Abstract
Theiler’s murine encephalomyelitis virus-induced demyelination (TMEVD) and experimental allergic encephalomyelitis (EAE) are the principal animal models of multiple sclerosis (MS). Previously we provided evidence that Tmevd2 and Eae3 may represent either a shared susceptibility locus or members of a gene complex controlling susceptibility to CNS inflammatory demyelinating disease. To explore the genetic relationship between Tmevd2 and Eae3, we generated a D2.C-Tmevd2 interval-specific congenic (ISC) line and three overlapping interval-specific recombinant congenic (ISRC) lines in which the Tmevd2 resistant allele from BALB/cByJ was introgressed onto the TMEVD-susceptible DBA/2J background. These mice, all H2d, were studied for susceptibility to EAE elicited by immunization with PLP180–199. Compared to DBA/2J mice, D2.C-Tmevd2 mice developed a significantly less severe clinical disease course and EAE pathology in the spinal cord, confirming the existence of Eae3 and its linkage to Tmevd2 in this strain combination. Compare to DBA/2J and D2.C-Tmevd2, all three ISRC lines exhibited clinical disease courses of intermediate severity. Neither differences in ex vivo antigen-specific cytokine nor proliferative responses uniquely cosegregated with differences in disease severity. These results indicate that multiple QTL within the Tmevd2/Eae3 interval influence EAE severity, one of which includes a homology region for a QTL found in MS by admixture mapping.
Keywords: EAE/MS, autoimmunity, neuroimmunology, genetics
INTRODUCTION
Multiple sclerosis (MS), a chronic inflammatory disease of the central nervous system (CNS) characterized by myelin loss, varying degrees of axonal damage, and progressive neurological dysfunction 1, is the most common disabling neurologic disease of young adults and adolescents affecting ˜350,000 individuals in the United States and more than 1 million individuals worldwide 2. The etiopathogenesis of MS is largely unknown; however, it involves both genetic and environmental factors 3–5. Importantly, recent genetic epidemiology studies implicate gene-by-environment interactions in susceptibility and more specifically, epigenetic modifications to germ line susceptibility due to environmental factors acting at the population level 6.
Theiler’s murine encephalomyelitis virus–induced demyelination (TMEVD) 7 and experimental allergic encephalomyelitis (EAE) 8 are the principal genetically controlled viral and autoimmune animal models of MS, respectively. Moreover, the effector molecules mediating demyelination and remyelination are the best characterized in these two models 9. TMEV is a murine picornavirus that is spread in natural and laboratory populations by the fecal/oral route. Following intracerebral inoculation, the virus establishes a persistent infection of CNS white matter in susceptible strains. EAE is elicited by sensitization of genetically susceptible animals with myelin antigens in conjunction with adjuvants. In TMEVD, virus-specific CD4+ T cells, and in EAE, autoreactive CD4+ T cells, initiate the disease by infiltrating the CNS and subsequently recruit additional lymphocytes, leading to inflammation and progressive demyelination. Clinical signs associated with TMEVD become apparent 35–40 days post-inoculation and show a progressive course characterized by gait abnormalities, limb spasms, and incontinence. The onset of EAE is between 10–15 days post-immunization with the clinical signs manifest as an ascending paresis followed by paralysis of the tail and hind limbs, frequently accompanied by fecal and urinary incontinence. Although EAE can occur as a chronic relapsing disease, the inducing antigen, induction protocol, and mouse strain all influence the disease 9.
Genetic control of TMEVD and EAE is polygenic in nature (www.informatics.jax.org). Susceptibility genes and quantitative trait loci (QTL) influencing TMEVD (Tmevd) and viral persistence (Tmevp) reside on Chrs 1, 3, 5, 6, 10, 11, 14, 15 and 17. Similarly, at least 35 loci controlling EAE have been mapped and in some instances, loci controlling susceptibility to TMEVD and EAE co-localize to the same general region. Previously, we presented evidence that Tmevd2 10 and Eae3 on Chr 3 may represent either a single, shared susceptibility gene or members of a gene complex involved in CNS immunopathology 11. However, the evidence supporting this hypothesis was based on comparative studies using data obtained from different crosses and mapping techniques.
In the present study, a D2.C-Tmevd2 interval-specific congenic (ISC) line and three overlapping interval-specific recombinant congenic (ISRC) lines possessing a resistant BALB/cByJ Tmevd2 allele 12 on the TMEVD-susceptible DBA/2J background were studied for susceptibility to EAE elicited by immunization with PLP180–199 13. The results of this study confirm the existence of Eae3 in the same strain combination that was used to map Tmevd2 10; 11; 14 and indicate that multiple QTL in this region control EAE severity; of interest, one of the ISRC lines and the parental ISC line contain a region that is directly homologous to a QTL found in MS by admixture mapping 15.
RESULTS
Multiple QTL within the Tmevd2 interval influence EAE severity
We hypothesized previously that Tmevd2 and Eae3 may represent either a shared locus or members of a gene complex controlling susceptibility to inflammatory diseases of the CNS 11. Because Tmevd2 was identified in genetic studies where DBA/2J was the sole susceptible parental strain 10; 11; 14, we were limited to studying EAE in genetic models using this strain. DBA/2J mice were reported to be resistant to EAE elicited by sensitization with encephalitogen-CFA emulsions prepared by syringe extrusion 16; however, they are susceptible to EAE induced by sensitization using encephalitogen-CFA emulsions prepared by sonication 16; 17 or by inclusion of pertussis toxin (PTX) as an ancillary adjuvant 18. Therefore, we conducted our studies by injecting a sonicated emulsion of PLP180–199-CFA + PTX into D2.C-Tmevd2 mice and three ISRC lines derived from TMEVD susceptible DBA/2J mice bearing the resistant BALB/cByJ Tmevd2 allele. Importantly, the congenic interval in D2.C-Tmevd2 mice encompassed the Eae3 interval (D3Mit64-D3Mit216) (49.9–123.1 Mb) (Table 1).
Table 1. Genotypes of congenic mice across the Tmevd2/Eae3 interval.
Map locations for marker loci are as reported by the Mouse Genome Informatics website (www.informatics.jax.org). Megabase (Mb) coordinates are from UniSTS annotation of NCBI Build 37. Proposed mouse Eae3 QTL are: Eae3.1 (in an interval bounded by D3Mit64 – D3Mit141), Eae3.2 (D3Mit12-D3Mit158), and Eae3.3 (D3Mit249- D3Mit318). The admixture interval in human studies has homology to the region between 97.7 Mb and 98.7 Mb in mice (see text).
| D2.C-Tmevd2/Eae3 | D2.C-Tmevd2/Eae3a | D2.C-Tmevd2/Eae3b | D2.C-Tmevd2/Eae3c | |||
|---|---|---|---|---|---|---|
| Marker | cM | Mb | ||||
| D3Mit 21 | 19.2 | 37.0 | D | D | D | D |
| D3Mit 64 | 23.3 | 49.9 |  |
 |
D | |
| D3Mit 25 | 29.5 | 56.6 | D | |||
| D3Mit 297 | 33.7 | 65.9 | D | |||
| D3Mit 52 | 34.4 | 71.2 | D | |||
| D3Mit 187 | 42.4 | 87.1 | D | |||
| D3Mit 29 | 45.2 | 90.3 | D | |||
| D3Mit 311 | 45.2 | 92.8 | D | |||
| D3Mit141 | 45.2 | 93.0 | D | |||
| rs30202387 | - | 98.17 | D | C | D | |
| rs31694597 | - | 98.64 | D | C | D | |
| D3Mit 12 | 49.2 | 100.2 | D |  |
||
| D3Mit 102 | 49.7 | 101.8 | D | |||
| D3Mit 284 | 51.1 | 103.6 | D | |||
| D3Mit 103 | 51.1 | 107.3 | D | |||
| D3Mit 158 | 52.5 | 109.1 | D | |||
| D3Mit 249 | 55.0 | 116.2 | D | D |  |
|
| D3Mit 216 | 58.8 | 123.1 | D | D | ||
| D3Mit 318 | 58.8 | 123.3 | D | D | ||
| D3Mit 110 | 64.1 | 131.6 | D | D | D | D |
| D3Mit 218 | 66.2 | 136.7 | D | D | D | D |
| D3Mit 127 | 70.3 | 142.8 | D | D | D | D |
Abbreviations: D, TMEVD-susceptible DBA/2J-derived alleles at the various marker loci; C, TMEVD-resistant BALB/cBy-derived alleles at the various marker loci.
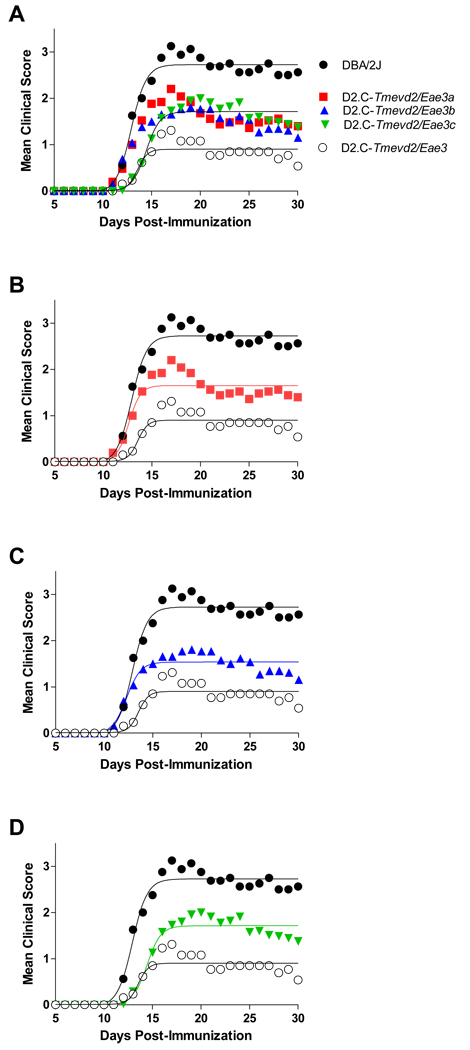
Age matched cohorts of male and female DBA/2J, D2.C-Tmevd2/Eae3, D2.C-Tmevd2/Eae3a, D2.C-Tmevd2/Eae3b, D2.C-Tmevd2/Eae3c mice were injected with PLP180–199-CFA + PTX 13. Given that sex specific QTL have been identified 10; 11; 14 that influence both susceptibility and disease outcome in TMEVD and EAE, we analyzed the data using a two factor design (Table 2). Neither a significant effect of sex nor strain-by-sex interactions were observed for any of the EAE trait variables studied. However, significant effects of strain were seen for incidence (p=0.05), CDS (p<0.0001), DO (p<0.0001), DA (p<0.0001), and PS (p=0.0008). Post-test comparisons for each trait variable revealed that the trait values for D2.C-Tmevd2/Eae3 mice were lower than for the susceptible DBA/2J parental strain, and that the trait values did not differ significantly among the three ISRC lines. Rather, they were all intermediate between DBA/2J and D2.C-Tmevd2/Eae3. Similarly, regression analysis revealed that the course of clinical disease was significantly more severe in DBA/2J mice compared with D2.C-Tmevd2/Eae3, and that the severity of the clinical disease courses seen with all three ISRC lines was significantly different from both DBA/2J and D2.C-Tmevd2/Eae3 but not significantly different from each other (Fig. 1). Thus all three ISRC lines were of intermediate phenotype.
Table 2.
Classical EAE clinical quantitative trait variable in parental and ISRC lines elicited by immunization with PLP180–199+CFA+PTX.
| Strain | Incidence | CDS | DO | DA | PS | SI |
|---|---|---|---|---|---|---|
| DBA/2J ♀A | 7/7 (100) | 29.4±4.6 | 13.7±1.0 | 17.2±1.0 | 2.7±0.4 | 2.2±0.2 |
| DBA/2J ♂ | 9/9 (100) | 32.0±3.7 | 12.8±0.6 | 17.6±0.7 | 2.8±0.2 | 1.9±0.1 |
| D2.C-Tmevd2/Eae3 ♀ | 7/12 (58) | 14.0±6.4 | 18.3±1.6 | 10.4±1.9 | 2.7±0.4 | 2.2±0.3 |
| D2.C-Tmevd2/Eae3 ♂ | 4/7 (57) | 8.2±3.1 | 18.6±1.7 | 10.7±1.8 | 2.7±0.2 | 1.8±0.2 |
| D2.C-Tmevd2/Eae3a ♀ | 11/14 (76) | 30.6±6.3 | 13.4±0.5 | 15.5±1.3 | 3.4±0.3 | 2.3±0.2 |
| D2.C-Tmevd2/Eae3a ♂ | 7/8 (88) | 29.6±5.7 | 14.5±1.4 | 16.5±1.4 | 4.0±0 | 2.3±0.2 |
| D2.C-Tmevd2/Eae3b ♀ | 10/12 (83) | 28.5±4.9 | 13.9±0.7 | 15.2±1.0 | 3.3±0.3 | 2.2±0.2 |
| D2.C-Tmevd2/Eae3b ♂ | 10/13 (77) | 27.5±5.8 | 13.5±0.7 | 15.0±1.8 | 3.6±0.3 | 2.3±0.2 |
| D2.C-Tmevd2/Eae3c ♀ | 20/25 (80) | 32.4±4.4 | 16.6±1.0 | 12.3±1.2 | 3.1±0.3 | 2.4±0.2 |
| D2.C-Tmevd2/Eae3c ♂ | 12/17 (71) | 39.8±5.1 | 15.6±0.9 | 14.6±1.1 | 3.7±0.2 | 2.6±0.2 |
| p-values | ||||||
| Strain | 0.05B | <0.0001C | <0.0001 | <0.0001 | 0.0008 | 0.07 |
| Sex | 1.0 | 0.89 | 0.81 | 0.37 | 0.09 | 0.49 |
| Interaction | 0.35 | 0.76 | 0.85 | 0.88 | 0.74 | 0.53 |
| DBA/2/J > D2.C-Tmevd2/Eae3 (p = 0.004 for incidence) | ||||||
| DBA/2J = D2.C-Tmevd2/Eae3a = D2.C-Tmevd2/Eae3b = D2.C-Tmevd2/Eae3c | ||||||
| D2.C-Tmevd2/Eae3a = D2.C-Tmevd2/Eae3b = D2.C-Tmevd2/Eae3c = D2.C-Tmevd2/Eae3 | ||||||
The significance of the difference in sex ratios from 1:1 was determined using the Fisher’s exact test and was not significantly different for any of the strains.
Significance of observed differences in disease incidence was determined using the Chi-square test.
Signficance of the observed differences in quantitative trait variables was determined by two-way ANOVA followed by Bonferroni post-tests. Means and SEM are shown.
Figure 1. Multiple QTL within the Tmevd2/Eae3 interval influence severity of clinical course of classical EAE elicited by immunization with PLP180–199-CFA plus PTX.
Regression analysis revealed that the disease courses elicited fit a sigmoid curve for all strains. The severity of the clinical disease courses were not significantly different among the three ISRC lines (single regression line for D2.C-Tmevd2/Eae3a, D2.C-Tmevd2/Eae3b, and D2.C-Tmevd2/Eae3c data); however, the clinical disease course among DBA/2J, D2.C-Tmevd2/Eae3 and three ISRC lines was significantly different (F = 28.8; p < 0.0001). Each ISRC line was significantly different from both parental strains: (A) D2.C-Tmevd2/Eae3a (F = 35.2, p < 0.0001), (B) D2.C-Tmevd2/Eae3b (F = 42.9, p < 0.0001) and (C) D2.C-Tmevd2/Eae3c (F = 36.0, p < 0.0001).
On day 30, EAE pathology in the SC and brain was quantified using the previously defined neuropathologic trait variables 19. Two factor analysis testing for the effect of strain and sex did not detect significant sex or sex-by-strain interactions for any of the SC EAE pathology trait variables; however, the effect of strain was significant for total pathology, suppuration, and lesion score (data not shown). EAE pathology in the SC did not differ significantly among the ISRC lines, nor was it significantly different from that seen in D2.C-Tmevd2/Eae3 mice; however, compared to DBA/2J mice, all four congenic lines had significantly less severe EAE pathology in the SC (Fig. 2). The incidence of axonal damage did not significantly differ among the strains studied (D2=23%; D2.C-Tmevd2/Eae3=32%; D2.C-Tmevd2/Eae3a=21%; D2.C-Tmevd2/Eae3b=26%; D2.C-Tmevd2/Eae3c=42%; χ2=4.5, df=4, p=0.3). Brain neuropathology was minimal with no significant differences detected for strain, sex, or strain-by-sex interaction (data not shown). Taken together, the clinical and neuropathology results confirm the existence of Eae3 and its linkage to Tmevd2 in congenic mice bred from this strain combination.
Figure 2. Multiple QTL within the Tmevd2/Eae3 interval influence severity of EAE pathology in the SC elicited by immunization with PLP180–199-CFA plus PTX.
Lesion severity, demyelination, suppuration, mononuclear cell infiltration and total pathology score were evaluated in each animal on day 30 post immunization and scored according to a semiquantitative scale as previously described 51. Lesion severity, suppuration, and total pathology score were significantly different among the strains (*, p<0.05) with DBA/2J > D2.C-Tmevd2/Eae3 = D2.C-Tmevd2/Eae3a = D2.C-Tmevd2/Eae3b = D2.C-Tmevd2/Eae3c (n = 14–46/group). Significance of differences was assessed using the Kruskal-Wallis test followed by Dunn’s multiple comparisons.
Correlation analysis was performed to determine if the CDS and EAE pathology trait variables vary together, and if this correlation could be used to distinguish the sub-congenic lines. When the CDS for all strains was included in the analysis, disease severity was significantly correlated with both brain and SC pathology trait variables (Table 3). However, when DBA/2J and D2.C-Tmevd2/Eae3 mice were analyzed separately, the CDS in DBA/2J mice was not significantly correlated with EAE pathology whereas the CDS in D2.C-Tmevd2/Eae3 mice was significantly correlated with SC lesion severity, mononuclear cell infiltration and total pathology score. These data suggest that the QTL within the Tmevd2/Eae3 interval may influence the severity of clinical disease as a consequence of EAE pathology. The DBA/2J results indicate that additional loci outside of the Tmevd2/Eae3 interval also contribute to differences in severity of clinical signs. Importantly, correlation analysis between the CDS and pathology trait variables among the ISRC lines varied significantly, with the CDS in D2.C-Tmevd2/Eae3a mice being correlated primarily with lesion score, mononuclear cell infiltration and overall pathology score in the brain and only overall pathology score in the SC. In contrast, in D2.C-Tmevd2/Eae3c mice clinical disease severity was more highly correlated with lesion score, axonal damage, demyelination, mononuclear infiltration and total pathology score in the SC. Finally, clinical disease severity in D2.C-Tmevd2/Eae3b mice did not significantly correlate with any of the EAE pathology trait variables.
Table 3.
Correlation of clinical EAE severity with EAE pathology trait variablesA.
| Brain | Spinal cord | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Lesion | Demy | Supp | Mono | PI | Lesion | Axonal Damage |
Demy | Supp | Mono | PI | CDSB |
| All strains | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.002 | 0.0004 | 0.001 | <0.0001 | <0.0001 | 28.8 | ||
| DBA/2J | 47.1 | |||||||||||
| D2.C-Tmevd2/Eae3 | 0.02 | 0.02 | 0.05 | 19.2 | ||||||||
| D2.C-Tmevd2/Eae3a | 0.02 | 0.02 | 0.02 | 0.04 | 29.8 | |||||||
| D2.C-Tmevd2/Eae3b | 28.1 | |||||||||||
| D2.C-Tmevd2/Eae3c | 0.02 | 0.01 | 0.02 | <0.0001 | <0.0001 | 0.002 | <0.0001 | <0.0001 | 27.5 | |||
For each strain Spearman’s nonparametric rank correlation procedure was used to evaluate the relationships between the CDS and each of the pathology quantitative trait variables.
Mean cumulative disease score is shown for each strain or all strains listed in left column.
These results indicate that at least two QTL controlling EAE severity reside within the Tmevd2/Eae3 interval, distributed in three segments (Table 1): Eae3.1 between 49.9 to 93.0 Mb in the D2.C-Tmevd2/Eae3a line; Eae3.2, between 100.2 to 109.1 Mb in the D2.C-Tmevd2/Eae3c and D2.C-Tmevd2/Eae3b lines, and an interval between these at 98.7–100 Mb. A third QTL, Eae3.3, may reside in the interval between 116.2–123.3 Mb, and is present in Tmevd2/Eae3c mice.
Ex vivo PLP180–199 specific immune responses
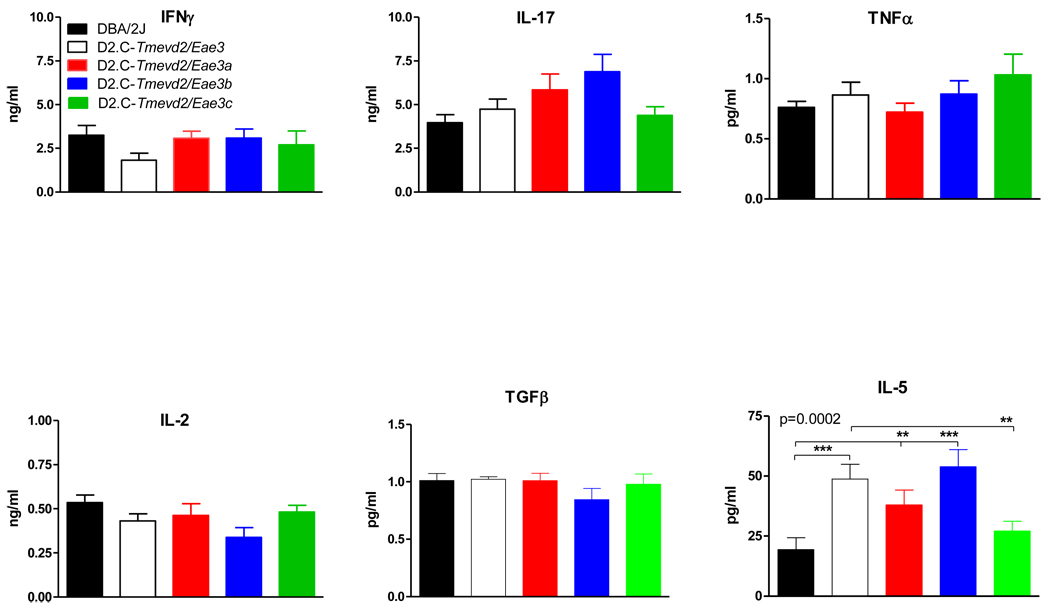
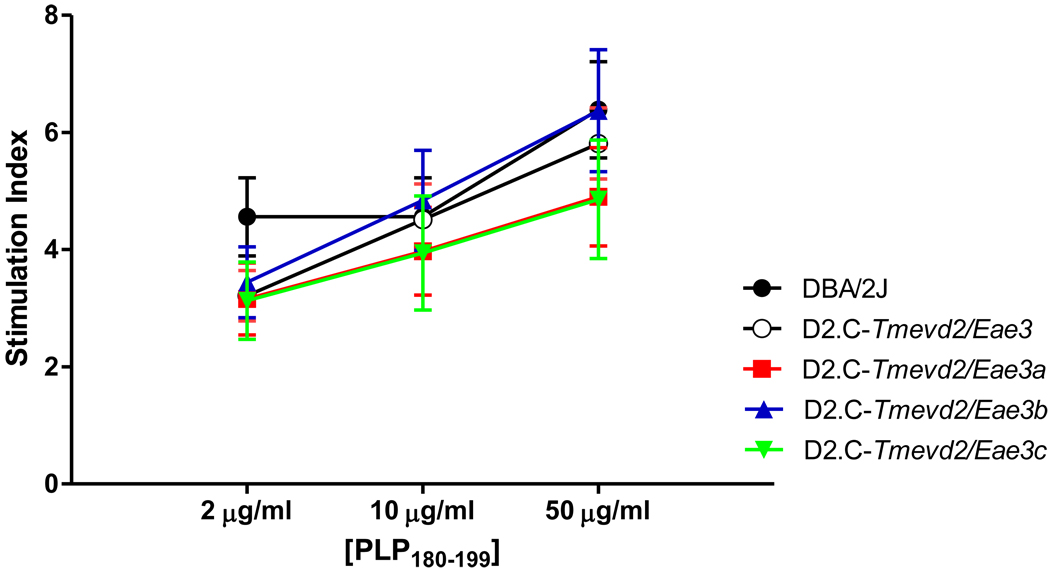
To explore the immune mechanism responsible for the suppressive effects of the EAE QTL within the Tmevd2/Eae3 interval, antigen-specific ex vivo cytokine and proliferative responses were compared among the strains. Of the 25 cytokines studies, IFNγ, IL-17, TNFα, IL-2, TGFβ (Fig. 3), G-CSF, GM-CSF, IL-1α, IL-1β, IL-3, IL-6, IL-10, IL-13, IP-10, KC, KIF, LIX, MCP-1, MIG, MIP-1α, MIP-1β, MIP-2, RANTES, VEGF, and TGFβ did not differ significantly among the strains (data not shown) while IL-4, IL-7, IL-9, IL-12(p40), IL-12(p70), IL-15, IL-23, eotaxin, and M-CSF were below the limits of detection. IL-5 was the only cytokine whose expression differed significantly between DBA/2J and D2.C-Tmevd2/Eae3 mice and segregated among the ISRC lines (Fig. 3). The data place the locus controlling differential IL-5 expression within the interval between D3Mit62 and D3Mit187 shared by the D2.C-Tmevd2/Eae3a and D2.C-Tmevd2/Eae3b lines (Table 1). The proliferative responses also did not differ significantly among the ISRC mice nor were they significantly different from the responses seen in DBA/2J and D2.C-Tmevd2/Eae3 mice (Fig. 4). The similarity in proliferative responses and cytokine production among the lines studied suggest that the QTL influencing EAE severity within the Tmevd2/Eae3 interval may do so by mechanisms other than regulating PLP180–199-specific proliferative responses and classical proinflammatory and/or regulatory cytokine responses.
Figure 3. Ex vivo PLP-specific responses of PLP180–199-CFA plus PTX immunized DBA/2J, D2.C Tmevd2/Eae3 and ISRC lines.
Cytokine, chemokine, and growth factor production were assessed by stimulating splenocytes and lymphocytes from draining lymph nodes on day 10 after immunization with PLP180–199-CFA + PTX. Single cell suspensions were stimulated with 50 µg PLP180–199 for 72 hr and the levels in supernatants were quantified using the Millipore MILLIPLEX™ MAP assay kit or ELISA (n = 10/group). The significance of differences among the strains was determined using a Friedman's test, an ANOVA based on ranked data. Significant differences in the main effect of strain were followed by a priori planned contrasts based on a multilevel congenic mapping strategy (DBA/2J vs. D2.C-Tmevd2/Eae3 followed by D2.C-Tmevd2/Eae3 vs. individual ISRC lines, and confirmed by comparison of individual ISRC lines with DBA/2J). Of the 25 cytokines assayed IL-5 singularly exhibited significant differences in the main effect of strain (p = 0.0002) that was significantly different between DBA/2J and D2.C-Tmevd2/Eae3 mice and segregated among the ISRC lines (**, p < 0.01; ***, p < 0.001).
Figure 4. Ex vivo PLP-specific proliferative responses of PLP180–199-CFA plus PTX immunized DBA/2J, D2.C-Tmevd2/Eae3 and ISRC lines.
Proliferation was assessed on day 10 post immunization by stimulating splenocytes and lymphocytes from draining lymph nodes with and without PLP180–199 for a total of 72 hrs and [3H]-thymidine incorporation measured in the final 18 h. Stimulation indices were calculated as average counts/background counts. The data (n = 8–11/group) were analyzed by two-way ANOVA (comparison: concentration of PLP180–199, p < 0.0001; strain p > 0.05; interaction, p > 0.05).
SNP haplotypes in the Tmevd2/Eae3 interval
Single nucleotide polymorphisms (SNPs) are found in ancestral blocks (or haplotypes) of linked genes. Haplotype association mapping (HAM) can distinguish blocks that are shared by susceptible vs. resistant strains of mice, and can be useful for narrowing the list of possible candidate genes underlying a QTL 20. With very large intervals, such as those in the congenics reported here, many genes could underlie the several QTL detected by clinical scoring. To narrow that list, we compared SNP blocks between TMEVD-susceptible (DBA/2) and TMEVD-resistant (BALB/cByJ) mice, reasoning that Chr 3 haplotype blocks shared between these two strains are unlikely to contain a gene that distinguishes these two strains. HAM is more powerful and accurate if only parental strains that have shown linkage to the same QTL region in previous mapping studies are used in the analysis 21, so we grouped the SNP haplotypes of BALB/cByJ and C57BL/6J vs. DBA/2J. B10.S vs. SJL/J were then added to the analysis, with the reasoning that these strains were used to map Tmevd2 and Eae3 to the same region of Chr 3 10; 11; 14; 22; 23 and were resistant or susceptible strains, respectively. The final HAM analysis thus sought to identify haplotype blocks that were shared by BALB/cByJ, C57BL/6J, and B10.S and were different from those shared by DBA/2J and SJL/J. Approximately 850 genes and genetic entities (defined in the Mouse Genome Database) exist between markers D3Mit64 (49.9 Mb) – D3Mit216 (123.1 Mb) in the original congenic mouse strain. Four hundred and seventeen of these are members of the Mammalian Gene collection of full-length protein-coding genes (http://mgc.nci.nih.gov/). The HAM result using the TMEVD strains (BALB/cByJ and C57BL/6J vs. DBA/2J) narrowed this list of genes bearing polymorphic UTR, intronic or exonic SNPs to 121 genes and unidentified but conserved cDNA transcripts in the entire region from 50–123 Mb of mouse Chr 3; eight microRNAs are also found in this region. Further narrowing of the list by the inclusion of B10.S and SJL/J in the HAM search for polymorphic genes resulted in 96 candidate genes for this large interval containing Tmevd2/Eae3a, Tmevd2/Eae3b and Tmevd2/Eae3c. A total of 47 of the polymorphic genes that reside in Tmevd2/Eae3a passed the HAM test; 91 in Tmevd2/Eae3b (76 when SJL and B10.S are included), and 55 in Tmevd2/Eae3c (30 when SJL and B10.S are included). Of interest, genes in the mouse that are homologues of the human Chr 1 MS admixture locus reside in a Chr 3 region between 97.7 Mb and 98.7 Mb. The original congenic and the ISRC line Tmevd2/Eae3b each contain the admixture homologue. Applying HAM to this region results in 8 polymorphic genes, and requiring DBA/2/J and SJL/J to share SNPs in this region did not reduce this number of candidate genes.
DISCUSSION
Tmevd2 was originally identified as a QTL controlling TMEVD in studies where DBA/2J was the susceptible parental strain 10; 11; 14. Eae3 was identified as a locus controlling EAE severity in genetic studies where SJL/J and NOD/LtJ mice were the susceptible strains 23–25. Both loci were mapped to central Chr 3. We have previously presented evidence that Tmevd2 and Eae3 may represent either a single, shared susceptibility gene or members of a gene complex involved in CNS immunopathology 11. However, since the evidence supporting this hypothesis was based on comparative studies using data from different crosses and mapping techniques, the possibility exists that they reflect unique loci that are unrelated both genetically and functionally. As the first step in assessing the relationship between Tmevd2 and Eae3, we generated an ISC line on the TMEVD susceptible DBA/2J background that has a resistance BALB/cByJ allele at Tmevd2 (D2.C-Tmevd2) and a congenic interval large enough to include Eae3. Additionally, ISRC lines were generated in order to map Eae3 using this strain combination.
When compared, DBA/2J mice had significantly more severe PLP180–199-induced EAE than D2.C-Tmevd2/Eae3 by clinical score, clinical disease traits, and pathology. Overall incidence of classical EAE was significantly greater in DBA/2J mice compared to D2.C-Tmevd2/Eae3 (Table 2). Further, the ISRC lines exhibited intermediate EAE severity and incidence compared to either parental strain; the ISRC lines themselves did not differ significantly from one another. Significant differences among all groups were seen, however, in the correlation or lack of correlation between CNS pathology and clinical severity. In examining these correlations, we noted that DBA/2J mice became highly significantly more affected with EAE signs than any other group, yet had little correlation between their clinical signs and brain or SC pathology. The sub-congenic lines, however, did show this type of correlation (Table 3). These data suggested that the QTL within the Tmevd2/Eae3a interval influence clinical disease and brain pathology whereas the QTL within the Tmevd2/Eae3c interval more likely influence clinical disease severity and SC pathology. Additionally, these results suggested that QTL within the Tmevd2b/Eae3b interval may influence clinical disease severity independent of the severity of EAE pathology. In this regard, cortical motor evoked potentials have been shown to correlate with clinical signs in TMEVD and EAE suggesting a possible role for myelin composition and/or synaptic transmission in susceptibility to clinical symptomatologies associated with inflammatory demyelinating diseases of the CNS 26–28, and QTL controlling these responses in EAE have been identified 28.
PLP180–199-specific proliferation and cytokine responses were not significantly different among the strains on day 10 post-immunization with the exception of IL-5. IL-5 expression differed significantly between DBA/2J and D2.C-Tmevd2/Eae3 and segregated among the ISRC lines (Fig. 3). The most parsimonious location for the QTL controlling differential IL-5 expression is between D3Mit62 and D3Mit187 within an interval shared by D2.C-Tmevd2/Eae3a and D2.C-Tmevd2/Eae3b mice. This finding is potentially interesting since IL-5 is a cytokine produced by a variety of cells including Th2 and mast cells 29 and increased levels could influence disease severity in both D2.C-Tmevd2/Eae3a and D2.C-Tmevd2/Eae3b mice. However, the majority of studies looking at the effects of IL-5 on EAE indicate that IL-5 is not involved in either the initiation or effector phase of the disease 30; 31 or that increased expression of IL-5 by T cells is capable of exacerbating EAE 31; 32. Moreover, increased expression of IL-5 and decreased severity of both clinical disease and EAE pathology did not uniquely segregate with increased disease resistance among the ISRC lines and therefore cannot fully account for the observed reduction in disease severity in all three ISRC lines. Importantly, other significant differences in cytokine responses were not seen between DBA/2J and D2.C-Tmevd2/Eae3 mice. Taken together, these results suggest that multiple QTL within the Tmevd2/Eae3 interval control the severity of EAE and the presence of susceptible DBA/2J alleles at all loci is required for severe EAE. Our study further suggests that the mechanism whereby these QTL influence disease severity is not through the regulation of IFNγ or IL-17 production, two cytokines of critical importance in EAE 33, or cytokines that are important in the generation and maintenance of Treg cells 34. However, tissue harvested at day 10 for in vitro analyses may miss an important window in the pathogenesis of EAE in mice, and because it did not reflect EAE status in the animals, it is not clear that such analyses can be used for defining the mode of action of a QTL.
To gain an appreciation of the role these intervals play in autoimmunity, and to define a suitable list of candidate genes for Tmedv2 and Eae3, it can be helpful to compare them with several other autoimmune disease susceptibility loci that have also been mapped to central Chr 3. An especially interesting QTL for collagen-induced arthritis (Cia5) has been mapped to the Eae3.1 region on Chr 3; it is known to interact with Cia26, 30, 31, and Cia32 within another QTL (Eae2) on Chr 15 35. Eae3 was found to interact with Eae2 25 in EAE models, and thus, the interval bearing Eae3.1 contains genes that modify CIA, EAE, and TMEV-induced demyelination. Of interest, there are two SLE QTL (Sles3 and Sle11) in the Eae3.1 region, and a modifier of T cell ratios (Trmq5). The placement of all four of these QTL is between 82 Mb to 87 Mb on Chr 3. Remarkably, HAM analysis identified only a few known genes that differ between BALB/c and DBA2/J in the 82–88 Mb interval allowing us to further refine the number of potential candidate genes within this region.
Tmevd2/Eae3b is one of several mouse shared QTL that control demyelination after viral infection or deliberate immunization. Both Tmevd6 28 and Tmevd9 14 co-localize with a broad interval containing eae30 on Chr 1. For mapping eae30, Ibrahim and colleagues used the trait of latency in cortical motor evoked potentials (cMEP), as cMEP provides a reproducible assessment of both myelin disturbance and subsequent functional impairment of the axons. The eae30 interval and the Tmevd6 and Tmevd9 regions overlap on proximal Chr 1 and also contain mouse CD28 and CTLA4, two genes important for T cell signaling. Another pair of shared QTL for demyelination include Tmevd5 36 and eae7 37 on distal Chr 11. Tmevd5 controls the severity of clinical disease without altering the viral load 36. Promising candidates for eae7 include Scya1 (TCA-3), Scya2 (monocyte chemoattractant protein (MCP)-1), and Scya12 (MCP-5), all of which are polymorphic between SJL and B10.S, the parental strains used for the linkage mapping of eae7 37.
The Tmevd2/Eae3b interval is of particular interest because part of it is syntenic with an MS locus on Chr 1 identified by admixture mapping 15. The significant genome-wide association of this interval extends from ˜114.9Mb to ˜147 Mb on human Chr 1 15. This human QTL had a LOD score of 4.0 with incidence and contributed to a 1.44 fold increased risk due to heterozygosity with respect to European ancestry. MS risk variants thus might be expected to have a dominant effect. More recent unpublished data suggest that the interval, when narrowed to 119.63 – 143.62 Mb of Build 35 (hg17) of the human genome, has a LOD score of 9.1 (David Reich, personal communication). Alleles of this human QTL are associated with MS susceptibility and are characteristically different between European American and African American populations. There were no interactions between the Chr 1 peak and the main known MHC genetic risk factors, HLA DRBI*1501 and HLA DRBI*1503, and this QTL has not been seen in solely European genome-wide association studies (David Reich, personal communication). While the GWAS result has been confirmed 38–40, no specific locus has yet been identified to be the gene underlying this strong admixture QTL, although several genes in the interval are attractive.
Human genes in the Chr 1 interval from ˜119.63 – 143.62 Mb have 11 homologous orthologues on mouse Chr 3 in Tmevd2/Eae3, from 96.7 Mb to 98.7 Mb. To narrow the list of genes for study, TMEV- or EAE-resistant mice (C57BL/6J, BALB/cByJ, C57BL/10-B10.S) vs. susceptible strains (DBA/2J, and SJL/J), were considered for HAM. This resulted in 9 genes with potentially informative polymorphisms. The list includes several mouse hydroxy-delta-5-steroid dehydrogenase (Hsd3b) gene isoforms, hydroxyacid oxidase 2 (Hao3), and 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (HMGCS2).
One strong human MS candidate gene, Cd58, has no mouse homologue, but has been implicated in MS susceptibility 40; 41, although it is no longer in the human admixture QTL region. CD58 (in humans) and CD48 (in mice and rats), are important for antigen recognition by T cells 42 and CD2, CD48 and CD58 are closely related members of the immunoglobulin superfamily and likely result from a duplication event 43. CD2 is known to bind to CD58 in humans, and in mice it likely binds to CD48 (encoded by a gene on Chr 1) 43; 44. CD2 is located on Chr 3 at 101 Mb, and is a homologue of several genes in the mouse Chr 1 SLAM/CD48 family; it has been proposed that autoimmunity in the NZM2410 mouse may be mediated by a combination of genes in the SLAM/”CD2” regions before the authentic mouse CD2 was cloned and located on Chr 3 45. The CD2 gene is polymorphic between DBA/2J vs. C57Bl/6J and BALB/cByJ, but SJL/J has not been typed for the distinguishing SNPs (rs31487864 and rs31093238). Interestingly, CD2 ligation has been reported to provide a strong helper signal for IL-5 production by T cells 46. CD2 is therefore a homologue of both human CD58 and an autoimmune QTL in mouse SLE, which makes mouse CD2 a very strong candidate gene for autoimmune disease of the CNS. Whether these QTLs represent the same gene or if this co-localization is fortuitous remains an area in need of exploration.
Of the 30 polymorphic genes between resistant BALB/cByJ and susceptible DBA/2J in the Eae3.3 locus, few candidates have been associated with altered MS risk. Potential candidates include Pde5a (122.5 Mb), a cGMP specific phosphodiesterase that alters CD11b+ myeloid-derived suppressor cell function. CD11b+ suppressor myeloid dendritic cells have been suggested to perform important regulatory functions including suppression of CD4+ and CD8+ T cells 47; 48. Importantly, suppressor CD11b+ monocytes are present in the CNS during EAE and these anti-inflammatory CD11b+ Type II monocytes can both prevent and reverse established EAE 49. Two additional candidates of interest are solute carrier family 35 member 3 (Slc35a3) (116.4 Mb) and Slc44a3 (121.2 Mb). Family members of these genes, Pde4B 40 and Slc25a36 38 respectively, have been associated with MS.
The overall conclusion from these studies is that Eae3 is equivalent to Tmevd2 in that susceptibility and resistance to EAE and TMEVD is determined by the same parental alleles at both QTL. Our data also indicate that at least two QTL within the Tmevd2/Eae3 interval influence EAE severity. The continuation of high resolution congenic mapping studies in which susceptibility to EAE and TMEVD are studied in parallel will further refine the EAE QTL candidate intervals and delineate the genetic relationship between Tmevd2 and Eae3a, Eae3b, and Eae3c. Ultimately, the goal of these studies is to positionally clone the gene or genes responsible for the differences in susceptibility to EAE and TMEVD.
MATERIALS and METHODS
Mice
Male and female DBA/2J and BALB/cByJ mice were purchased from The Jackson Laboratory (Bar Harbor, ME). (BALB/cByJ × DBA/2J) F1 hybrids (CD2) mice were bred to DBA/2J mice to produce backcross 1 (BC1 or N2) mice. Mice that were heterozygous across the Tmevd2 (D3Mit64-D3Mit216) interval at BC1 (N2) and each successive BC generation were selected for continued breeding. At N15, heterozygous D2.C-Tmevd2 mice were brother-sister mated and homozygous male and female progeny used to fix a D2.C-Tmevd2 (D3Mit64-D3Mit216) (49.9–123.1 Mb) ISC line 11. D2.C-Tmevd2 ISRC lines were subsequently generated by identifying recombinant haplotypes across the Tmevd2 interval among (D2.C-Tmevd2 × DBA/2J) × DBA/2J BC1 mice and then fixed as inbred lines as described above. The following ISRC lines were generated: D2.C-Tmevd2/Eae3D3Mit64-D3Mit1141 (49.9–93.0 Mb) [D2.C-Tmevd2/Eae3a]; D2.C-Tmevd2/Eae3D3Mit64-D3Mit158 (49.9–109.1 Mb) [D2.C-Tmevd2/Eae3b]; D2.C-Tmevd2/Eae3D3Mit12-D3Mit318 (100.2–123.1 Mb) [D2.C-Tmevd2/Eae3c] (Table 1). All mice were housed at 25°C with 12:12h light-dark cycles and 40–60% humidity. Naïve, age-matched male and female mice were used throughout. The experimental procedures performed in this study were approved by the Animal Care and Use Committee of the University of Vermont.
Genotyping
Genomic DNA was isolated from 3 mm of mouse tail and genotyping was performed by PCR using polymorphic microsatellite markers distinguishing BALB/cByJ and DBA/2J mice 11; 14 as in our previous publications 17. In addition to microsatellite marker analyses, single nucleotide polymorphisms (SNPs) were typed on Chr 3 using the phototyping technique 50. Two alternate allele-specific SNP-primers and one common opposite-strand primer were designed for each SNP using Primer3 to have a Tmelt of 58–60° C, where the allele-specific primer has a 3’ nucleotide that matches one of the two SNP alleles. SNP haplotypes were assembled in an Excel database and used to delineate the breakpoints of the recombinant Chr 3 in the three ISRC lines. Only SNPs that distinguish the two parental strains, DBA/2J and BALB/cByJ, are shown in this report.
Induction and Evaluation of EAE
Because DBA/2J mice possess the H2d haplotype, the H2d class II restricted proteolipid protein peptide 180–199 (PLP180–199) was used as the encephalitogen in this study 13. EAE was elicited by subcutaneously injecting a sonicated emulsion of 100 µg PLP180–199 and 200 µg Mycobacterium tuberculosis H37Ra in CFA (Difco Laboratories, Detroit, MI, USA) in 0.2 ml on the posterior right and left flank and the scruff of the neck 17; 51. Each mouse received 200 ng PTX (List Biological Laboratories, Campbell, CA, USA) in 0.2 ml by intravenous injection immediately after and again on day 2 13. EAE was evaluated daily beginning at day 10 through day 30 as follows: 0, no clinical expression of disease; 1, flaccid tail without hind limb weakness; 2, hind limb weakness; 3, complete hind limb paralysis and floppy tail; 4, hind leg paralysis accompanied by a floppy tail and urinary or fecal incontinence; 5, moribund. Clinical quantitative trait variables were generated as previously described 22; 52. Mice were considered positive for incidence if they showed any clinical signs ≥1 for two or more days. The severity index is the cumulative disease score per days affected. All animals that died or were euthanized prior to day 30 were excluded from the analysis which is based on classical EAE clinical and pathologic quantitative trait variables 22; 52–54. The significance of differences in the severity of the clinical disease courses was determined by regression analysis 51; 55.
Cell culture conditions and lymphokine assays
For ex vivo cytokine and chemokine analysis, spleens and draining lymph nodes were obtained from mice immunized 10 days earlier for EAE. Single cell suspensions at 1 × 106 cells/ml in RPMI 1640 media (Cellgro Mediatech, Inc., Manassas, VA, USA) plus 5% FBS (HyClone, Logan, UT, USA) were stimulated with 50 µg PLP180–199. Cell culture supernatants were recovered at 72 hours and tested for cytokine levels by ELISA as previously described 55 using primary mAbs specific for IFNγ, IL-2, IL-4, and IL-17 and their corresponding biotinylated secondary mAbs (BD Biosciences Pharmingen, San Diego, CA, USA). Other ELISA reagents included horseradish peroxidase-conjugated avidin D (Vector Laboratories, Burlingame, CA, USA), TMB microwell peroxidase substrate and stop solutions (Kirkegaard and Perry Laboratories, Gaithersburg, MD, USA) and recombinant IFNγ, IL-2, IL-4, and IL-17 (R&D Systems, Minneapolis, MN, USA) were used as standards. TNFα was measured using BD OptEIA mouse TNF ELISA set (BD Biosciences). IL-23 (p19/p40) and TGFβ were measured using ELISA Ready-SET-Go! Kits (eBioscience, San Diego, CA, USA). Additionally, eotaxin, G-CSF, GM-CSF, IL-1α, IL-1β, IL-3, IL-5, IL-6, IL-7, IL-9, IL-12(p40), IL-12(p70), IL-13, IL-15, IP-10, KC, KIF, LIX, MCP-1, M-CSF, MIG, MIP-1α, MIP-1β, MIP-2, RANTES, and VEGF levels were quantified using the Millipore MILLIPLEX™ MAP assay kit (#MPXMCYTO-10K; Millipore, Billerica, MA, USA) and run on the Bio-Rad Bio-Plex instrument (Bio-Rad, Hercules, CA, USA) 51. For ex vivo proliferation, single cell suspensions were prepared and 5 × 105 cells/well in RPMI 1640 media (5% FBS) were plated on standard 96-well U-bottom tissue culture plates and stimulated with 0, 2, 10 or 50 µg PLP180–199 for 72 h at 37°C. During the last 18 h of culture, 1µCi of [3H]-thymidine (PerkinElmer, Stelton, CT, USA) was added. Cells were harvested onto glass fiber filters and thymidine uptake was determined by liquid scintillation.
Histology
Histological assessment of EAE neuropathology was done as previously described 54. Briefly, brains and spinal cords (SC) were dissected from calvarias and vertebral columns, respectively, and fixed by immersion in phosphate buffered (pH 7.2) 10% formalin. Representative areas of the brain and SC, including brainstem, cerebrum, cerebellum, and the cervical, thoracic, and lumbar segments of the SC, were selected for histopathological evaluation. EAE pathology reflects the overall severity of the lesions observed, extent and degree of myelin loss and tissue injury (swollen axon sheathes, swollen axons, and reactive gliosis), severity of the acute inflammatory response (predominantly neutrophils), and the severity of the chronic inflammatory response (lymphocytes/macrophages). The presence of axonal damage was noted, although not graded, if groups of axonal spheroids were seen in areas of inflammation on H&E stained sections.
Haplotype association mapping (HAM)
Single nucleotide polymorphisms (SNPs) reflecting haplotypes of linked genes were compared for BALB/cByJ, and C57BL/6J vs. DBA/2J, with the reasoning that these strains were used in crosses that mapped Tmevd2 and were resistant or susceptible, respectively. The HAM list was further limited by polymorphisms in C57BL/10 (the parental background strain of EAE-resistant B10.S-H2s/SgMcdJ (B10.S) mice), and SJL/J, representing polymorphic alleles in Eae3 in the same region 21. Polymorphic genes and gene elements were defined by the detection of SNPs that are categorized as introns, exons or untranslated regions (UTR) by the Mouse Phenome Database (www.jax.org/phenome).
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 5 (GraphPad Software, San Diego, CA, USA). The specific tests used are detailed in the Figure legends and Table footnotes. A p-value of ≤0.05 was considered significant.
Table 4.
Selected QTL and genes within Tmevd2/Eae3 intervalA.
| Mb | Gene | QTL | D2.C- Tmevd2/Eae3 |
D2.C- Tmevd2/Eae3a |
D2.C- Tmevd2/Eae3b |
D2.C- Tmevd2/Eae3c |
|
|---|---|---|---|---|---|---|---|
| Eae3.1 | 49.9 | C | C | C | D | ||
| Region | ˜54 | Trpc4 | C | C | C | D | |
| ˜55 | Nbea | C | C | C | D | ||
| ˜69 | Ppm1l | C | C | C | D | ||
| 78.6 | Sles3 | C | C | C | D | ||
| 82.9 | Cia5 | C | C | C | D | ||
| 87.06 | Trmq5 | C | C | C | D | ||
| 87.1 | Sle11 | C | C | C | D | ||
| Human | 97.7–98.7 | C | D | C | D | ||
| Admixture | 98.2 | Zfp697 | C | D | C | D | |
| Orthologous | 98.37- 98.67 |
Hsdb1,6 | C | D | C | D | |
| Region | 98.68 | Hao3 | C | D | C | D | |
| Eae3.2 | 99–100 | Idd10 | C | D | C | C | |
| Region | 100.5 | Cia21 | C | D | C | C | |
| 100.7 | Vtcn1 | C | D | C | C | ||
| 100.7 | Trim45 | C | D | C | C | ||
| 101 | Cd2 | C | D | C | C | ||
| 101.2 | Igsf3 | C | D | C | C | ||
| 107.2 | Cia22 | C | D | C | C | ||
| 107.2 | Trmq6 | C | D | C | C | ||
| 109.1 | C | D | C | C | |||
| Eae3.3 | 116.4 | Slc35a3 | C | D | D | C | |
| Region | 121.2 | Slc44a3 | C | D | D | C | |
| 122.5 | Pde5a | C | D | D | C | ||
| 123.1 | C | D | D | C | |||
| 131.6 | D | D | D | D | |||
Map locations for genes, QTL and marker loci are as reported by the Mouse Genome Informatics website (www.informatics.jax.org) or as previously cited. Megabase (Mb) coordinates are from UniSTS annotation of NCBI Build 37.
Abbreviations: D, TMEVD-susceptible DBA/2J-derived alleles at the various marker loci; C, TMEVD-resistant BALB/cByJ-derived alleles at the various marker loci.
ACKNOWLEDGEMENTS
We wish to thank Laura Cort and Meena Subramanian for technical support. The authors have no conflicting financial interests. This work was supported by the National Institutes of Health grants NS36526 (CT), AI41747 (CT), AI058052 (CT), NS061014 (CT), NS060901 (CT), and AI45666 (CT) and by National Multiple Sclerosis Society grant RG3575 (EPB) and RG3575A5/1 (EPB).
Abbreviations
- Chr
chromosome
- CDS
cumulative disease score
- EAE
experimental allergic encephalomyelitis
- ISC
interval specific congenic
- ISRC
interval specific recombinant congenic
- MS
multiple sclerosis
- PLP180–199
proteolipid protein peptide 180–199
- PTX
pertussis toxin
- QTL
quantitative trait locus
- TMEVD
Theiler’s murine encephalomyelitis virus-induced demyelination
Footnotes
The authors declare no conflict of interest.
REFERENCES
- 1.Noseworthy JH, Lucchinetti C, Rodriguez M, Weinshenker BG. Multiple sclerosis. N Engl J Med. 2000;343(13):938–952. doi: 10.1056/NEJM200009283431307. [DOI] [PubMed] [Google Scholar]
- 2.Courtney AM, Treadaway K, Remington G, Frohman E. Multiple sclerosis. Med Clin North Am. 2009;93(2):451–476. doi: 10.1016/j.mcna.2008.09.014. ix–x. [DOI] [PubMed] [Google Scholar]
- 3.Dyment DA, Ebers GC, Sadovnick AD. Genetics of multiple sclerosis. Lancet Neurol. 2004;3(2):104–110. doi: 10.1016/s1474-4422(03)00663-x. [DOI] [PubMed] [Google Scholar]
- 4.Ramagopalan SV, Dyment DA, Ebers GC. Genetic epidemiology: the use of old and new tools for multiple sclerosis. Trends Neurosci. 2008;31(12):645–652. doi: 10.1016/j.tins.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Ebers GC. Environmental factors and multiple sclerosis. Lancet Neurol. 2008;7(3):268–277. doi: 10.1016/S1474-4422(08)70042-5. [DOI] [PubMed] [Google Scholar]
- 6.Lincoln MR, Ramagopalan SV, Chao MJ, Herrera BM, Deluca GC, Orton SM, et al. Epistasis among HLA-DRB1, HLA-DQA1, and HLA-DQB1 loci determines multiple sclerosis susceptibility. Proc Natl Acad Sci U S A. 2009;106(18):7542–7547. doi: 10.1073/pnas.0812664106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipton HL, Kumar AS, Trottier M. Theiler's virus persistence in the central nervous system of mice is associated with continuous viral replication and a difference in outcome of infection of infiltrating macrophages versus oligodendrocytes. Virus Res. 2005;111(2):214–223. doi: 10.1016/j.virusres.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Stromnes IM, Goverman JM. Active induction of experimental allergic encephalomyelitis. Nat Protoc. 2006;1(4):1810–1819. doi: 10.1038/nprot.2006.285. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez M. Effectors of demyelination and remyelination in the CNS: implications for multiple sclerosis. Brain Pathol. 2007;17(2):219–229. doi: 10.1111/j.1750-3639.2007.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melvold RW, Jokinen DM, Miller SD, Dal Canto MC, Lipton HL. Identification of a locus on mouse chromosome 3 involved in differential susceptibility to Theiler's murine encephalomyelitis virus-induced demyelinating disease. J Virol. 1990;64(2):686–690. doi: 10.1128/jvi.64.2.686-690.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teuscher C, Rhein DM, Livingstone KD, Paynter RA, Doerge RW, Nicholson SM, et al. Evidence that Tmevd2 and eae3 may represent either a common locus or members of a gene complex controlling susceptibility to immunologically mediated demyelination in mice. J Immunol. 1997;159(10):4930–4934. [PubMed] [Google Scholar]
- 12.Nicholson SM, Peterson JD, Miller SD, Wang K, Dal Canto MC, Melvold RW. BALB/c substrain differences in susceptibility to Theiler's murine encephalomyelitis virus-induced demyelinating disease. J Neuroimmunol. 1994;52(1):19–24. doi: 10.1016/0165-5728(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 13.Lyons JA, Ramsbottom MJ, Trotter JL, Cross AH. Identification of the encephalitogenic epitopes of CNS proteolipid protein in BALB/c mice. J Autoimmun. 2002;19(4):195–201. doi: 10.1006/jaut.2002.0619. [DOI] [PubMed] [Google Scholar]
- 14.Butterfield RJ, Roper RJ, Rhein DM, Melvold RW, Haynes L, Ma RZ, et al. Sex-specific quantitative trait loci govern susceptibility to Theiler's murine encephalomyelitis virus-induced demyelination. Genetics. 2003;163(3):1041–1046. doi: 10.1093/genetics/163.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;(37):10. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 16.Maatta JA, Nygardas PT, Hinkkanen AE. Enhancement of experimental autoimmune encephalomyelitis severity by ultrasound emulsification of antigen/adjuvant in distinct strains of mice. Scand J Immunol. 2000;51(1):87–90. doi: 10.1046/j.1365-3083.2000.00686.x. [DOI] [PubMed] [Google Scholar]
- 17.Fillmore PD, Brace M, Troutman SA, Blankenhorn EP, Diehl S, Rincon M, et al. Genetic analysis of the influence of neuroantigen-complete Freund's adjuvant emulsion structures on the sexual dimorphism and susceptibility to experimental allergic encephalomyelitis. Am J Pathol. 2003;163(4):1623–1632. doi: 10.1016/S0002-9440(10)63519-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz JJ, Mackay IR. Production of experimental allergic encephalomyelitis with the aid of pertussigen in mouse strains considered genetically resistant. J Neuroimmunol. 1984;7(2–3):91–96. doi: 10.1016/s0165-5728(84)80009-0. [DOI] [PubMed] [Google Scholar]
- 19.Blankenhorn EP, Butterfield RJ, Rigby R, Cort L, Giambrone D, McDermott P, et al. Genetic analysis of the influence of pertussis toxin on experimental allergic encephalomyelitis susceptibility: an environmental agent can override genetic checkpoints. J Immunol. 2000;164(6):3420–3425. doi: 10.4049/jimmunol.164.6.3420. [DOI] [PubMed] [Google Scholar]
- 20.Burgess-Herbert SL, Cox A, Tsaih SW, Paigen B. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics. 2008;180(4):2227–2235. doi: 10.1534/genetics.108.090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manenti G, Galvan A, Pettinicchio A, Trincucci G, Spada E, Zolin A, et al. Mouse genome-wide association mapping needs linkage analysis to avoid false-positive Loci. PLoS Genet. 2009;5(1) doi: 10.1371/journal.pgen.1000331. e1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butterfield RJ, Sudweeks JD, Blankenhorn EP, Korngold R, Marini JC, Todd JA, et al. New genetic loci that control susceptibility and symptoms of experimental allergic encephalomyelitis in inbred mice. J Immunol. 1998;161(4):1860–1867. [PubMed] [Google Scholar]
- 23.Encinas JA, Lees MB, Sobel RA, Symonowicz C, Greer JM, Shovlin CL, et al. Genetic analysis of susceptibility to experimental autoimmune encephalomyelitis in a cross between SJL/J and B10.S mice. J Immunol. 1996;157(5):2186–2192. [PubMed] [Google Scholar]
- 24.Encinas JA, Weiner HL, Kuchroo VK. Inheritance of susceptibility to experimental autoimmune encephalomyelitis. J Neurosci Res. 1996;45(6):655–669. doi: 10.1002/(SICI)1097-4547(19960915)45:6<655::AID-JNR2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 25.Sundvall M, Jirholt J, Yang HT, Jansson L, Engstrom A, Pettersson U, et al. Identification of murine loci associated with susceptibility to chronic experimental autoimmune encephalomyelitis. Nat Genet. 1995;10(3):313–317. doi: 10.1038/ng0795-313. [DOI] [PubMed] [Google Scholar]
- 26.Iuliano BA, Schmeizer JD, Thiemann RL, Low PA, Rodriguez M. Motor and somatosensory evoked potentials in mice infected with Theiler's murine encephalomyelitis virus. J Neurol Sci. 1994;123(1–2):186–194. doi: 10.1016/0022-510x(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 27.McGavern DB, Murray PD, Rivera-Quinones C, Schmelzer JD, Low PA, Rodriguez M. Axonal loss results in spinal cord atrophy, electrophysiological abnormalities and neurological deficits following demyelination in a chronic inflammatory model of multiple sclerosis. Brain Pathol. 2000;123(3):519–531. doi: 10.1093/brain/123.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazon Pelaez I, Vogler S, Strauss U, Wernhoff P, Pahnke J, Brockmann G, et al. Identification of quantitative trait loci controlling cortical motor evoked potentials in experimental autoimmune encephalomyelitis: correlation with incidence, onset and severity of disease. Hum Mol Genet. 2005;14(14):1977–1989. doi: 10.1093/hmg/ddi203. [DOI] [PubMed] [Google Scholar]
- 29.Takatsu K, Kouro T, Nagai Y. Interleukin 5 in the link between the innate and acquired immune response. Adv Immunol. 2009;101:191–236. doi: 10.1016/S0065-2776(08)01006-7. [DOI] [PubMed] [Google Scholar]
- 30.Weir C, Bernard CC, Backstrom BT. IL-5-deficient mice are susceptible to experimental autoimmune encephalomyelitis. Int Immunol. 2003;15(11):1283–1289. doi: 10.1093/intimm/dxg127. [DOI] [PubMed] [Google Scholar]
- 31.Wensky A, Marcondes MC, Lafaille JJ. The role of IFN-gamma in the production of Th2 subpopulations: implications for variable Th2-mediated pathologies in autoimmunity. J Immunol. 2001;167(6):3074–3081. doi: 10.4049/jimmunol.167.6.3074. [DOI] [PubMed] [Google Scholar]
- 32.Bell JJ, Divekar RD, Ellis JS, Cascio JA, Haymaker CL, Jain R, et al. In trans T cell tolerance diminishes autoantibody responses and exacerbates experimental allergic encephalomyelitis. J Immunol. 2008;180(3):1508–1516. doi: 10.4049/jimmunol.180.3.1508. [DOI] [PubMed] [Google Scholar]
- 33.Dardalhon V, Korn T, Kuchroo VK, Anderson AC. Role of Th1 and Th17 cells in organ-specific autoimmunity. J Autoimmun. 2008;31(3):252–256. doi: 10.1016/j.jaut.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoyer KK, Dooms H, Barron L, Abbas AK. Interleukin-2 in the development and control of inflammatory disease. Immunol Rev. 2008;226:19–28. doi: 10.1111/j.1600-065X.2008.00697.x. [DOI] [PubMed] [Google Scholar]
- 35.Karlsson J, Johannesson M, Lindvall T, Wernhoff P, Holmdahl R, Andersson A. Genetic interactions in Eae2 control collagen-induced arthritis and the CD4+/CD8+ T cell ratio. J Immunol. 2005;174(1):533–541. doi: 10.4049/jimmunol.174.1.533. [DOI] [PubMed] [Google Scholar]
- 36.Aubagnac S, Brahic M, Bureau JF. Viral load and a locus on chromosome 11 affect the late clinical disease caused by Theiler's virus. J Virol. 1999;73(10):7965–7971. doi: 10.1128/jvi.73.10.7965-7971.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teuscher C, Butterfield RJ, Ma RZ, Zachary JF, Doerge RW, Blankenhorn EP. Sequence polymorphisms in the chemokines Scya1 (TCA-3), Scya2 (monocyte chemoattractant protein (MCP)-1), and Scya12 (MCP-5) are candidates for eae7, a locus controlling susceptibility to monophasic remitting/nonrelapsing experimental allergic encephalomyelitis. J Immunol. 1999;163(4):2262–2266. [PubMed] [Google Scholar]
- 38.Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;18(11):2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39(11):1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 41.Rubio JP, Stankovich J, Field J, Tubridy N, Marriott M, Chapman C, et al. Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun. 2008;9(7):624–630. doi: 10.1038/gene.2008.59. [DOI] [PubMed] [Google Scholar]
- 42.Kato K, Koyanagi M, Okada H, Takanashi T, Wong YW, Williams AF, et al. CD48 is a counter-receptor for mouse CD2 and is involved in T cell activation. J Exp Med. 1992;176(5):1241–1249. doi: 10.1084/jem.176.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong YW, Williams AF, Kingsmore SF, Seldin MF. Structure, expression, and genetic linkage of the mouse BCM1 (OX45 or Blast-1) antigen. Evidence for genetic duplication giving rise to the BCM1 region on mouse chromosome 1 and the CD2/LFA3 region on mouse chromosome 3. J Exp Med. 1990;171(6):2115–2130. doi: 10.1084/jem.171.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selvaraj P, Plunkett ML, Dustin M, Sanders ME, Shaw S, Springer TA. The T lymphocyte glycoprotein CD2 binds the cell surface ligand LFA-3. Nature. 1987;326(6111):400–403. doi: 10.1038/326400a0. [DOI] [PubMed] [Google Scholar]
- 45.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, et al. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21(6):769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Peng X, Kasran A, Bullens D, Ceuppens JL. Ligation of CD2 provides a strong helper signal for the production of the type 2 cytokines interleukin-4 and -5 by memory T cells. Cell Immunol. 1997;181(1):76–85. doi: 10.1006/cimm.1997.1197. [DOI] [PubMed] [Google Scholar]
- 47.Gabrilovich D. Mechanisms and functional significance of tumour-induced dendritic-cell defects. Nat Rev Immunol. 2004;4(12):941–952. doi: 10.1038/nri1498. [DOI] [PubMed] [Google Scholar]
- 48.Serafini P, Meckel K, Kelso M, Noonan K, Califano J, Koch W, et al. Phosphodiesterase-5 inhibition augments endogenous antitumor immunity by reducing myeloid-derived suppressor cell function. J Exp Med. 2006;203(12):2691–2702. doi: 10.1084/jem.20061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber MS, Prod'homme T, Youssef S, Dunn SE, Rundle CD, Lee L, et al. Type II monocytes modulate T cell-mediated central nervous system autoimmune disease. Nat Med. 2007;13(8):935–943. doi: 10.1038/nm1620. [DOI] [PubMed] [Google Scholar]
- 50.Bunce M, O'Neil CM, Barnardo MC, Krausa P, Browning MJ, Morris PJ, et al. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP) Tissue Antigens. 1995;46(5):355–367. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 51.Teuscher C, Noubade R, Spach K, McElvany B, Bunn JY, Fillmore PD, et al. Evidence that the Y chromosome influences autoimmune disease in male and female mice. Proc Natl Acad Sci U S A. 2006;103(21):8024–8029. doi: 10.1073/pnas.0600536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butterfield RJ, Blankenhorn EP, Roper RJ, Zachary JF, Doerge RW, Sudweeks J, et al. Genetic analysis of disease subtypes and sexual dimorphisms in mouse experimental allergic encephalomyelitis (EAE): relapsing/remitting and monophasic remitting/nonrelapsing EAE are immunogenetically distinct. J Immunol. 1999;162(5):3096–3102. [PubMed] [Google Scholar]
- 53.Muller DM, Pender MP, Greer JM. A neuropathological analysis of experimental autoimmune encephalomyelitis with predominant brain stem and cerebellar involvement and differences between active and passive induction. Acta Neuropathol. 2000;100(2):174–182. doi: 10.1007/s004019900163. [DOI] [PubMed] [Google Scholar]
- 54.Butterfield RJ, Blankenhorn EP, Roper RJ, Zachary JF, Doerge RW, Teuscher C. Identification of genetic loci controlling the characteristics and severity of brain and spinal cord lesions in experimental allergic encephalomyelitis. Am J Pathol. 2000;157(2):637–645. doi: 10.1016/S0002-9440(10)64574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noubade R, Milligan G, Zachary JF, Blankenhorn EP, del Rio R, Rincon M, et al. Histamine receptor H1 is required for TCR-mediated p38 MAPK activation and optimal IFN-gamma production in mice. J Clin Invest. 2007;117(11):3507–3518. doi: 10.1172/JCI32792. [DOI] [PMC free article] [PubMed] [Google Scholar]