Abstract
Membrane protein structure and function can be studied by two powerful and highly complementary electron cryomicroscopy (cryo-EM) methods: electron crystallography of two-dimensional (2D) crystals and single particle analysis of detergent-solubilized protein complexes. To obtain the highest-possible resolution data from membrane proteins, whether prepared as 2D crystals or single particles, cryo-EM samples must be vitrified with great care. Grid preparation for cryo-EM of 2D crystals is possible by back-injection, the carbon sandwich technique, drying in sugars before cooling in the electron microscope, or plunge-freezing. Specimen grids for single particle cryo-EM studies of membrane proteins are usually produced by plunge-freezing protein solutions, supported either by perforated or a continuous carbon film substrate. This review outlines the different techniques available and the suitability of each method for particular samples and studies. Experimental considerations in sample preparation and preservation include the protein itself and the presence of lipid or detergent. The appearance of cryo-EM samples in different conditions is also discussed.
Keywords: Cryo-EM, electron cryomicroscopy, membrane protein, electron crystallography, single particle analysis, grid preparation
1. Introduction
Membrane proteins currently pose one of the biggest challenges in structural biology as evidenced by the small number of atomic models of membrane proteins in the Protein Data Bank (PDB; www.pdb.org). While X-ray crystallography of three-dimensional (3D) crystals has contributed the majority of the atomic-resolution membrane protein structures available, a large part of the difficulty in structural studies can be attributed to producing quantities of protein sufficient to allow for the substitution of the phospholipid bilayer with detergent and 3D crystallization of the resulting complex system.
In some cases, electron cryomicroscopy (cryo-EM) provides an alternative to X-ray crystallography, while in other cases it serves as a complementary method of structural analysis. Cryo-EM not only allows for the structural study of membrane proteins that can be obtained only in small quantities or that are difficult to crystallize in 3D (Hankamer et al., 2007; Hite et al., 2007; Fujiyoshi and Unwin, 2008; Taylor and Glaeser, 2008; Glaeser, 2008a, 2008b; Müller et al., 2008; Reichow and Gonen, 2009), but combined with X-ray crystallography of 3D crystals, cryo-EM can also contribute to an improved understanding of function (Appel et al., 2009) as well as information on larger protein complex formation (Abrahams et al., 1994; Stock et al., 1999; Gibbons et al., 2000; Vonck et al., 2002; Rubinstein et al., 2003; Meier et al., 2005). Taylor and Glaeser (1974) initially demonstrated the power of this method. In 1975, Henderson and Unwin determined the first structure of a membrane protein to 7 Å resolution, an effort that culminated in the determination of the atomic structure of bacteriorhodopsin (Henderson et al., 1990). Most recently, Gonen et al. (2005) obtained the highest resolution cryo-EM map to date by solving the structure of aquaporin-0 at 1.9 Å resolution.
Cryo-EM data of membrane proteins can be obtained from electron crystallography of two-dimensional (2D) crystals or single particle analysis of detergent-solubilized protein complexes. While methods to collect cryo-EM data are well established, specimen grid preparation is a crucial step in obtaining high-resolution data and the details of specimen preparation procedures can vary significantly from laboratory to laboratory. This concise review focuses on the most critical factors in grid preparation and sample preservation for freezing samples of both membrane protein 2D crystals and single particles.
Here we describe the key points in cryo-EM grid preparation both to best preserve the membrane protein structure and to obtain the highest possible resolution data of 2D crystals and single particles. 2D crystals do not usually contain detergents. However, detergents can play a significant role in the appearance of images in single particle EM experiments and the properties of detergents in this context are discussed. Methods and protocols are outlined for optimization of grid preparation of 2D crystals with a variety of different techniques.
2.1 Introduction to electron crystallography of two-dimensional crystals of membrane proteins
Two-dimensional crystals of membrane proteins can be produced through various approaches. The most common method is reconstitution into a lipid bilayer of the purified and detergent-solubilized protein by dialysis (Stahlberg et al., 2001; Kühlbrandt, 2003; Schmidt-Krey, 2007a). Incubation of detergent-protein solutions with detergent-adsorbing polystyrene ‘BioBeads’ and the monolayer technique, where 2D crystals are grown at an air-water interface (Auer et al., 1998, 1999) or with the help of interactions with lipid monolayers (Fromherz, 1971; Uzgiris and Kornberg, 1983; Kubalek et al., 1991; Avila-Sakar et al., 1994; Chiu et al., 1997; Vénien-Bryan et al.,1998; Brisson et al., 1999; Lebeau et al., 2001; Isoda et al., 2004; Richter and Brisson, 2005; Mancheño et al., 2006; Dryden et al., 2009; Fezoua-Boubegtiten et al., 2010), are frequently used when only small quantities of protein are available. BioBeads and monolayers generally have the advantage of short times until crystal formation, usually varying from hours to 1 – 2 days, while reconstitution by dialysis can take considerably longer.
Reconstitution through dialysis requires mixing of the detergent-solubilized protein with detergent-solubilized lipid, after which the detergent is slowly removed from the detergent-protein-lipid mixture by dialysis against detergent-free buffer. Upon reconstitution of the membrane protein into a phospholipid bilayer, the 2D crystals, in the form of stable proteoliposomes, possess exceptional mechanical strength. In fact, they are often able to withstand several minutes of sonication without being disrupted, and even eukaryotic membrane proteins, which are often quite unstable, retain their functional integrity at room temperature within proteoliposomes (Schmidt-Krey, 2007a). Crystal formation within native membranes, such as observed for bacteriorhodopsin (Oesterhelt and Stoeckenius, 1971; Henderson et al., 1990), is a more unusual case that, for cryo-EM grid preparation, can be treated in the same manner as reconstituted 2D crystals. Both dialyzed specimens and 2D crystals from native membranes are usually stable for at least several months (Schmidt-Krey, 2007a). Furthermore, the 2D crystal stock is not sensitive to removal of small samples for grid preparation. On the other hand, the entire balance of the system of 2D crystals grown with the help of monolayers or BioBeads can change upon removal of a sample for grid preparation. In the case of monolayers, the small volumes used for 2D crystal growth are an added complication as the monolayer usually spans a few millimeters within a well, allowing for preparation of only 1–2 grids from each monolayer.
2.2 Back-injection
Since it was first described (Kühlbrandt and Downing, 1989; Wang and Kühlbrandt, 1991), back-injection has become the most frequently used technique for preparing 2D crystal specimens that has ultimately lead to high-resolution 3D models and maps that have revealed the secondary structure of membrane proteins. The back-injection method also provides the foundation for the carbon sandwich technique outlined in section 2.3.
Wang and Kühlbrandt (1991) provide a detailed description of the back-injection technique with tannin and other media. Briefly, an approximately grid-sized piece of carbon film is floated from mica onto a drop (~150 µl) of buffered tannin solution and picked up from underneath the air-water interface with a grid held by anti-capillary forceps (Figure 1). The grid is turned by 180 ° so that the carbon side faces down. The upwards-facing bare metal side of the grid is then wiped with the side of a pipette tip to remove any excess carbon film that might wind around the side of the grid. Next, 1 – 2 µl of the crystal suspension is placed on the grid surface and mixed by pipetting slowly approximately ten times from the buffer side. After approximately 1 minute of incubation, the grid is again turned by 180 ° and placed onto two pieces of Whatman #4 filter paper to remove excess liquid from the bare metal side of the grid. The grid is picked up and air-dried for between 10 – 15 s, followed by hand-plunging into liquid nitrogen and transfer of the grid to a cryo-holder for data collection.
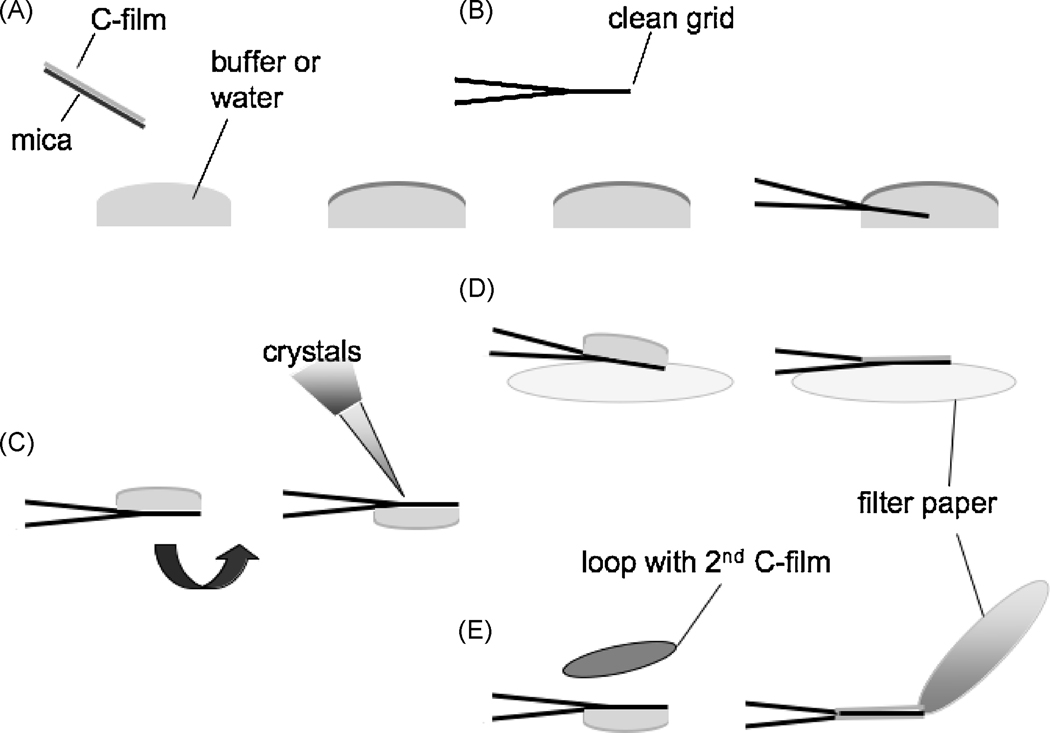
Figure 1.
Back-injection and carbon sandwich grid preparation involve the same initial steps. A, carbon film is floated off of a piece of mica. B, The carbon film is then picked up with a grid held by forceps. C, The grid is inverted by 180 ° so that the carbon film faces down, and the 2D crystal suspension is pipetted onto the grid. D, For the back-injection preparation, the grid is again inverted by 180 °. Thus the carbon faces up, and the grid is placed onto two pieces of filter paper for removal of the bulk of the liquid. After air-drying for 10–15 s to produce a thin layer of 2D crystal suspension, the grid is hand plunged into liquid nitrogen. E, The carbon sandwich preparation method requires a second carbon film to be picked up with a loop and placed over the grid. Excess liquid is removed from the side of the grid. After extensive air-drying, sometimes for minutes, the grid is hand-plunged into liquid nitrogen.
2D crystals prepared by back injection are preserved for cryo-EM data collection by a thin layer of precipitated tannin covering the sample (Wang and Kühlbrandt, 1991). The superior preservation of LHCII (light harvesting complex II) 2D crystals with tannin, in contrast to other media, has been proposed to be due to an interaction of the tannin layer with the protein (Wang and Kühlbrandt, 1991). The LHCII experiments used an unusually high salt concentration of 300 mM KCl, which is known to precipitate tannin. Similar precipitation can be observed with other samples; in the case of human vitamin K-dependent γ-glutamyl carboxylase the precipitation was attributed to the high salt concentration of 250 mM NaCl, which was subsequently confirmed by precipitation studies with increasing salt concentrations (Schmidt-Krey et al., 2007b; Schmidt-Krey unpublished observations). Wang and Kühlbrandt (1991), as well as Koning et al. (2003), found the precipitation of tannin and its interaction with the 2D crystal to be critical to sample preservation. The tannin in the back-injection technique can be substituted either by 1 – 10 % glucose or trehalose (Fujiyoshi, 1998) as well as other sugars, depending on the sensitivity of the sample to different media and their concentration. The presence of 10 – 20 % glycerol in a 2D crystal suspension could be sufficient for sample preservation (Wang and Kühlbrandt, 1991).
2.3 Carbon sandwich technique
With the back-injection technique as a foundation, the carbon sandwich technique was recently developed to increase flatness of carbon films and of 2D crystals (Koning et al., 2003), as well as to compensate for charging effects of highly-tilted specimens (Gyobu et al., 2004). This method has had a dramatic impact on the success-rate for data collection at high tilt angles, probably due to the compensation for charge-induced sample movement by a second carbon film. Consequently, the carbon sandwich technique has been a major factor in minimizing time and effort invested in data collection and structure determination by electron crystallography.
For this technique, specimen grids are prepared in a nearly identical fashion to that used with the back-injection technique up to the point where the solution of 2D crystals is pipetted onto the grid (Figure 1). At this step, rather than turning by 180 ° and blotting the grid, a second carbon film of approximately 3 × 3 mm is picked up from a drop of water or buffer (~150 µl) with a loop that has a slightly larger diameter than the grid. The second carbon film is placed on the back-injection grid on top of the crystal droplet. The carbon sandwich technique by Koning et al. (2003) describes blotting with the loop in place over the grid. In comparison, Gyobu et al. (2004; http://www.2dx.unibas.ch/download/movies/Gyobu-Sandwich.mov/view) depict blotting after the loop with the second carbon film has been pulled over both the grid and part of the forceps. In the ISK laboratory, identical grid quality was observed when preparing carbon sandwich grids by both methods with longer blotting times required for the Gyobu procedure (Schmidt-Krey, unpublished observations). In addition, multiple blotting steps around the edge of the grid may be necessary to decrease the partial air drying time and/or decrease the thickness of the ice. After partial drying, which can take up to several minutes depending on humidity conditions in the laboratory, the grid is manually plunged into liquid nitrogen and transferred to a cryo-holder for data collection.
A crucial difference between the carbon sandwich technique and the back-injection technique is that, for the former method, thin grids with large grid squares must be used, otherwise too much liquid is retained on the grid. Small grid squares (ie., a large mesh number) and thick grids, in combination with the carbon film, form deeper wells, from which the buffer cannot be sufficiently removed, resulting in thicker vitreous ice. Koning et al. (2003) found 200-mesh grids to be optimal, while Gyobu et al. (2004) used custom-made grids with a thickness of 10 µm and 100 µm diameter circular holes. Both authors noted that it is important for the second carbon film to be smaller than the first, which will prevent the second film from folding around the edge of the grid and creating more than two carbon layers. In the ISK laboratory, it was also found that drawing the side of a pipette tip across the surface of both regular back-injection and carbon sandwich grids (before placing the second carbon film) removes any pieces of carbon film that might have folded around the edge of the grid. Furthermore, to reduce charging effects, the second carbon film should have the same thickness as the first film; this matching of thickness can most easily be accomplished by using adjacent areas of carbon-coated mica for the two layers of the carbon sandwich grid.
The second carbon film is most easily placed on the grid with a loop that has a larger diameter than the grid. In the ISK laboratory, a loop with an inner diameter of 4 mm, handmade by winding platinum wire with a thickness of 0.2 mm once around a 4 mm metal rod, is regularly used. The shorter end of the wire is twisted around the longer end (approximately 2 – 4 cm) to form the loop. The long wire is then melted firmly into a broken off narrow end of a Pasteur pipette. We tested and used the technique with readily available 200-mesh grids and found it to be easily reproducible. The key factors in reproducibility were the large mesh as well as extensive blotting. Even at room humidity levels far below 20 %, when carbon sandwich grids were blotted excessively with a torn filter paper around the entire rim of the grid, the grid still required minutes of partial air-drying before a sufficiently thin layer of vitreous ice could be obtained. Thus, during a first trial of the method it is recommended to aim for at least 3 – 5 minutes of air-drying.
Koning et al. (2003) also described a second carbon sandwich preparation technique that requires a large volume of sample (100 µl) to be placed in a well, from which a large piece of carbon film is picked up with the edge of a grid from underneath. While grid preparation is convenient with this method, it is not feasible for most samples because it requires excessive volumes of crystal suspension,
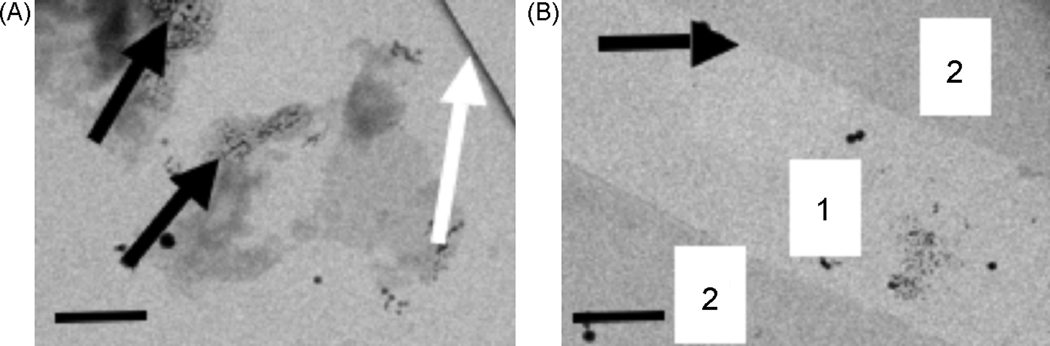
During inspection of the grid, distinguishing an intact carbon sandwich from single carbon films can require some experience. In the case of two entirely flat and unfolded carbon films separated by a thin layer of ice, this distinction can be difficult or impossible. However, the second carbon film tends to fold and break more easily than the first layer. This difference is likely due to the additional manipulation of the second film when it is picked up with the loop to place it onto the grid. A partially broken second carbon film is a clear indication of a carbon sandwich, even if only a partial one for a particular grid square is observed. Figure 2 illustrates this point, where Figure 2a shows an overview of 2D crystals on a carbon sandwich grid after data collection. Diagonally crossing the upper right corner of the image (Figure 2a, white arrow) is a fold in the second layer of the carbon film illustrating that a carbon sandwich has successfully been prepared. The appearance of the radiation damage on the parts of the specimen used for focusing the microscope (Figure 2a, black arrows) shows the presence of vitreous ice rather than complete drying of the sample.
Figure 2.
Typical appearance of a carbon sandwich grid. A, The second carbon layer of the carbon sandwich grid can be identified by the fold (white arrow). The black arrows indicate focus areas that were excessively exposed to show that the grid contains vitreous ice. B, Grid areas containing two carbon layers can, at times, only be distinguished when a broken second carbon film area is present. The numbers of carbon layers are indicated, and the transition from one film to the carbon sandwich is indicated by an arrow. The scale bars represent 1 µm.
In addition to the advantages in preparing flat carbon films and compensating for charging effects at high tilt angles, we have found the carbon sandwich to have a third crucial advantage. The carbon sandwich is of tremendous benefit for membrane proteins that are sensitive to various parts of the cryo-EM grid preparation procedure (Schmidt-Krey et al., 2007b; Schmidt-Krey, unpublished observations), such as membrane proteins with hydrophilic domains that are sensitive to contact with an air-water interface or changing buffer conditions during partial drying. The second carbon film protects the membrane protein from contact with the air-water interface. However, upon inspection of focus areas after imaging (Figure 2), we have observed that the carbon-sandwiched areas usually retain a slightly larger amount of buffer, and thus vitreous ice, than a back-injection grid, which speaks against excessive drying of a carbon sandwich grid. In addition to preventing contact with the air-water interface, one could speculate that the rapid blotting from the side might minimize evaporation of volatile components from the buffer so that concentration of the buffer components does not occur.
In the back-injection technique the flat surface of the filter paper does not absorb the buffer as quickly as blotting from the side with a torn piece of filter paper. The paper fibers of the torn paper absorb the buffer at a significantly higher rate than the surface of or the side of a cut piece of filter paper. Thus, for improved sample preservation it is not only critical to avoid contact with the air-water interface, but also to prevent the concentration of buffer components by a faster buffer removal (Schmidt-Krey, 2007a, 2007b, Schmidt-Krey, unpublished observations).
2.4 Regular grid preparation with complete drying prior to cryo-cooling
A method very similar to negative stain grid preparation of 2D crystals can be performed using tannin and various sugars. This approach is similar to the preparation used by Unwin and Henderson (1975), who mixed their 2D crystal suspension of bacteriorhodopsin with glucose, and subsequently dried the sample on the grid. Once cryoholders were developed, dried samples were cooled in the electron microscope to reduce the effects of radiation damage (Henderson et al., 1986, 1990). A general grid preparation protocol equivalent to negative stain can be followed for other samples, but using trehalose, tannic acid, or glucose instead of a heavy metal salt stain. Use of a torn piece of filter paper is critical for fast removal of the buffer as described in 2.3. This method has been applied to robust 2D crystals, such as bacteriorhodopsin and thin catalase crystals, and has also been successful for preserving high-resolution information from crystals of largely hydrophobic membrane proteins such as microsomal glutathione transferase 1 (MGST1) and leukotriene C4 synthase. In the case of MGST1, electron diffraction data from dried test samples showed that the crystalline order was preserved to 3 Å resolution (Schmidt-Krey, unpublished observations). The technique is usually most successful for highly hydrophobic proteins, such as porins (Jap et al., 1990). However, with the development of vitrification techniques applied before transfer to the electron microscope, this method is used less frequently.
2.5 Plunge-freezing
Plunging of grids with a guillotine-like device into liquid ethane cooled by liquid nitrogen is frequently used for monolayer 2D crystals because back-injection or carbon sandwich techniques cannot be employed for these types of samples (Norville et al., 2007). The reason for this constraint is that the 2D monolayer needs to be picked up from the sample well with a carbon-coated grid. Norville et al. (2007) achieved a vast improvement in data quality by using a loop to transfer the monolayer crystals to the grid before plunge-freezing. Unless preserving media (e.g., tannin, trehalose, or glucose) is added to the sample well before 2D crystal formation, these specimens will also require the rapid cooling only possible with fast immersion in cryogen by a guillotine. Briefly, the sample is placed on the grid, blotted, and plunged into ethane with the help of gravity, a spring, or pressurized air. Unger, Yeager, and co-workers (Unger et al., 1999) have described a protocol in which grids were first glow-discharged in amylamine, allowing gap junction 2D crystals to adhere optimally to the carbon film before plunging. The amylamine protocol was somewhat modified and adapted for plunging of vitamin K-dependent γ-glutamyl carboxylase (Schmidt-Krey, 2007a). Ren et al. (2000) used plunging for the 3D data collection of aquaporin 1, resulting in the first atomic model of 2D crystals of a membrane protein vitrified without the use of any additives. It should be noted that this method has been particularly important for the structure determination of helical crystals such as of the nicotinic acetylcholine receptor (Unwin, 2005) and P-type ATPases (Pomfret et al., 2007).
Structure-function studies may benefit from time resolved cryo-EM studies where a reaction is stopped before it comes to equilibrium. This type of experiment was demonstrated elegantly for studies of the activation of the nicotinic acetylcholine receptor in helical arrays with acetylcholine (Berriman and Unwin, 1994) and conformational changes in 2D crystals of bacteriorhodopsin induced by different wavelengths of light (Subramaniam and Henderson, 2000). The 2D crystal suspension on the grid can either be sprayed with a substrate, or illuminated, as for bacteriorhodopsin, before the grid is plunged. For this type of experiment, the reaction rate must be considered and the speed and timing of plunging carefully controlled. Freezing devices are commercially available and plans for plunging devices that can be built in a workshop are available from many cryo-EM laboratories.
2.6 Data collection from tilted specimens
Grid preparation for data collection from tilted specimens can require more optimization than for untilted specimens. The carbon film must be particularly flat for tilted specimens as subsequent grid preparation steps rely on the best possible carbon film. A pre-evaporation step in the evaporation of the carbon film (Fujiyoshi, 1998) eliminates or greatly reduces the formation of sparks during the initial degassing of the carbon rod or wire, thus resulting in flatter carbon films. With the use of the carbon sandwich and completely dried samples, Koning et al. (2003) could achieve an optimally reduced curvature of flat carbon film of less than 0.2 ° per µm. Specimen curvature can also occur due to the method by which the sample is placed on the grid. Various techniques might result in 2D crystals that appear flat upon visual inspection, but which contain a large degree of curvature that is detrimental to data collection, especially for data of tilted specimens (Henderson, 1990).
Gyobu and coworkers (2004) successfully overcame one of the most severe problems associated with collecting data from tilted samples by dramatically minimizing and/or removing the charge build-up and subsequent image drift through the use of the second carbon film in the carbon sandwich technique. While specimen charging effects and thus movement will only cause a small change in focus for data of untilted specimens, the charging effect can make a large fraction of tilted data unusable. With the carbon sandwich technique, successful data collection at tilts of 60 ° and 70 ° could be improved from 2 % to 90 % for several proteins (Gyobu et al., 2004).
2.7 Carbon film and grids
The thickness of the carbon film required depends on the grid preparation method used. Plunging and larger mesh sizes will usually necessitate thicker carbon film. Thinner carbon film is better for the carbon sandwich technique, because the two carbon films act to mutually stabilize each other and their combined thickness must be taken into account. It is particularly important to consider carbon thickness for the collection of data from tilted specimens, which requires the beam to pass through increasing sample thicknesses at larger tilt angles. Once the optimal thickness of the carbon film is determined experimentally, ideally by collection of electron diffraction patterns or images from highly tilted samples, it should be kept constant for further data collection. This consistency can be achieved either by measuring the thickness of the carbon film during evaporation or by visually comparing filter paper adjacent to the mica with the carbon on paper from mica that previously gave good results.
A thorough investigation of the optimal carbon film preparation (Han et al., 1994) evaluated a range of factors in the preparation of carbon films and found the only notable factor was the stock of carbon used. Thus, the authors recommended testing of carbon stocks in the event of specimen flatness problems. Glaeser (1992) studied grid preparation techniques for sample flatness and found molybdenum grids to have a more favorable thermal expansion coefficient compared to copper grids for preventing wrinkling of 2D crystals. Long-range wrinkling effects of at least one layer of the carbon sandwich were investigated by reflection light microscopy (Koning et al., 2003).
2.8 Choice of protocols
The recent success of the carbon sandwich technique will likely make this the method of choice for cryo-EM grid preparation for 2D crystals of reconstituted membrane proteins. The improved flatness of 2D crystals due to the stretching described by Glaeser (1992) may constitute an additional factor in the success of carbon sandwich data collection from tilted specimens. An important alternative to the carbon sandwich technique is the plunging method, which can be used for both monolayers as well as for structure-function studies requiring careful timing. A disadvantage of plunging is that only one carbon film is used, and charging effects can reduce the success rate for data collection from tilted specimens (Gyobu et al., 2004).
Standard low-dose conditions need to be applied with electron exposure dependent on the temperature used for data collection, accelerating voltage of the microscope, and the resolution at which the experiment aims to optimize the signal to noise ratio (Baker et al., 2010). Image collection by the spot-scan technique results in dramatic reduction of both beam-induced charging and specimen movement (Downing et al., 1986; Henderson et al., 1990; Downing, 1991). For spot-scan imaging, the beam size is focused to approximately 1000 Å (described as a ‘spot’), and an image-sized area is exposed as the beam is deflected spot by spot with a slight overlap of exposed areas to cover the entire film (Figure 3). If available, the combination of spot scan and carbon sandwich grids will result in the largest yield of high quality images. Should the carbon sandwich preparation not be an option, use of spot-scanning will still greatly reduce charging problems and be an effective method in combination with any type of grid preparation.
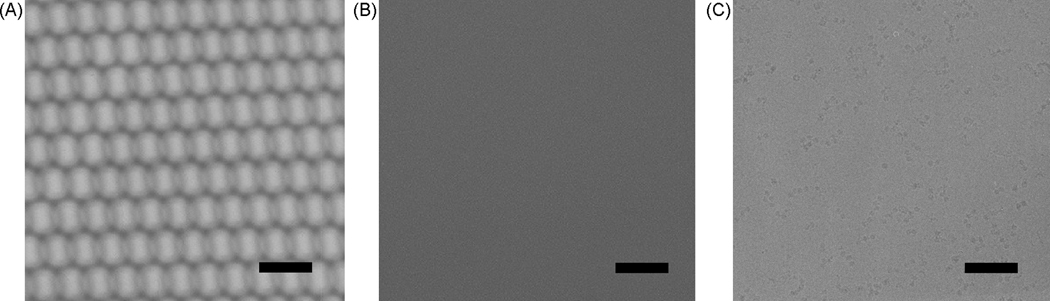
Figure 3.
Cryo-EM images of 2D crystals and single molecules. A, Flood-beam and B spot-scan images of 2D crystals. The 2D crystal lattice cannot be discerned visually under cryo-EM conditions. The location and quality of each 2D crystal requires identification by optical diffraction and/or image processing. C, Single particle images are collected under conditions that allow for visual identification of the membrane protein complexes. The scale bars represent 100 nm.
3. Introduction to single particle EM of detergent-solubilized membrane proteins
Membrane proteins may be extracted from a phospholipid bilayer with detergents and purified to homogeneity in detergent-containing solutions. In principle, these protein-detergent micelles could be analyzed by single particle EM, as with soluble protein complexes. In practice, there are several complications that occur when attempting to image detergent-solubilized membrane protein complexes.
A detergent micelle differs from the native phospholipid environment of membrane proteins due to the structure of detergent molecules. Phospholipids, consisting of a polar headgroup and two hydrophobic tails, have approximately the same cross section through the polar headgroup and the end of the hydrophobic tails distal from the headgroup. This geometry leads phospholipids to self-assemble into relatively flat bilayers in aqueous media. In contrast, detergents have a polar headgroup and a single hydrophobic tail, giving them an overall conical shape. In aqueous solution at concentrations above their critical micelle concentration (CMC), detergents self-assemble into micelles, which have a more strongly curved surface than a phospholipid bilayer. This difference in structure leads to differences in behavior that can complicate single particle EM studies.
3.1 Problems with imaging proteins in detergents and some alternatives
Potential problems with detergent-solubilized membrane protein complexes can be illustrated with a few examples. WzzE, the polysaccharide co-polymerase from the inner membrane of E. coli, forms a thimble shaped oligomer facing the periplasm (Tocilj et al., 2008). However, as seen in micrographs of detergent solubilized WzzE complexes, the assembly can further oligomerize to form dimers of thimble structures with their openings facing each other (Tocilj et al., 2008). This non-biological structure is clearly a detergent-induced artifact. A second type of artifact can arise due to instability of membrane protein complexes in detergents. In the detergents Brij-35 and dodecyl maltoside (DDM), the Saccharomyces cerevisiae ATP synthase loses two membrane-bound subunits, subunits e and g (Arnold et al., 1998), while the Bos taurus (bovine) assembly retains these subunits in either detergent (Rubinstein et al., 2003). Naïve structural analysis of the two different complexes by cryo-EM would suggest that they have dramatically different structures for their membrane regions, while in truth the complexes are probably nearly identical in their native phospholipid environment (Lau et al., 2008). Clearly, working with detergents presents some undesirable complications. Furthermore, vitrifying solutions that contain a high-concentration of detergents for single particle EM can produce ice with a strong texture that resembles protein particles, even if protein particles are not present in the buffer. Images like these lead to the possibility of serious misinterpretation by subjecting images of non-protein structures to analysis by single particle EM software.
One alternative to the use of detergents is the use of amphipols to maintain membrane protein complexes as single particles in solution. Amphipols, large polymers containing both hydrophilic and hydrophobic moieties, function by binding irreversibly to the hydrophic transmembrane regions of membrane proteins making their outer surface hydrophilic (Tribet et al., 1996). There has been some success in using amphipols for both negative stain EM (Tribet et al., 1996) and cryo-EM (Wilkens, 2000). A second alternative is to maintain the membrane protein in a phospholipid bilayer, by reconstituting it into a small unilamellar vesicle. Membrane proteins thus prepared can be subjected to structural analysis by the technique known as random spherically constrained EM (Wang and Sigworth, 2009), offering the possibility of building models of a non-crystalline membrane protein in its native environment. However, for the most part, 3-D single particle analysis of membrane proteins has relied on the use of detergents, and therefore it is worthwhile to know how to prepare optimal grids of detergent-solubilized membrane proteins and how to interpret images acquired from these specimens. Detailed protocols for preparing both negative stain and cryo-EM specimens for single particle analysis of detergent-solubilized membrane protein complexes are described elsewhere (Rubinstein, 2007). The following sections describe the rationale for these protocols.
3.2 Negative stain EM of single particles
Preparing negative stain EM grids of membrane proteins does not differ significantly from preparing grids for soluble protein complexes. One minor difference is that it is necessary to wash the grid several times in order to remove detergents before applying stain. During the staining process, the membrane protein complex, still in the detergent-containing buffer, is adsorbed to the continuous carbon substrate of a grid. At this stage, the detergent-protein micelle is still in equilibrium with monomers, which in turn are in equilibrium with pure detergent micelles in the bulk solution. Once the protein particles have adsorbed to the carbon film and the grid has been transferred to the wash water, pure detergent micelles and monomers are removed and monomers of detergent begin to leave the protein-detergent micelle. The extent of this depletion will depend on how long the washing takes and the chemical properties of the detergent used. It is almost certain that, regardless of which detergent is used, some depletion will occur but the depletion process will not go to completion. Detergent will exclude aqueous stain as effectively as protein, making it indistinguishable from protein in an image of negatively stained protein. Therefore, the amount of detergent that remains bound to the protein will dramatically affect the appearance of a negatively stained membrane protein complex.
3.3 Cryo-EM of single particles
Cryo-EM of single particles of membrane-proteins can be performed with both perforated (Serysheva et al., 2008) and continuous carbon supports (Mitra et al., 2005) (Figure 4). In the latter case, just like with negative stain, some of the detergent can be washed away once the protein is adsorbed to the carbon surface. Alternatively, if using a perforated carbon substrate, the solution containing detergent-protein micelles, detergent, and buffer can be blotted to the correct thickness and vitrified. In the laboratory of one of the authors (JLR), it was found that in order to produce a good distribution of particles over holes, one must subject the grids to extensive glow discharge in air prior to specimen deposition. For cryo-EM over holes in a perforated carbon film, the presence of detergent may increase the thickness of the sample by contributing a monolayer of detergent at both air-water interfaces of the specimen (Figure 4a). However, the most notable effect of detergents is that membrane proteins are required at much higher concentrations than soluble protein complexes for cryo-EM with perforated carbon. For example, while one can prepare negative stain EM grids with sample concentrations as low as 20 nM and cryo-EM grids over continuous carbon at similar concentrations, concentrations closer to 5 µM are usually required in order to freeze membrane proteins over holes (Rubinstein and Walker, 2002; Rubinstein et al., 2005; Bueler et al., 2008; Rubinstein et al., 2003; Lau et al., 2008; Lau and Rubinstein, 2001). In comparison, when working with soluble protein complexes applied to perforated carbon, concentrations closer to 20 nM than 5 µM can be employed (for example, see Williams et al., 2008). It may be that this requirement for high concentrations of protein is due to the extensive glow discharge required to spread hydrophobic proteins across unsupported ice. Alternatively, the requirement for high concentrations of protein may be a result of the presence of detergents. However, the concentration seems to reflect the actual density of particles in solution, whereas the lower concentration that can be used for soluble proteins indicates a concentrating effect for soluble proteins on the grid, probably at the air-water interface.
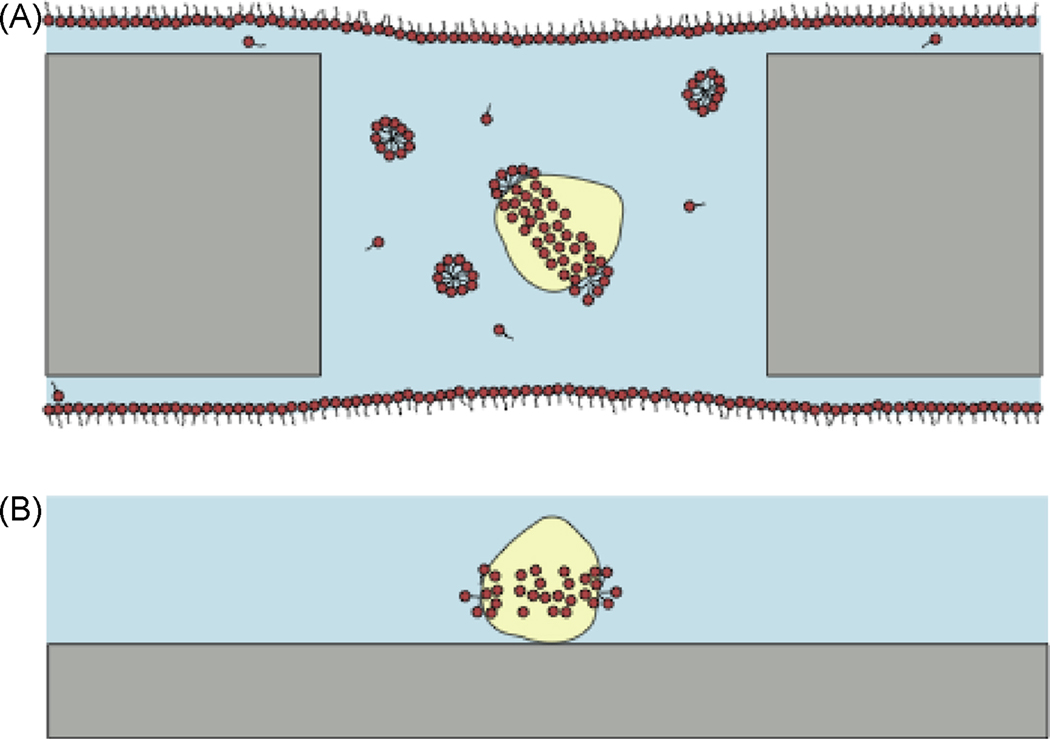
Figure 4.
Behavior of detergent-solubilized membrane proteins on perforated and continuous carbon support films in single particle cryo-EM. A, With a detergent-solubilized membrane protein complex prepared on a perforated carbon film coated (dark grey) EM grid, the detergent-protein micelle will exist in equilibrium with free monomers of detergent (found at the critical micelle concentration), micelles of detergent, and monolayers of detergent at the air-water interfaces. The contrast of the detergent bound to the protein will depend on the density of the detergent used. B, In comparison, detergent-solubilized membrane proteins immobilized on a continuous carbon film coated EM grid may be washed with detergent-free buffer. This washing step will remove the monolayer of detergent at the air-water interface, the free monomers and micelles of detergent, and will partially deplete the protein of bound detergent. In cryo-EM, the appearance of the remaining bound detergent will depend on which detergent was employed, while in negative stain EM, detergent will exclude stain and appear indistinguishable from protein.
Therefore, when choosing a substrate for cryo-EM of detergent-solubilized membrane proteins, the low background noise and random orientations of particles usually obtained with perforated grids is offset by the requirement for higher concentrations of proteins and the concern that additional charging may occur with ice that is not supported by a carbon layer (Brink et al., 1998). Alternatively, continuous carbon films may be used (Figure 3), with the advantages that less protein is required, that the detergent may be removed or buffer changed in the sample once the protein is adsorbed to the grid, and that the carbon may help to reduce beam induced charging in the microscope (Figure 4b). On the other hand, a continuous carbon support may induce preferred orientations and will contribute additional structural noise to the image. One approach that may offer some of the benefits of each of the above, is binding histidine-tagged membrane protein complexes to a phospholipid monolayer that contains Ni-NTA (Kelly et al., 2008a; Kelly et al., 2008b) thereby concentrating the protein and allowing it to be imaged over a hole in a perforated carbon film coated EM grid. Whereas contrast in negative stain images comes from the strong difference in scattering cross-section between protein or detergent and the heavy metal salt stain, the phase contrast in images of ice embedded specimens arises due to the subtle difference in scattering cross section between ice, protein, and detergent. Detergents that are more dense than ice (e.g. DDM and CHAPS, see Table 1) will resemble protein in ice embedded specimens (Figure 5), while detergents that are around the density of ice (e.g. Brij-35 at 1.05 g/ml) will effectively be invisible. Therefore, the appearance of the detergent micelle in a cryo-EM image depends critically on which detergent was used, the size of the detergent micelle, whether or not the grid was washed to remove detergent before freezing the specimen and, if so, how much detergent depletion occurred during that washing. Several good reviews allow for the interested investigator to look up the density of a specific detergents (le Maire et al., 2000; Timmins et al., 1988) and compare them to the known density of ~0.94 g/ml for vitreous ice (Mishima et al., 1985) and the approximation of 1.36 g/ml for protein (Fischer et al., 2004). Phospholipids, particularly their hydrophobic tail-regions, usually have a lower density than ice and therefore bound phospholipids are unlikely to be confused with protein. A recent map of the V-type ATPase from Thermus thermophilus (Lau and Rubinstein, 2010) demonstrated these expectations dramatically, with the membrane region of the complex being enclosed by an obvious micelle of dodecylmaltoside that appeared to be of similar density to protein (Figure 5). Table 1 gives the density of several commonly used detergents.
Table 1.
Density of ice, protein, and various detergents affects the contrast in different regions of single particle cryo-EM maps of membrane proteins.
| Substance | Density (g/ml) | Reference |
|---|---|---|
| Amorphous ice | 0.94 | Mishima et al., 1985 |
| Protein | ~1.36 | Fischer et al., 2004 |
| Dodecyl maltoside (DDM) | 1.19 | Timmins et al., 1988 |
| Lauryldimethylamine-oxide (LDAO) | 0.882 | Timmins et al., 1988 |
| Triton X-100 | 1.10 | Ganong et al., 1989 |
| Octyl glucoside (OG) | 1.16 | le Maire et al., 2000 |
| 3-[(3-Cholamidopropyl)dimethylammonio]-1- propanesulfonate (CHAPS) |
1.23 | le Maire et al., 2000 |
| Deoxycholic acid | 1.29 | le Maire et al., 2000 |
| Sodium dodecyl sulfate (SDS) | 1.16 | le Maire et al., 2000 |
Figure 5.
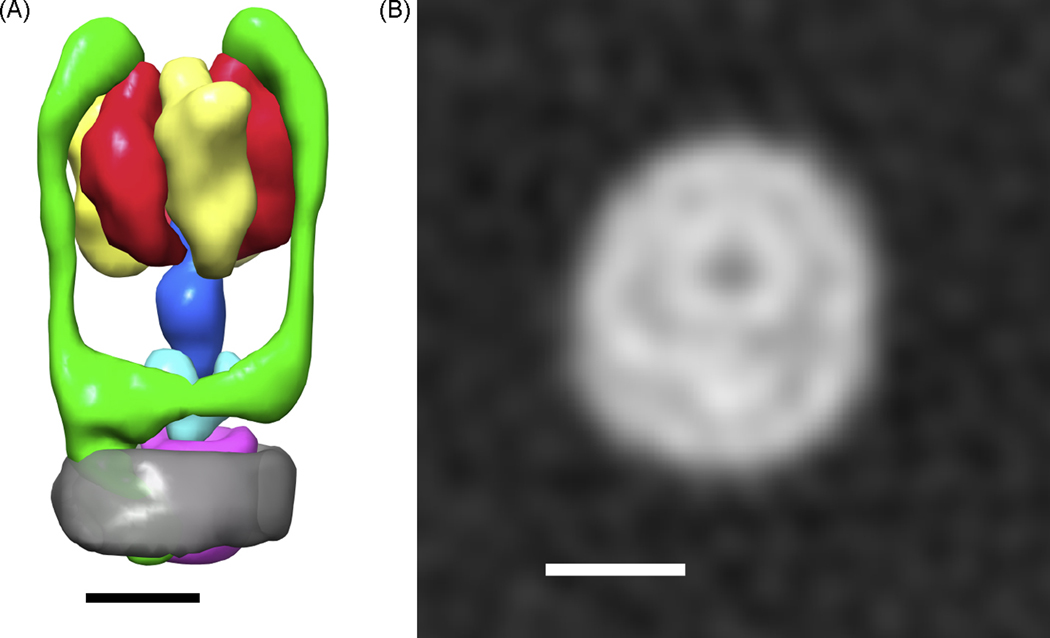
Three-dimensional models from single particle cryo-EM of a detergent-solubilized protein complexes can show density from the detergent micelle. A, A map from single particle analysis of dodecyl maltoside (DDM) solubilized Thermus thermophilus V-type ATPase (Lau and Rubinstein, 2010) resolved the two different hydrophobic subunits, I and L, from the detergent micelle used to keep the complex soluble. A 3D model depicting segmented subunits and subcomplexes shows a segment corresponding to the DDM micelle in semi-transparent dark grey near the bottom of the model. The other segments depict the A-subunits (yellow), B-subunits (red), the D and F subunit (dark blue), the C-subunit (light blue), the membrane bound I-subunit with the peripheral stalks consisting of subunits E and G (green), and the ring of L-subunits (pink). B, A cross section through the region of the complex usually embedded in the membrane resolves the ring of L-subunits, the I-subunit, and the micelle of DDM. The scale bars in both parts A and B represent 50 Å.
4. Resolution and sample preservation
“High” and “low” resolution for cryo-EM is continuously re-defined as methods improve and resolution barriers are broken. Visualizing α-helices as elongated densities is possible with 6–10 Å resolution and currently constitutes intermediate resolution for 2D crystals, while it is still an impressive achievement with single particles, especially of asymmetric particles. Currently, a range of 2.5–4 Å resolution is considered high resolution for a membrane protein structure solved by any means, while the aquaporin-0 structure at 1.9 Å resolution (Gonen et al., 2005) could then easily be defined as exceptionally high resolution. Any structure by single particle EM at better than 5 Å resolution would likely fall in the exceptionally high-resolution range. The term “high resolution” is bound to be redefined with time as specimen and grid preparation, cryo-EM data collection, and image processing are improved. While resolution is seen as one of the main means of gauging the quality of a structure, it should be emphasized that at times a structure at lower resolution, even with artificial removal of the high resolution data (Schmidt-Krey et al., 2004), will answer the relevant questions. Whatever the aim of the study is, the greatest possible care needs to be taken in optimally preserving the structure through the means of grid preparation.
5. Discussion
Negative staining is still a critical experimental approach for EM as it provides the best means of screening for 2D crystallization conditions and single particle preparations and can provide important data on single particles as discussed in this issue by DeCarlo and Harris. As the information limit from 2D crystals in negative stain is usually approximately 12–15 Å resolution after image processing, this experiment supplies an approximate value for the unit cell dimensions of 2D crystals, a first impression of the crystal packing and contacts, and initial information on the number of subunits. Negative stain also provides a first insight into the symmetry of a crystal, even though this can be misleading and should be confirmed with cryo-EM data. One can employ negative stain to substitute for tannin during back-injection both as a control for the method and to obtain a cleaner background for negative staining (Schmidt-Krey laboratory, unpublished observations). Negative stain used for single particles provides information on the overall molecular shape, and possibly conformational flexibility. It may also allow for the structural analysis of proteins that are too small to be studied by single particle cryo-EM.
Due to flatness issues, first tests of 2D crystals should always be attempted with back-injection. Currently, it appears advisable to immediately couple this technique with the carbon sandwich preparation method, as it will avoid charging problems at high tilt, render the sample more stable and prevent crystalline disorder at the air-water interface, and prevent evaporation from increasing the concentration of buffer components. Cryo-EM studies of membrane proteins as single particles will always require plunging, and the choice of continuous versus perforated carbon film requires testing.
Gonen et al. (2005) and Tani et al. (2009) were particularly successful in their 2D crystal experiments with aquaporin-0 largely because of the crystal quality, likely due to the high order (crystal contacts), and the stabilization of the 2D crystal flatness caused by the two membrane layers. Other contributing factors were the careful optimization of the cryo-EM grid preparation suitable for this particular sample. The use of a 4k × 4k CCD camera for collection of electron diffraction patterns improved the quality of the data further. The structure of aquaporin-0 (Gonen et al., 2005) was solved by molecular replacement. Thus, not only is the high resolution of the structure notable, but avoiding the collection of experimental image phases meant that the cryo-EM structure determination became comparable in timeframe to an X-ray crystal structure determination that also used molecular replacement for phasing. In general, it is important to note that X-ray crystallography and electron microscopy are complementary, and data from both experiments have been combined for important insights (e.g. Appel et al., 2009).
6. Conclusions and perspectives on future developments
Despite a few complications in preparing specimens of membrane protein complexes for single particle EM and in interpreting images from these experiments, these assemblies behave much like soluble protein complexes in single particle EM. Therefore, the prospects for single particle EM of membrane proteins is strongly tied to the prospects for single particle EM in general and the excitement surrounding this technique is equally merited. In fact, because membrane protein complexes are highly refractory to most other forms of structural analysis, the excitement about single particle methods is particularly warranted for these important protein complexes.
There continue to be critical method developments in data collection and image processing methods for 2D crystals (Henderson et al., 1990; Downing, 1991; Crowther et al., 1996; Fujiyoshi, 1998; Gipson et al., 2007; Cheng et al., 2007), which help to extend the resolution obtained as evidenced by the aquaporin-0 structure at 1.9 Å resolution (Gonen et al., 2005). The carbon sandwich is one of the most significant recent developments in sample preparation and preservation for 2D crystals. The technique may be one of the critical factors in eventually making electron crystallography even more productive and complementary with X-ray crystallography, especially when considering the shorter time required for 2D crystallization. To extend the carbon sandwich method to time-resolved structure-function studies, a combination of the carbon sandwich technique with plunging will be desirable. A device could be developed for reproducible carbon sandwiching, which would ideally be coupled to freezing in liquid ethane.
The importance of the carbon support film as demonstrated by Glaeser (1992) and colleagues has shown the need of further characterization of carbon and possibly additional support materials (Rhinow and Kühlbrandt, 2008). Grids might benefit from further optimization for the carbon sandwich technique, possibly with thinner molybdenum grids as used by Gyobu et al. (2004). Introducing angles or wells into grid bars would allow for faster removal of buffer and possibly faster introduction of substrate for structure-function studies.
To obtain information at the highest-possible resolution, it should be emphasized that careful image processing is essential to extract the maximal amount of information from images. Otherwise, the most careful sample preparation at the level of the protein production, grid preparation, and cryo-EM data collection will be in vain. Samples themselves, particularly 2D crystals, will benefit greatly from a deeper understanding of how 2D crystals form as well as extensive, general screening guidelines. Less well-ordered 2D crystals and heterogeneous single particle samples will benefit from further methods development in image processing.
Acknowledgements
We would like to thank our lab members and colleagues for valuable discussions throughout the years and Tina Dreaden for critical reading of the manuscript. Funding for part of the studies was provided by NIH grant HL090630 (ISK) and CIHR grant MOP 81294 (JLR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ingeborg Schmidt-Krey, Email: Ingeborg.Schmidt-Krey@biology.gatech.edu.
John L. Rubinstein, Email: John.Rubinstein@utoronto.ca.
References
- Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1 ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Appel M, Hizlan D, Vinothkumar KR, Ziegler C, Kühlbrandt W. Conformations of NhaA, the Na/H exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J. Mol. Biol. 2009;388:659–672. doi: 10.1016/j.jmb.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schagger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer M, Scarborough G, Kühlbrandt W. Three-dimensional Map of the Plasma Membrane H+ATPase in the Open Conformation. Nature. 1998;392:840–843. doi: 10.1038/33967. [DOI] [PubMed] [Google Scholar]
- Auer M, Scarborough G, Kühlbrandt W. Surface crystallisation of the plasma membrane H+ATPase on a carbon support film for electron crystallography. J. Mol. Biol. 1999;287:961–968. doi: 10.1006/jmbi.1999.2652. [DOI] [PubMed] [Google Scholar]
- Avila-Sakar AJ, Chiu W. Visualization of beta-sheets and side-chain clusters in two-dimensional periodic arrays of streptavidin on phospholipids monolayers by electron crystallography. Biophys. J. 1994;70:57–68. doi: 10.1016/S0006-3495(96)79597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LA, Smith EA, Bueler SA, Rubinstein JL. The resolution dependence of optimal exposures in liquid nitrogen temperature electron cryomicroscopy of catalase crystals. J. Struct. Biol. 2010;169:431–437. doi: 10.1016/j.jsb.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Berriman J, Unwin N. Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy. 1994;56:241–252. doi: 10.1016/0304-3991(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Brink J, Sherman MB, Berriman J, Chiu W. Evaluation of charging on macromolecules in electron cryomicroscopy. Ultramicroscopy. 1998;72:41–52. doi: 10.1016/s0304-3991(97)00126-5. [DOI] [PubMed] [Google Scholar]
- Brisson A, Bergsma-Schutter W, Oling F, Lambert O, Reviakine I. Two-dimensional crystallization of proteins on lipid monolayers at the air–water interface and transfer to an electron microscopy grid. J. Cryst. Growth. 1999;196:456–470. [Google Scholar]
- Bueler SA, Rubinstein JL. Location of subunit d in the peripheral stalk of the ATP synthase from Saccharomyces cerevisiae. Biochemistry. 2008;47:11804–11810. doi: 10.1021/bi801665x. [DOI] [PubMed] [Google Scholar]
- Cheng A, Leung A, Fellmann D, Quispe J, Suloway C, Pulokas J, Abeyrathne PD, Lam JS, Carragher B, Potter CS. Toward automated screening of two-dimensional crystals. J. Struct. Biol. 2007;160:324–331. doi: 10.1016/j.jsb.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Avila-Sakar AJ, Schmid MF. Electron crystallography of macromolecular periodic arrays on phospholipid monolayers. Adv. Biophys. 1997;34:161–172. doi: 10.1016/s0065-227x(97)89638-4. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM. MRC Image Processing Programs. J. Struct. Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Downing KH, Glaeser RM. Improvement in high resolution image quality of radiation-sensitive specimens achieved with reduced spot size of the electron beam. Ultramicroscopy. 1986;20:269–278. doi: 10.1016/0304-3991(86)90191-9. [DOI] [PubMed] [Google Scholar]
- Dryden KA, Crowley CS, Tanaka S, Yeates TO, Yeager M. Two-dimensional crystals of carboxysome shell proteins recapitulate the hexagonal packing of three-dimensional crystals. Protein Sci. 2009;18:2629–2635. doi: 10.1002/pro.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fezoua-Boubegtiten Z, Desbat B, Brisson A, Lecomte S. Determination of molecular groups involved in the interaction of Annexin A5 with lipid membrane models at the air-water interface. Biochim. Biophys. Act. 2010 doi: 10.1016/j.bbamem.2010.03.014. In Press. [DOI] [PubMed] [Google Scholar]
- Fischer H, Polikarpov I, Craievich AF. Average protein density is a molecular-weight-dependent function. Protein Sci. 2004;13:2825–2828. doi: 10.1110/ps.04688204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromherz P. Electron microscopic studies of lipid protein films. Nature. 1971;231:267–268. doi: 10.1038/231267a0. [DOI] [PubMed] [Google Scholar]
- Fujiyoshi Y. The structural study of membrane proteins by electron crystallography. Adv. Biophys. 1998;35:25–80. doi: 10.1016/s0065-227x(98)90004-1. [DOI] [PubMed] [Google Scholar]
- Fujiyoshi Y, Unwin N. Electron crystallography of proteins in membranes. Curr. Opin. Struct. Biol. 2008;18:587–592. doi: 10.1016/j.sbi.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganong BR, Lu C-M. Application of nonionic polyoxyethylebe detergents in studies of phosphatidylionsitol kinase. Anal. Biochem. 1989;179:66–71. doi: 10.1016/0003-2697(89)90201-7. [DOI] [PubMed] [Google Scholar]
- Gibbons C, Montgomery MG, Leslie AGW, Walker JE. The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nature Struct. Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- Gipson B, Zeng X, Zhang ZY, Stahlberg H. 2dx—User-friendly image processing for 2D crystals. J. Struct. Biol. 2007;157:64–72. doi: 10.1016/j.jsb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Glaeser RM. Specimen flatness of thin crystalline arrays: influence of the substrate. Ultramicroscopy. 1992;46:33–43. doi: 10.1016/0304-3991(92)90006-6. [DOI] [PubMed] [Google Scholar]
- Glaeser RM. Retrospective: Radiation damage and its associated “Information Limitations”. J. Struct. Biol. 2008a;163:271–276. doi: 10.1016/j.jsb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Glaeser RM. Cryo-electron microscopy of biological nanostructures. Phys. Today. 2008b;61:48–54. [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid–protein interactions in double layered two dimensional crystals of AQP0. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyobu N, Tani K, Hiroaki Y, Kamegawa A, Mitsuoka K, Fujiyoshi Y. Improved specimen preparation for cryo-electron microscopy using a symmetric carbon sandwich technique. J. Struct. Biol. 2004;146:325–333. doi: 10.1016/j.jsb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Han B-G, Wolf SG, Vonck J, Glaeser RM. Specimen flatness of glucose-embedded biological materials for electron crystallography is affected significantly by the choice of carbon evaporation stock. Ultramicroscopy. 1994;55:1–5. doi: 10.1016/0304-3991(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Hankamer B, Glaeser R, Stahlberg H. Electron crystallography of membrane proteins. J. Struct. Biol. 2007;160:263–264. doi: 10.1016/j.jsb.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Unwin PNT. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Downing KH, Lepault J, Zemlin F. Structure of purple membrane from halobacterium halobium: recording, measurement and evaluation of electron micrographs at 3.5Å resolution. Ultramicroscopy. 1986;19:147–178. [Google Scholar]
- Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Hite RK, Raunser S, Walz T. Revival of electron crystallography. Curr. Opin. Struct. Biol. 2007;17:389–395. doi: 10.1016/j.sbi.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda S, Miyamoto M, Inatomi K-I, Ueyama S. A self-assembled molecular rectifier: two-dimensional crystals of cytochrome c formed on a flavolipid monolayer. Thin Solid Films. 2004;468:255–261. [Google Scholar]
- Jap BK, Walian PJ, Gehring K. Structural architecture of an outer membrane channel as determined by electron crystallography. Nature. 1991;350:167–170. doi: 10.1038/350167a0. [DOI] [PubMed] [Google Scholar]
- Kelly DF, Abeyrathne PD, Dukovski D, Walz T. The Affinity Grid: a pre-fabricated EM grid for monolayer purification. J. Mol. Biol. 2008a;382:423–433. doi: 10.1016/j.jmb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. Monolayer purification: a rapid method for isolating protein complexes for single-particle electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 2008b;105:4703–4708. doi: 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koning RI, Oostergetel GT, Brisson A. Preparation of flat carbon support films. Ultramicroscopy. 2003;94:183–191. doi: 10.1016/s0304-3991(02)00263-2. [DOI] [PubMed] [Google Scholar]
- Kubalek EW, Kornberg RD, Darst SA. Improved transfer of two-dimensional crystals from the air/water interface to specimen support grids for high-resolution analysis by electron microscopy. Ultramicroscopy. 1991;35:295–299. doi: 10.1016/0304-3991(91)90082-h. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W, Downing KH. Two-dimensional structure of plant light-harvesting complex at 3.7 Å resolution by electron crystallography. J. Mol. Biol. 1989;207:823–826. doi: 10.1016/0022-2836(89)90247-7. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W. In: Membrane Protein Purification and Crystallization: A Practical Approach. Schägger H, Hunte C, editors. San Diego: Academic Press; 2003. pp. 253–284. [Google Scholar]
- Lau WCY, Baker LA, Rubinstein JL. Cryo-EM structure of the yeast ATP synthase. J. Mol. Biol. 2008;382:1256–1264. doi: 10.1016/j.jmb.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Lau WCY, Rubinstein JL. Structure of intact Thermus thermophilus V-ATPase by cryo-EM reveals organization of the membrane-bound VO motor. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1367–1372. doi: 10.1073/pnas.0911085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Maire M, Champeil P, Moller JV. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- Lebeau L, Lach F, Vénien-Bryant C, Renault A, Dietrich J, Jahn T, Palmgren MG, Kühlbrandt W, Mioskowski C. Two-dimensional crystallization of a membrane protein on a detergent-resistant monolayer. J. Mol. Biol. 2001;308:639–647. doi: 10.1006/jmbi.2001.4629. [DOI] [PubMed] [Google Scholar]
- Mancheño JM, Martín-Benito J, Gavilanes JG, Vázquez L. A complementary microscopy analysis of Sticholysin II crystals on lipid films: Atomic force and transmission electron characterizations. Biophys Chem. 2006;119:219–223. doi: 10.1016/j.bpc.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-type Na+ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- Mishima O, Calvert LD, Whalley E. An apparent first-order transition between two amorphous phases of ice induced by pressure. Nature. 1985;314:76–78. [Google Scholar]
- Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, 3rd, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller SA, Aebi U, Engel A. What transmission electron microscopes can visualize now and in the future. J. Struct. Biol. 2008;163:235–245. doi: 10.1016/j.jsb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Norville JE, Kelly DF, Knight TF, Jr, Belcher AM, Walz T. 7 Å projection map of the S-layer protein sbpA obtained with trehalose-embedded monolayer crystals. J. Struct. Biol. 2007;160:313–323. doi: 10.1016/j.jsb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D, Stoeckenius W. Rhodposin-like protein from the purple membrane of Halobacterium halobium. Nature New Biol. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- Pomfret AJ, Rice WJ, Stokes DL. Application of the iterative helical real-space reconstruction method to large membranous tubular crystals of P-type ATPases. J. Struct. Biol. 2007;157:106–116. doi: 10.1016/j.jsb.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichow SL, Gonen T. Lipid–protein interactions probed by electron crystallography. Curr. Opin. Struct. Biol. 2009;19:560–565. doi: 10.1016/j.sbi.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Cheng A, Reddy V, Melnyk P, Mitra AK. Three-dimensional fold of the human AQP1 water channel determined at 4 Å resolution by electron crystallography of two-dimensional crystals embedded in ice. J. Mol. Biol. 2000;301:369–387. doi: 10.1006/jmbi.2000.3949. [DOI] [PubMed] [Google Scholar]
- Rhinow D, Kühlbrandt W. Electron cryo-microscopy of biological specimens on conductive titanium–silicon metal glass films. Ultramicroscopy. 2008;108:698–705. doi: 10.1016/j.ultramic.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Richter RP, Brisson AR. Following the Formation of Supported Lipid Bilayers on Mica: A Study Combining AFM, QCM-D, and Ellipsometry. Biophys. J. 2005;88:3422–3433. doi: 10.1529/biophysj.104.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein JL, Dickson VK, Runswick MJ, Walker JE. ATP synthase from Saccharomyces cerevisiae: location of subunit h in the peripheral stalk region. J. Mol. Biol. 2005;345:513–520. doi: 10.1016/j.jmb.2004.10.060. [DOI] [PubMed] [Google Scholar]
- Rubinstein JL, Walker JE. ATP synthase from Saccharomyces cerevisiae: location of the OSCP subunit in the peripheral stalk region. J. Mol. Biol. 2002;321:613–619. doi: 10.1016/s0022-2836(02)00671-x. [DOI] [PubMed] [Google Scholar]
- Rubinstein JL, Walker JE, Henderson R. Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J. 2003;22:6182–6192. doi: 10.1093/emboj/cdg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein JL. Structural analysis of membrane protein complexes by single particle electron microscopy. Methods. 2007;41:409–416. doi: 10.1016/j.ymeth.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Serysheva II, Ludtke SJ, Baker ML, Cong Y, Topf M, Eramian D, Sali A, Hamilton SL, Chiu W. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9610–9615. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Krey I, Kanaoka Y, Mills DJ, Irikura D, Haase W, Lam BK, Austen KF, Kühlbrandt W. Human leukotriene C4 synthase at 4.5Å resolution in projection. Structure. 2004;12:2009–2014. doi: 10.1016/j.str.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Schmidt-Krey I. Electron crystallography of membrane proteins: Two-dimensional crystallization and screening by electron microscopy. Methods. 2007a;41:417–426. doi: 10.1016/j.ymeth.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Schmidt-Krey I, Mutucumarana V, Haase W, Stafford DW, Kühlbrandt W. Two-dimensional crystallization of human vitamin K-dependent γ-glutamyl carboxylase. J. Struct. Biol. 2007b;157:437–442. doi: 10.1016/j.jsb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Stahlberg H, Fotiadis D, Scheuring S, Remigy H, Braun T, Mitsuoka K, Fujiyoshi Y, Engel A. Twod-imensional crystals: a powerful approach to assess structure, function and dynamics of membrane proteins. FEBS Lett. 2001;504:166–172. doi: 10.1016/s0014-5793(01)02746-6. [DOI] [PubMed] [Google Scholar]
- Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Henderson R. Crystallographic analysis of protein conformational changes in the bacteriorhodopsin photocycle. Biochim. Biophys. Acta. 2000;1460:157–165. doi: 10.1016/s0005-2728(00)00136-5. [DOI] [PubMed] [Google Scholar]
- Tani K, Mitsuma T, Hiroaki Y, Kamegawa A, Nishikawa K, Tanimura Y, Fujiyoshi Y. Mechanism of Aquaporin-4's Fast and Highly Selective Water Conduction and Proton Exclusion. J. Mol. Biol. 2009;389:694–706. doi: 10.1016/j.jmb.2009.04.049. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Electron diffraction of frozen hydrated protein crystals. Science. 1974;186:1036–1037. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J. Struct. Biol. 2008;163:214–223. doi: 10.1016/j.jsb.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins PA, Leonhard M, Welzien HU, Wacker T, Welte W. A physical characterization of some detergents of potential use for membrane protein crystallization. FEBS Lett. 1988;238:361–368. [Google Scholar]
- Tocilj A, Munger C, Proteau A, Morona R, Purins L, Ajamian E, Wagner J, Papadopoulos M, Van Den Bosch L, Rubinstein JL, Fethiere J, Matte A, Cygler M. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 2008;15:130–138. doi: 10.1038/nsmb.1374. [DOI] [PubMed] [Google Scholar]
- Tribet C, Audebert R, Popot JL. Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999;283:1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- Unwin PNT, Henderson R. Molecular structure determination by electron microscope of unstained specimens. J. Mol. Biol. 1975;94:425–440. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4Å resolution. J. Mol. Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Uzgiris EE, Kornberg R. Two-dimensional crystallization technique for imaging macromolecules, with application to antigen-antibody-complement complexes. Nature. 1983;301:125–127. doi: 10.1038/301125a0. [DOI] [PubMed] [Google Scholar]
- Vénien-Bryan C, Lenne PF, Zakri C, Renault A, Brisson A, Legrand JF, Berge B. Characterization of the growth of 2D protein crystals on a lipid monolayer by ellipsometry and rigidity measurements coupled to electron microscopy. Biophys. J. 1998;74:2649–2657. doi: 10.1016/S0006-3495(98)77970-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonck J, Krug von Nidda T, Meier T, Matthey U, Mills DJ, Kühlbrandt W, Peter Dimroth P. Molecular Architecture of the Undecameric Rotor of a Bacterial Na+ATP Synthase. J. Mol. Biol. 2002;321:307–316. doi: 10.1016/s0022-2836(02)00597-1. [DOI] [PubMed] [Google Scholar]
- Wang DN, Kühlbrandt W. High-resolution electron crystallography of light-harvesting chlorophyll a/b-protein complex in three different media. J. Mol. Biol. 1991;217:691–699. doi: 10.1016/0022-2836(91)90526-c. [DOI] [PubMed] [Google Scholar]
- Wang L, Sigworth FJ. Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature. 2009;461:292–295. doi: 10.1038/nature08291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens S. F1F0-ATP synthase-stalking mind and imagination. J. Bioenerg Biomembr. 2000;32:333–339. doi: 10.1023/a:1005567717821. [DOI] [PubMed] [Google Scholar]
- Williams DR, Lee K-J, Shi J, Chen DJ, Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals α helices and insight into DNA binding. Strucutre. 2008;15:468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Gipson B, Zheng ZY, Renault L, Stahlberg H. Automatic lattice determination for two-dimensional crystal images. J. Struct. Biol. 2007;160:353–361. doi: 10.1016/j.jsb.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]