avidin, has a strong absorbance at 500 nm, whereas free HABA has an absorbance peak at 348 nm. Because of the relatively weak affinity binding between HABA and avidin (Kd = 5.8 × 10−6) and the strong affinity binding between avidin and biotin (Kd = 1 × 10−15), HABA bound to avidin can be easily displaced by biotin, leading to a decrease in absorbance of the reaction mixture at 500 nm (or an increase at 348 nm). Thus the amount of HABA released from the HABA/avidin complex is highly proportional to the biotin in the biotinylated (carbonylated) proteins. A fluorescence version of this assay is also available [69].
2.3.4 Application of biotin probes
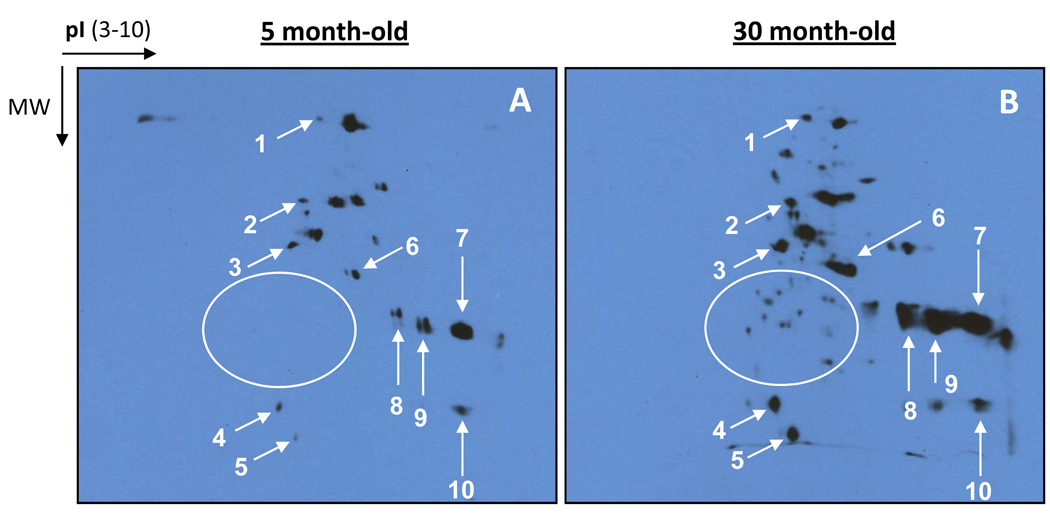
Based on the apparent advantages discussed above, our laboratories have recently applied biotin probes in lieu of our previous approach involving DNPH. In particular, we were interested in 2D profiling of aging-related protein carbonylation using biotin-hydrazide. In this study, rat liver mitochondria were used and were prepared as previously described [70] from animals aged either 5 or 30 months. The first dimensional isoelectric focusing was performed according to standard procedures using Bio-Rad PROTEAN IEF cell [23] while the second dimension was 7.5% Laemmli SDS-PAGE [71]. Results in Fig. 3 show clearly an age-dependent increase in protein carbonyls (Fig. 3, arrow indicated). Moreover, many carbonylated proteins detected in the old animals could not be detected in those aged 5 months (the circled area in Fig. 3), indicating that more proteins were carbonylated in the old than in the young.
Fig. 3.
Representative 2D Western blot detection of carbonylated proteins derivatized with biotin-hydrazide. Shown are profiles of age-related carbonylated proteins in rat liver mitochondria prepared as previously described [70]. Protein carbonyl labeling with biotin-hydrazide was performed also as previously described [19]. Two dimensional polyacrylamide gel electrophoresis and Western blot transfer were performed using Bio-Rad PROTEAN IEF cell and Mini-PROTEAN III cell according to standard procedures [23]. The second dimension was 7.5% Laemmli SDS-PAGE [71]. Protein amount loaded on each IPG strip in the first dimension was 40 µg. Rats aged 5 or 30 months were used in this study. Arrows indicate the selected gel spots showing age-related increases in protein carbonyls; gel spots inside the circled area in Panel B indicate carbonylated proteins that were not detected in the young animals.
2.4 Fluorescence probes
2.4.1 Fluorescein-5-thiosemicarbazide (FTC) and fluorescence hydroxylamine (FHA)
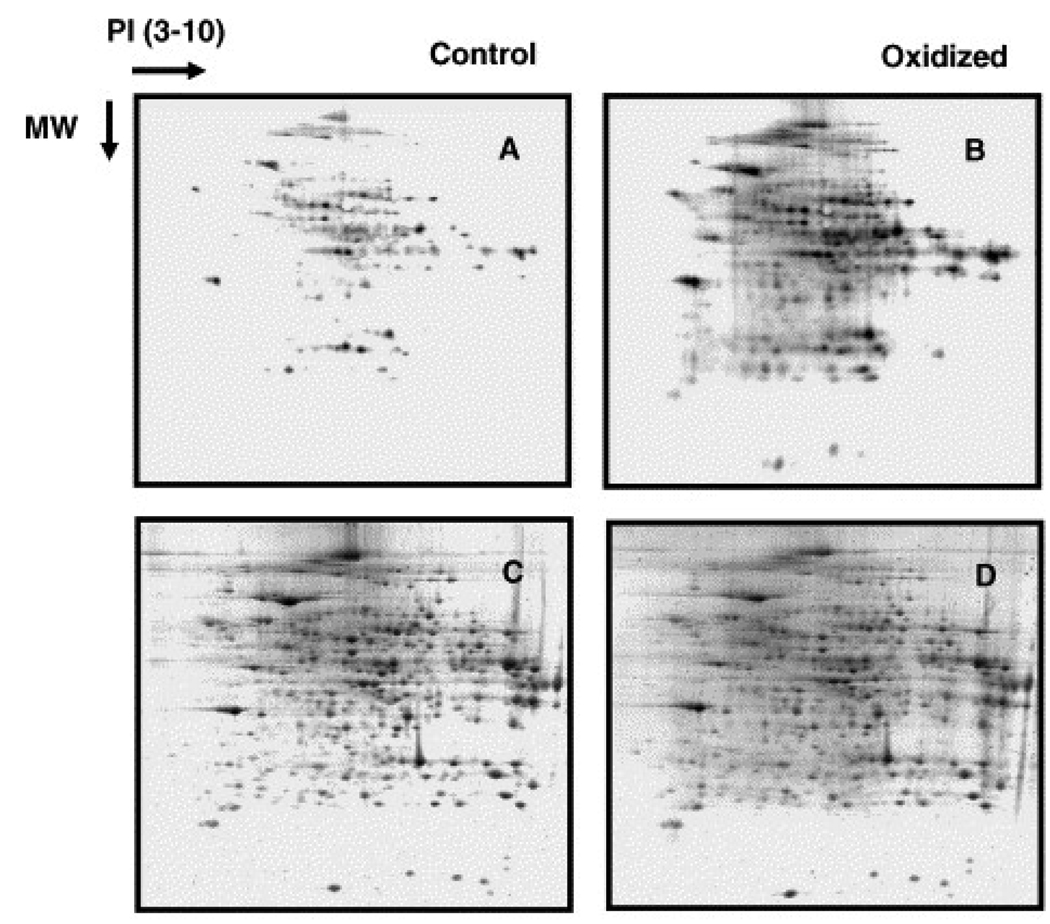
Two visible-wavelength fluorescence probes have been reported in the literature for the analysis of protein carbonyls: fluorescein-5-thiosemicarbazide (FTC) [21,72,73] and Alexa 488 fluorescence hydroxylamine (FHA, Invitrogen) [22]. Both probes are commercially available and have been successfully used in 2D gel-based proteomic analysis of carbonylated proteins. Application of these probes to quantify carbonyl content in protein mixtures has been limited, presumably because of the fact that it is difficult to completely remove the excess probes following TCA precipitation or gel filtration [21]. However, this difficulty is not encountered when fluorescence probes are used in gel-based analysis, as the unbound probes are separated from proteins during gel electrophoresis. As shown in Fig. 4 (comparing Panels A and B), Chaudhuri and co-workers used an FTC probe to identify a number of proteins in the mouse liver that showed an oxidation-dependent increase in their carbonyl content (21). Similarly, Poon et al. (22) used an FHA probe to identify 9 proteins that underwent carbonylation in mouse brains (Fig. 5).
Fig. 4.
Two dimensional polyacrylamide gel electrophoresis of FTC-labeled control and oxidized liver cytosolic protein. Proteins (1 mg/ml) were oxidized with or without FeSO4 (12 µM) in the presence of ascorbic acid (3 mM) One hundred microgram proteins were applied in the first dimension. Panels A and B show the fluorescence of FTC binding to the unoxidized (A) and oxidized protein extracts (B), respectively. Panels C and D show the Sypro Ruby fluorescence of the identical gels shown in A and B. (Figure reproduced with permission from Ref. [21].)
Fig. 5.
(A) Representative 2D gel image of protein carbonyl groups detected by FHA labeling. (B) Coomassie blue staining of the identical gel. Numbered spots indicate corresponding spots between fluorescence imaging and Coomassie blue staining. (Figure reproduced with permission from Ref. [22].)
2.4.2 N-aminoperylene-3, 4, 9, 10-tetracarboxylic-bismides (APTB)
APTB is unique in that the probe itself is not fluorescent but becomes fluorescent upon reaction with carbonyl groups. While applications of this chemical have been explored by chemists [74], it has not as yet been used by biologists for the detection of protein carbonyls. However, if APTB were used instead of FTC or FHA, protein carbonyl content could then be read directly from protein mixtures with a fluorometer, because the unconjugated, excess APTB is non-fluorescent. This probe should be equally applicable in 2D gel-based proteomic analysis of carbonylated proteins.
2.4.3 Near infrared (IR) fluorescence probes
Besides the visible wavelength fluorescence probes described above, near infrared (IR, 670 – 1000 nm) fluorescence dyes or probes have also found increasing applications in measurements of protein carbonyls [75–79]. Both carbonyl-reactive near IR fluorescence probes and antibody- or streptavidin-linked near IR fluorescence dyes are now commercially available (Molecular Probes/Invitrogen). The use of near IR fluorescence probes offers the following advantages. First, as cellular macromolecules lack near IR fluorescence due to reduced light scattering, there is virtually no auto-fluorescence background in the near IR region [55,80]. Hence, the IR detection method is highly sensitive and accurate. Second, near IR fluorescence signal is quite stable when protected from light, which makes quantification flexible and reproducible [55]. This is in contrast to the conventional Western blot measurement whereby substrate is used for a time sensitive development of chemiluminescence signals. For quantitative measurement, the near IR fluorescence signal is simply quantified by a near IR fluorescence scanner such as the Odyssey infrared imaging system made by Li-Cor Biosciences (Lincoln, NE).
2.4.4 Overall advantages of fluorescence probes
Regardless of the fluorescent wavelength region (be it visible or infrared), a fluorescence probe offers at least two advantages when used in conjunction with gel-based analysis of carbonylated proteins. First, there are no Western blot experiments to be carried out, so the whole process, from gel running to image documenting, can be completed in a much shorter time. Second, the same 2D (or 1D) gel can be used for both protein staining and protein carbonyl imaging [21,22], which reduces error associated with gel spot identification and excision for subsequent mass spectrometric analysis. This is in contrast to that of DNPH- or biotin-based 2D gel analysis of carbonylated proteins, whereby gels are used for total protein staining and Western blot membranes are used for imaging of the carbonylated proteins [28,29]. Additionally, it is worth noting that with the use of near infrared fluorescence probes/dyes and the finding that Coomassie blue-stained proteins fluoresce in the near infrared region, both protein content and carbonyl content can also be measured on a same membrane blot after gel transfer [55].
3. Potential applications of 2D blue native-PAGE/SDS-PAGE in the analysis of carbonylated proteins
In conventional 2D IEF/SDS-PAGE approach, the use of immobilized pH gradient (IPG) strips in the first IEF dimension has greatly improved the resolution of 2D gel spots. Nevertheless, the inherent limitations of 2D IEF/SDS-PAGE have largely remained, including the difficulty in focusing highly basic proteins and certain membrane proteins that precipitate at the basic end of the IPG strips [81].
The development of BN-PAGE [82], in particular, nongradient BN-PAGE [83], may offer a better approach to 2D gel-based analysis of carbonylated proteins. On a 12% nongradient blue native gel, proteins ranging from 50–1000 kDa, including very basic proteins and highly hydrophobic membrane proteins, can be resolved [83]. Hence, this technique should cover more proteins. Fluorescence-based probes, as well as the biotin-containing probe ARP, can be used because labeling with these probes does not involve removal of the excess probes and the reaction mixtures can be directly loaded onto blue native gels.
When BN-PAGE is performed as the first dimension, a potential problem is that sample preparation requires non-denaturing conditions that may lead to inefficient labeling of those carbonylated amino acid side chains that are not accessible. Indeed, we have found that when protein samples were labeled with ARP in a BN-PAGE sample buffer containing 75 mM aminocaproic acid and 15 mM Bis-Tris (pH 7.0), only a few proteins could be labeled (unpublished data, the authors’ laboratories), indicating a low efficiency in labeling of the oxidized proteins. This problem of inefficient labeling can be solved via in-gel labeling involving incubation of the post-run blue native gel strips (or gel plugs) in a labeling solution supplemented with 1% SDS [84,85], the presence of which would denature proteins and facilitate probe access to the carbonylated sites [84,85]. After incubation for 20–30 min, the gel strips (or gel plugs) can then be directly placed onto a second dimensional SDS-PAGE followed by Western blot detection of carbonylated proteins. Alternatively, as has been reported [86], proteins can be first separated by 2D BN-PAGE/SDS-PAGE, and then derivatized with probes after transfer to immunoblot membranes.
4. Mass spectrometric identification of carbonylated proteins
All the probes described in this review should be applicable to mass spectrometric identification of carbonylated proteins. For gel-based approach, gel bands or spots, whether detected by chemiluminescence, radioactivity, or fluorescence, can be processed for peptide sequencing using liquid chromatography-tandem mass spectrometry (LC-MS/MS) techniques following trypsin digestion and HPLC separation of the tryptic peptides. It should be noted that gel-based approach will only identify protein targets that are putatively carbonylated [29,66] unless peptides bearing carbonylated amino acid residues happen to be sequenced leading to identification of the site of modification. Unfortunately, this is usually not the case for samples that do not receive prior selective enrichment. Therefore, for identification of only the proteins that undergo carbonylation under given experimental conditions, a bottom-up shotgun proteomic approach may be performed [87,88]. This is a gel-free approach, in which protein carbonyls are derivatized with biotin-containing probes, followed by trypsin digestion, affinity capture of the carbonylated/tryptic peptides using streptavidin beads, and mass spectrometric identification of the carbonylated proteins [59,62,63,87]. The use of biotin-containing probes not only facilitates isolation and enrichment of the carbonylated peptides, but also tags the carbonylated sites that can be distinguished by mass spectrometry. If both identification of the carbonylated sites and quantification of the carbonyl content need to be achieved, a technique called stable isotope coded affinity tagging (ICAT) [89,90] may be used. An improved approach of the ICAT technique, isobaric Tag for Relative and Absolute Quantification (iTRAQ), is also available [91,92].
5. Summary
The chemical probes covered in this review are summarized in Table 1, whereby their applications, advantages, and potential disadvantages are also given. Based on our discussions, a general guideline for analyzing protein carbonylation may be formed as the following. (1) For spectrophotometric measurement of protein carbonyls, DNPH is the probe of choice; (2) for gel-based 2D analysis of carbonylated proteins, either biotin-linked probes or fluorescence probes can be used. However, fluorescence probes may be preferred given that a single gel can be used for both protein staining and fluorescence imaging of carbonylated proteins. In particular, near IR fluorescence probes may be preferred for blot-based carbonyl quantification because both protein content and carbonyl content can be measured on a same blot following gel transfer. Finally, for capture and enrichment of carbonylated proteins to be analyzed by either gel-based or gel-free proteomic approaches, biotin-linked probes should be used.
Table I.
Summary of the carbonyl probes discussed in this review
| Probe | Applications and/or advantages | Disadvantages | Refs. |
|---|---|---|---|
| DNPH | Good for colorimetric assay, WB1, and Immunofluorescent imaging |
Labeling efficiency affected by pH Subject to interference by nucleic acid. DNP-derivatives may be unstable |
[48,49] |
| NaB3H4 | Very sensitive May replace DNPH colorimetric assay when needed Good for gel-based assay |
Radioactive | [17,18] |
| Biotin-hydrazide | Good for WB, fluorescent imaging, affinity purification, and shotgun proteomics |
Labeling needs further reduction | [19,59,61] |
| ARP | Same as biotin-hydrazide No further reduction needed |
Possible false positive labeling | [20] |
| FTC, FHA | Good for gel-based assay WB may be obviated |
Unsuitable for fluorometric assay | [21,22] |
| APTB | Good for fluorometric assay No need to remove excess probe |
Not yet commercially available | [74] |
| NIR2 fluorescence probes |
Excellent for blot-based analysis Sensitive, accurate, and stable No background fluorescence |
High-cost equipments needed | [55,80] |
Western blot;
near infrared.
Acknowledgements
The work was supported by the National Institute on Aging, grant number PO1AG022550.
Abbreviations
- APTB
N-aminoperylene-3,4,9,10-tetracarboxylic-bismides
- ARP
aldehyde reactive probe (N’-aminooxymethylcarbonylhydrazino-D-biotin)
- DNPH
2,4-dinitrophenylhydrazine
- ELISA
enzyme-linked immunosorbent assay
- FHA
fluorescence hydroxylamine
- FTC
fluorescein-5-thiosemicarbazide
- HABA
4’hydroxyazobenzene-2-carboxylic acid
- IEF
isoelectric focusing
- IR
infrared
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCA
trichloroacetic acid
- UV
ultraviolet
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sies H, Cadenas E. Philos Trans R Soc Lond B Biol Sci. 1985;311:617. doi: 10.1098/rstb.1985.0168. [DOI] [PubMed] [Google Scholar]
- 2.Stadtman ER. Science. 1992;257:1220. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 3.Shacter E. Methods Enzymol. 2000;319:428. doi: 10.1016/s0076-6879(00)19040-8. [DOI] [PubMed] [Google Scholar]
- 4.Stadtman ER. Ann N Y Acad Sci. 2001;928:22. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 5.Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. Free Radic Biol Med. 2008;45:667. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Wong CM, Cheema AK, Zhang L, Suzuki YJ. Circ Res. 2008;102:310. doi: 10.1161/CIRCRESAHA.107.159814. [DOI] [PubMed] [Google Scholar]
- 7.Wong CM, Marcocci L, Liu L, Suzuki YJ. Antioxid Redox Signal. 2010;12:393. doi: 10.1089/ars.2009.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shacter E. Drug Metab Rev. 2000;32:307. doi: 10.1081/dmr-100102336. [DOI] [PubMed] [Google Scholar]
- 9.Sultana R, Perluigi M, Newman SF, Pierce WM, Cini C, Coccia R, Butterfield DA. Antioxid Redox Signal. 2010;12:327. doi: 10.1089/ars.2009.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stadtman ER. Free Radic Res. 2006;40:1250. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 11.Uchida K, Stadtman ER. Proc Natl Acad Sci U S A. 1992;89:5611. doi: 10.1073/pnas.89.12.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uchida K, Stadtman ER. Proc Natl Acad Sci U S A. 1992;89:4544. doi: 10.1073/pnas.89.10.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szweda LI, Uchida K, Tsai L, Stadtman ER. J Biol Chem. 1993;268:3342. [PubMed] [Google Scholar]
- 14.Morgan PE, Dean RT, Davies MJ. Arch Biochem Biophys. 2002;403:259. doi: 10.1016/s0003-9861(02)00222-9. [DOI] [PubMed] [Google Scholar]
- 15.Picklo MJ, Montine TJ, Amarnath V, Neely MD. Toxicol Appl Pharmacol. 2002;184:187. doi: 10.1006/taap.2002.9506. [DOI] [PubMed] [Google Scholar]
- 16.Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, Ahn BW, Shaltiel S, Stadtman ER. Methods Enzymol. 1990;186:464. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 17.Lenz AG, Costabel U, Shaltiel S, Levine RL. Anal Biochem. 1989;177:419. doi: 10.1016/0003-2697(89)90077-8. [DOI] [PubMed] [Google Scholar]
- 18.Yan LJ, Sohal RS. Anal Biochem. 1998;265:176. doi: 10.1006/abio.1998.2868. [DOI] [PubMed] [Google Scholar]
- 19.Yoo BS, Regnier FE. Electrophoresis. 2004;25:1334. doi: 10.1002/elps.200405890. [DOI] [PubMed] [Google Scholar]
- 20.Chung WG, Miranda CL, Maier CS. Electrophoresis. 2008;29:1317. doi: 10.1002/elps.200700606. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhuri AR, de Waal EM, Pierce A, Van Remmen H, Ward WF, Richardson A. Mech Ageing Dev. 2006;127:849. doi: 10.1016/j.mad.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Poon HF, Abdullah L, Reed J, Doore SM, Laird C, Mathura V, Mullan M, Crawford F. Biol Proced Online. 2007;9:65. doi: 10.1251/bpo134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan LJ. Curr Protoc Protein Sci. 2009;Chapter 14 doi: 10.1002/0471140864.ps1404s55. Unit14 4. [DOI] [PubMed] [Google Scholar]
- 24.Keller RJ, Halmes NC, Hinson JA, Pumford NR. Chem Res Toxicol. 1993;6:430. doi: 10.1021/tx00034a007. [DOI] [PubMed] [Google Scholar]
- 25.Shacter E, Williams JA, Lim M, Levine RL. Free Radic Biol Med. 1994;17:429. doi: 10.1016/0891-5849(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 26.Yan LJ, Levine RL, Sohal RS. Proc. Natl. Acad. Sci. USA. 1997;94:11168. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan LJ, Sohal RS. Proc Natl Acad Sci USA. 1998;95:12896. doi: 10.1073/pnas.95.22.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura A, Goto S. J Biochem. 1996;119:768. doi: 10.1093/oxfordjournals.jbchem.a021306. [DOI] [PubMed] [Google Scholar]
- 29.Choi J, Forster MJ, McDonald SR, Weintraub ST, Carroll CA, Gracy RW. Free Radic Biol Med. 2004;36:1155. doi: 10.1016/j.freeradbiomed.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Boyd-Kimball D, Castegna A, Sultana R, Poon HF, Petroze R, Lynn BC, Klein JB, Butterfield DA. Brain Res. 2005;1044:206. doi: 10.1016/j.brainres.2005.02.086. [DOI] [PubMed] [Google Scholar]
- 31.Mello CF, Sultana R, Piroddi M, Cai J, Pierce WM, Klein JB, Butterfield DA. Neuroscience. 2007;147:674. doi: 10.1016/j.neuroscience.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reverter-Branchat G, Cabiscol E, Tamarit J, Ros J. J Biol Chem. 2004;279:31983. doi: 10.1074/jbc.M404849200. [DOI] [PubMed] [Google Scholar]
- 33.Sohal RS, Weindruch R. Science. 1996;273:59. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebrin I, Forster MJ, Sohal RS. Brain Res. 2007;1127:10. doi: 10.1016/j.brainres.2006.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakravarti B, Chakravarti DN. Gerontology. 2007;53:128. doi: 10.1159/000097865. [DOI] [PubMed] [Google Scholar]
- 36.Hyun DH, Emerson SS, Jo DG, Mattson MP, de Cabo R. Proc Natl Acad Sci U S A. 2006;103:19908. doi: 10.1073/pnas.0608008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barreiro E, Gea J, Di Falco M, Kriazhev L, James S, Hussain SN. Am J Respir Cell Mol Biol. 2005;32:9. doi: 10.1165/rcmb.2004-0021OC. [DOI] [PubMed] [Google Scholar]
- 38.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Science. 2003;299:1751. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 39.Desnues B, Cuny C, Gregori G, Dukan S, Aguilaniu H, Nystrom T. EMBO Rep. 2003;4:400. doi: 10.1038/sj.embor.embor799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernebring M, Brolen G, Aguilaniu H, Semb H, Nystrom T. Proc Natl Acad Sci U S A. 2006;103:7700. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erjavec N, Larsson L, Grantham J, Nystrom T. Genes Dev. 2007;21:2410. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank J, Pompella A, Biesalski HK. Free Radic Biol Med. 2000;29:1096. doi: 10.1016/s0891-5849(00)00395-6. [DOI] [PubMed] [Google Scholar]
- 43.Frank J, Pompella A, Biesalski HK. Methods Mol Biol. 2002;196:35. doi: 10.1385/1-59259-274-0:35. [DOI] [PubMed] [Google Scholar]
- 44.Buss H, Chan TP, Sluis KB, Domigan NM, Winterbourn CC. Free Radic Biol Med. 1997;23:361. doi: 10.1016/s0891-5849(97)00104-4. [DOI] [PubMed] [Google Scholar]
- 45.Winterbourn CC, Buss IH. Methods Enzymol. 1999;300:106. doi: 10.1016/s0076-6879(99)00118-4. [DOI] [PubMed] [Google Scholar]
- 46.Alamdari DH, Kostidou E, Paletas K, Sarigianni M, Konstas AG, Karapiperidou A, Koliakos G. Free Radic Biol Med. 2005;39:1362. doi: 10.1016/j.freeradbiomed.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 47.Hensley K, Abdel-Moaty H, Hunter J, Mhatre M, Mou S, Nguyen K, Potapova T, Pye QN, Qi M, Rice H, Stewart C, Stroukoff K, West M. J Neuroinflammation. 2006;3:2. doi: 10.1186/1742-2094-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo S, Wehr NB. Redox Rep. 2009;14:159. doi: 10.1179/135100009X392601. [DOI] [PubMed] [Google Scholar]
- 49.Wang P, Powell SR. Free Radic Biol Med. 2010;49:119. doi: 10.1016/j.freeradbiomed.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Drug Metab Rev. 2006;38:651. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 51.Reznick AZ, Packer L. Methods Enzymol. 1994;233:357. doi: 10.1016/s0076-6879(94)33041-7. [DOI] [PubMed] [Google Scholar]
- 52.Dalle-Donne I, Carini M, Orioli M, Vistoli G, Regazzoni L, Colombo G, Rossi R, Milzani A, Aldini G. Free Radic Biol Med. 2009;46:1411. doi: 10.1016/j.freeradbiomed.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 53.Pipinos SA, II, Swanson Z, Zhu AA, Nella DJ, Weiss TL, Gutti RD, McComb BT, Baxter TG, Lynch GP. Casale, Am J Physiol Regul Integr Comp Physiol. 2008;295:R290. doi: 10.1152/ajpregu.90374.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson CE, Keshavarzian A, Pasco DS, Frommel TO, Winship DH, Holmes EW. Anal Biochem. 1999;266:48. doi: 10.1006/abio.1998.2932. [DOI] [PubMed] [Google Scholar]
- 55.Luo S, Wehr NB, Levine RL. Anal Biochem. 2006;350:233. doi: 10.1016/j.ab.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 56.Harrigan JA, Piotrowski J, Di Noto L, Levine RL, Bohr VA. J Biol Chem. 2007;282:36403. doi: 10.1074/jbc.M706107200. [DOI] [PubMed] [Google Scholar]
- 57.Gitlin G, Bayer EA, Wilchek M. Biochem J. 1987;242:923. doi: 10.1042/bj2420923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chavez J, Wu J, Han B, Chung WG, Maier CS. Anal Chem. 2006;78:6847. doi: 10.1021/ac0607257. [DOI] [PubMed] [Google Scholar]
- 59.Madian AG, Regnier FE. J Proteome Res. 2010;9:1330. doi: 10.1021/pr900890k. [DOI] [PubMed] [Google Scholar]
- 60.Mirzaei H, Regnier F. J Chromatogr A. 2007;1141:22. doi: 10.1016/j.chroma.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 61.Mirzaei H, Baena B, Barbas C, Regnier F. Proteomics. 2008;8:1516. doi: 10.1002/pmic.200700363. [DOI] [PubMed] [Google Scholar]
- 62.Roe MR, Xie H, Bandhakavi S, Griffin TJ. Anal Chem. 2007;79:3747. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 63.Chavez J, Chung WG, Miranda CL, Singhal M, Stevens JF, Maier CS. Chem Res Toxicol. 2010;23:37. doi: 10.1021/tx9002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maier CS, Chavez J, Wang J, Wu J. Methods Enzymol. 2010;473:305. doi: 10.1016/S0076-6879(10)73016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan LJ, Christians ES, Liu L, Xiao X, Sohal RS, Benjamin IJ. Embo J. 2002;21:5164. doi: 10.1093/emboj/cdf528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prokai L, Yan LJ, Vera-Serrano JL, Stevens SM, Forster MJ. J. Mass Spectrom. 2007;42:1583. doi: 10.1002/jms.1345. [DOI] [PubMed] [Google Scholar]
- 67.Hofstetter H, Morpurgo M, Hofstetter O, Bayer EA, Wilchek M. Anal Biochem. 2000;284:354. doi: 10.1006/abio.2000.4617. [DOI] [PubMed] [Google Scholar]
- 68.Landar A, Oh JY, Giles NM, Isom A, Kirk M, Barnes S, Darley-Usmar VM. Free Radic Biol Med. 2006;40:459. doi: 10.1016/j.freeradbiomed.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 69.Batchelor RH, Sarkez A, Cox WG, Johnson I. Biotechniques. 2007;43:503. doi: 10.2144/000112564. [DOI] [PubMed] [Google Scholar]
- 70.Navarro A, Boveris A. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1244. doi: 10.1152/ajpregu.00226.2004. [DOI] [PubMed] [Google Scholar]
- 71.Laemmli UK. Nature. 1970;227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 72.Jung Y, Chay K, Song D, Yang S, Lee M, Ahn B. Arch Biochem Biophys. 1997;345:311. doi: 10.1006/abbi.1997.0268. [DOI] [PubMed] [Google Scholar]
- 73.Ahn B, Rhee SG, Stadtman ER. Anal Biochem. 1987;161:245. doi: 10.1016/0003-2697(87)90448-9. [DOI] [PubMed] [Google Scholar]
- 74.Langhals H, Jona W. Chem. Eur. J. 1998;4:2110. [Google Scholar]
- 75.Khan MA, Chock PB, Stadtman ER. Proc Natl Acad Sci U S A. 2005;102:17326. doi: 10.1073/pnas.0508120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arai H, Berlett BS, Chock PB, Stadtman ER. Proc Natl Acad Sci U S A. 2005;102:10472. doi: 10.1073/pnas.0504685102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo S, Levine RL. Faseb J. 2009;23:464. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miyoshi N, Oubrahim H, Chock PB, Stadtman ER. Proc Natl Acad Sci U S A. 2006;103:1727. doi: 10.1073/pnas.0510346103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shonsey EM, Eliuk SM, Johnson MS, Barnes S, Falany CN, Darley-Usmar VM, Renfrow MB. J Lipid Res. 2008;49:282. doi: 10.1194/jlr.M700208-JLR200. [DOI] [PubMed] [Google Scholar]
- 80.Calvert VS, Tang Y, Boveria V, Wulfkuhle J, Schutz-Geschwender A, Oliver DM, Liotta LA, Petricoin EF., III Clinical Proteomics Journal. 2004;1:81. [Google Scholar]
- 81.Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, Bogdanova A, Robinson M. Electrophoresis. 2000;21:3427. doi: 10.1002/1522-2683(20001001)21:16<3427::AID-ELPS3427>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 82.Schagger H, von Jagow G. Anal Biochem. 1991;199:223. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 83.Yan LJ, Forster MJ. Anal Biochem. 2009;389:143. doi: 10.1016/j.ab.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choksi KB, Boylston WH, Rabek JP, Widger WR, Papaconstantinou J. Biochim Biophys Acta. 2004;1688:95. doi: 10.1016/j.bbadis.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 85.Wen JJ, Garg N. Free Radic Biol Med. 2004;37:2072. doi: 10.1016/j.freeradbiomed.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 86.Choksi KB, Papaconstantinou J. Free Radic Biol Med. 2008;44:1795. doi: 10.1016/j.freeradbiomed.2008.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soreghan BA, Lu BW, Thomas SN, Duff K, Rakhmatulin EA, Nikolskaya T, Chen T, Yang AJ. J Alzheimers Dis. 2005;8:227. doi: 10.3233/jad-2005-8302. [DOI] [PubMed] [Google Scholar]
- 88.McDonagh B, Ogueta S, Lasarte G, Padilla CA, Barcena JA. J Proteomics. 2009;72:677. doi: 10.1016/j.jprot.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 89.Han B, Stevens JF, Maier CS. Anal Chem. 2007;79:3342. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 90.Madian AG, Regnier FE. J Proteome Res. 2010 [Google Scholar]
- 91.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Proteomics. 2007;7:1150. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 92.Feng J, Xie H, Meany DL, Thompson LV, Arriaga EA, Griffin TJ. J Gerontol A Biol Sci Med Sci. 2008;63:1137. doi: 10.1093/gerona/63.11.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]