Abstract
With MRI possibly becoming a modality of choice for detection and staging of prostate cancer, fast and accurate outlining of the prostate is required in the volume of clinical interest. We present a semi-automatic algorithm that uses a priori knowledge of prostate shape to arrive at the final prostate contour. The contour of one slice is then used as initial estimate in the neighboring slices. Thus we propagate the contour in 3D through steps of refinement in each slice. The algorithm makes only minimum assumptions about the prostate shape. A statistical shape model of prostate contour in polar transform space is employed to narrow search space. Further, shape guidance is implicitly imposed by allowing only plausible edge orientations using template matching. The algorithm does not require region-homogeneity, discriminative edge force, or any particular edge profile. Likewise, it makes no assumption on the imaging coils and pulse sequences used and it is robust to the patient's pose (supine, prone, etc.). The contour method was validated using expert segmentation on clinical MRI data. We recorded a mean absolute distance of 2.0 ± 0.6 mm and dice similarity coefficient of 0.93 ± 0.3 in midsection. The algorithm takes about 1 second per slice.
Keywords: Prostate biopsy, prostate, segmentation, MRI
1. INTRODUCTION
With an approximate annual incidence of 220,000 new cases and 33,000 deaths, prostate cancer continues to be the most common cancer in men in western hemisphere. The definitive diagnostic method of prostate cancer is core needle biopsy. While TRUS imaging has been the gold standard for guiding prostate biopsy, but MRI may also become an attractive choice, owing to its high sensitivity and excellent soft tissue contrast [1]. The biopsy workflow involves fusion of pre-operative and intra-operative MRI acquired in different body positions and with different endorectal coils and pulse sequences. Owing to such unfavorable conditions, outlining the clinical target volume (i.e. the prostate gland) becomes a prerequisite step. Currently, researchers perform the tedious task of manually outlining cross-sectional MRI slices before registering the two datasets [2, 3], which is suboptimal for routine clinical use. Hence the focus of our work is outlining of prostate in intra-operative MRI while accommodating for the variability of conditions and parameters.
Current literature on prostate segmentation focuses on ultrasound [4–11], but works also cater to MRI [12–15]. Pathak et al. [4] presented an edge-guided delineation of prostate in 2D ultrasound images. Abolmaesumi et al. [5] and later, Badiei et al. [6] used spatial Kalman filter with Interacting Multiple Modes Probabilistic Data Association (IMMPDA) filters to detect closed contours of prostate. Deformable models like snakes, level-sets, deformable mesh were proposed [7, 8] to overcome some limitations of purely edge-based algorithms. To further improve and constrain the curve or model evolution, deformable models with shape guidance were introduced [9–12]. These methods are computationally expensive [9]; require considerable user interaction [10]; rely on region or texture homogeneity [11, 12]. Among the works done on MRI, Zwiggelaar et al. [13] presented contouring from 2D MRI slices in polar-transform space using ridge extraction and non-maximal suppression techniques. Zhu et al. [14] employed 2D active shape model in combination with 3D statistical shape modeling to achieve segmentation. Allen et al. [15] combined 3D shape modeling and voxel classification.
With such a significant body of research available, we first attempted to adapt existing techniques to solve our problem. However, they had little success due to: 1) Lack of discriminative image force to drive the model to lock onto true contour as too many contours are present near the true boundary; 2) Absence of reliable region homogeneity and texture; 3) Missing boundary segments in regions of low contrast, particularly in the end slices, where the boundaries of the prostate gland blend into surrounding tissues; 4) Variable edge profile within slice and across slices. To add to an already exigent task, we needed an algorithm that can cope with great variability of imaging parameters across datasets, primarily caused by different endorectal coils and pulse sequences used pre- and intra-operatively. The algorithm sought should meet critical intra-operative requirements: fast, accurate, needs minimal user intervention at initialization, and allows for flexible correction of the results.
The prostate gland, for practical biopsy purpose, consists of the apex, midsection, and base. The midsection encompasses about 90 percent of the prostate. The apex and base, that cap the gland from the bottom and top, respectively, have practically no features or gradients to be seen in MRI. Although some existing segmentation methods attempt to propagate contours from midsection using some statistical shape model, in the absence of discriminating gray level gradient they are always erroneous to some extent and thus need significant manual correction by the clinician. The extent of manual corrections varies among the different methods, but there is always some manual labor at the console. As the apex and base are quite small and in most imaging protocols do not run over more than 2 or 3 anatomically relevant transverse slices, we are not particularly concerned about automatically contouring them and fixing the contour lines manually. We thus concentrated our efforts on contouring the midsection of the gland which, under the aforementioned harsh clinical conditions, is a very challenging task in itself.
2. METHODS
We chose to build upon the ideas of Pathak et al. [4], further automating it, for determination of initial outline of a midsection slice and then proceed in 3D similarly to Wang et al. [8] by iteratively propagating the middlemost slice to the adjacent slices and then repeating the same process toward the base and apex. Pathak et al. [4] requires manual delineation from a set of probable edges on each 2D image, resulting in excessive user intervention. The two approaches [4] and [8] do not suffice alone independently for the reasons mentioned. We, therefore, combined the merits of both, implicitly imposing shape guidance and thus obtaining a significantly more automated process with less user interaction in the end. The complete algorithm is as follows.
The segmentation is initiated with clicking at the approximate center of the prostate in the approximately middlemost slice. We then integrate this information with a priori knowledge of prostate shape in several steps to arrive at the final contour. Then the contour of the middlemost slice is used as the initial estimate for the neighboring slices, where the same steps are repeated all over, as described below.
2.1 A-priori knowledge: Shape model
We incorporate the prostate shape information in a systematic manner by representing contour in polar transform space. Since the prostate is walnut shaped closed object, each point on contour can be represented in terms of radial distance r from the prostate center (xc, yc) and angle θ ranging in interval [0°–360°. Instead of capturing contour radial statistics in four quadrants as in [4], we use angular sections of 9 degrees resulting in statistics for 40 angular sections for each prostate contour. The small angular section was chosen to capture the variation of prostate shape under varying deformations. A mean midsection prostate contour in polar representation is calculated from 54 manually contoured midsection slices, by recording the average radii (ravg(θ)) and its standard deviation (σ(θ)).
2.2 Contrast enhancement and edge enhancement
In our MRI datasets, speckle-like grainy noise arises due to motion artifacts of image acquisition. Conventional edge-detection algorithms on these images result in an excessive number of false edges. Using line segments (called “sticks”) in different angular orientations as templates and selecting the orientation at each point that is most likely to represent a line in the image, it is possible to significantly reduce speckle-like noise and improve edge information, making them more suitable for edge detection [4]. Figure 1b illustrates the effect of noise suppression and contrast enhancement with `sticks'.
Figure 1.
Algorithm steps for processing single slice: contrast enhancement; forming canny edge image; narrowing contour search space using shape model; enforcing edge orientation constraints; clearing overlaps to enforce continuity; filling missing information from shape model; fitting closed spline to get final contour for the slice.
2.3 Initialization
To initiate segmentation, user clicks prostate center in roughly the central slice of prostate. The bounding slices of the prostate are determined using a-priori available information about average prostate volume. The center of average shape model (described earlier) is aligned with user's input. The central slice is set to be the first slice to be segmented using the average shape model as initial estimate. In the next step, a canny edge filter is used to obtain edge-feature image (Figure 1c).
2.4 A-priori knowledge based removal of false contours
entails the following three consecutive steps:
2.4.1 Narrow contour search
In this step, starting from prostate center, a radial search in polar transform space is carried out. The pixels falling at the radial distance from prostate center in the range of [ravg(θ) + 2*σ(θ), ravg(θ) − 2*σ(θ) are kept, and the rest are eliminated. The values for ravg(θ) and σ(θ), which represent average radius of prostate contour from prostate center and corresponding standard deviation at specific angular section determined by θ respectively, are given by the polar representation of the initial estimate. This leaves us with a region of interest with possible prostate candidate edges (Figure 1d).
2.4.2 Enforce edge orientation constraints using templates
After narrowing down to region of interest, further a priori information is incorporated into the algorithm about the expected edge orientation of the prostate contour at different angular sections. The intuition behind this step is that the prostate contour has specific orientation at specific angles from the center. For example, consider angular section in first quadrant; in this section, prostate contour is rising and towards left. Similarly, each angular zone has specific allowed orientation templates. Therefore, to enforce edge-orientation constraint, an angular sweep is made from 0 to 360 degrees, and only those pixels are kept, which are in allowed orientations for that angle. This step results in getting rid of many false contours and breaking connectivity of prostate contour with ill-oriented contours (Figure 1e).
2.4.3 Enforce continuity constraints
The intuition behind this step is that the prostate contour should be continuous and hence cover more angular extent. Thus, when there is more than one contour in a particular angular region overlapping each other, the contour that covers greater angular extent is more likely to be true prostate contour and the smaller overlapping edge fragments are discarded. Also, a contour that does not cover a significant angular extent is likely to be noise and is discarded. After this step, we are left with edge fragments that are strong candidates to be part of true prostate contour (Figure 1f).
2.5 Fill missing information and fit closed spline to data
A final angular sweep is made to see if there is huge angular chunk missing; in which case, the missing information is filled by putting pixels at ravg (θ) from the initial estimate (Figure 1g). The rationale behind this step is to model human way of segmenting who uses the estimate from average shape in places where the information is not so strong or is completely missing. Finally, a closed spline is approximated out of the remaining fragments (Figure 1h).
2.6 Propagation to 3D
The contour estimate obtained of the central slice thus obtained is taken as initial estimate for the next 2D slice in the series. This process (steps 2.4–2.5) is then moved iteratively in both forward and backward directions, using the calculated contour from each slice as initial estimate for the next slice, until all the slices containing prostate are contoured.
2.7 User Interaction
The contour estimates are overlaid on original slices and presented to user for editing. The user is given the ability to quickly snap the contour into exact location through an intuitive interface. We provide intelligent and efficient user interaction in the following manner. If a user corrects the contour in one slice, the correction is iteratively propagated to adjacent slices to calculate new corrected contour estimates. In this way, correction in one slice may in turn, correct many slices. Once the user is satisfied, he approves the contours.
2.8. Stack slices to form volume
In the final step after user approval, the contoured slices are stacked to form the prostate volume.
3. RESULTS AND DISCUSSION
The algorithm was developed in VC++ and executed on Intel Pentium PC running at 1.4 GHz with 1024 MB RAM. Three 3D axial MRI prostate datasets (patient positions: two prone and one supine) acquired using different endorectal coils (coil diameter = 13 mm for two datasets, coil diameter = 26 mm for third) were used for algorithm evaluation. The scans were performed on Philips Intera 3T MRI system; T2-weighted images acquired using Spin Echo (SE) sequence with following parameters: SENSE protocol with acceleration factor of 1; TE/TR = 180 / 7155 ms for two datasets, TE/TR = 120 / 7155 ms for third; matrix 256 × 256; field of view 140 × 140 mm; voxel size 0.55 × 0.55 mm; slice thickness 3 mm; 26 slices. It should be noted that not all 26 slices contain prostate, typically prostate showed up in 13 slices. SENSE technique was used only as a means of obtaining intensity corrected images. Algorithm's performance is compared against the manual segmentations of these datasets by two experts agreeing on the ground truth contour. The results are presented as the output of the algorithm without user correction.
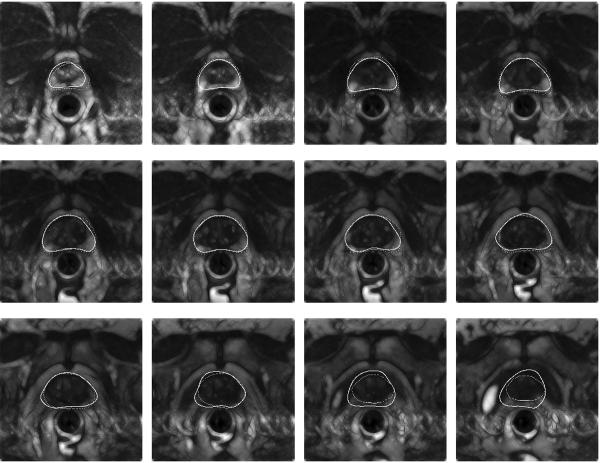
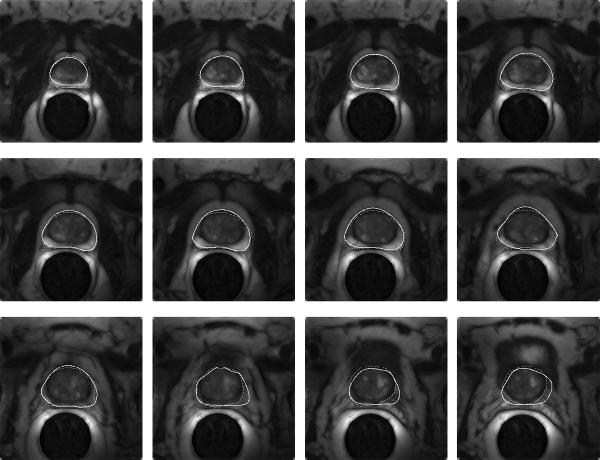
Figure 2 shows the comparison of algorithm-based contouring (solid contour) with manual segmentation (dashed contour) for one of the datasets. Here the algorithm took about 23 seconds to run, of which 10 sec were spent on contrast improvement and then the segmentation of each slice took about 1 sec. Figure 3 shows another dataset with overlaid contours generated by algorithm (solid) and manual (dashed) segmentation.
Figure 2.
Comparison by visual overlay of expert segmentation (dashed) with algorithmic outlining (solid contour) on contrast enhanced slices. The images are consecutive transverse slices ordered from apex (first slice) through midgland, to base (last slice).
Figure 3.
Comparison by visual overlay of expert segmentation (dashed) with algorithmic outlining (solid contour) on contrast enhanced slices. The images are consecutive transverse slices ordered from apex (first slice) through midgland, to base (last slice).
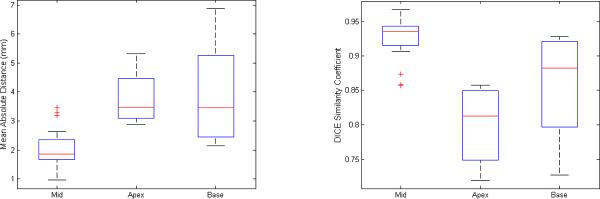
Distance and area based metrics [16] were used to quantitatively assess the accuracy of the algorithm-based segmentation. Distance based metrics measure the difference between two contours, specified as a set of points, calculating Euclidean distances between corresponding points. We used Mean Absolute Distance (MAD) which estimates disagreement averaged over two contours. We used Dice Similarity Coefficient (DSC) as area-based metric, which measures the amount of overlap between the two segmentations (manual and algorithmic). The DSC is appropriate in comparison-of-agreement studies. The results are summarized in box plots of Figure 4. To generate the box plots, the 39 slices over three patients (13×3) were divided into three categories: mid (27), apex (6), and base (6). The average disagreement (MAD) in midsection slices is 2.0 ± 0.6 mm, in apex slices is 3.8 ± 0.9 mm, and in base slices is 3.9 ± 1.8 mm. The overall agreement of expert segmentation with algorithmic is DSC value of 0.93 ± 0.03 in midsection, 0.80 ± 0.05 in apex and 0.86 ± 0.08 in base slices (DSC > 0.7 represents excellent agreement [17]). The results obtained indicate that some user correction will be further required on algorithm's results mostly on apex/base slices to get satisfactory outlines. On the whole, the new algorithm was able to outline the prostate in clinical MRI with sufficient veracity, although there was significant variability of imaging parameters, patient position across the datasets.
Figure 4.
Box plots of Mean Absolute Distance (left) and Dice Similarity Coefficient (right), comparing each for midsection, apex, and base slices. The plots clearly indicate better performance for mid-section slices.
As we expected, the output of the algorithm and the manual ground truth differ in the apex and base. The algorithm (that we specifically designed for the contouring the midsection) performs less accurately in the apex and base, which is apparent from Figure 2, Figure 3 and from the MAD/DSC values in Figure 4. This is consistent with common practice and confirms our initial observation: towards the apex and base, prostatic tissues blend seamlessly into surrounding tissues, causing deterioration and finally complete disappearance of any discriminative grey level information. But again, lack of fidelity in auto-contouring the apex and base is of a lesser concern, because the clinician must always check and modify the contours, which is an unavoidably subjective task. Nonetheless, a stronger shape model could undoubtedly propagate the contours from midgland to apex/base somewhat better. This, in fact, is an issue under current investigation, with the aim of finding a good balance for the strictness of the shape model.
Our algorithm implicitly imposes shape guidance at two steps: 1) Looking at statistically probable radial distance from prostate center at corresponding angles 2) Correction of edge orientation by template matching. The method of propagation of result of a 2D slice into three dimensions by using calculated contour as initial estimate is able to capture large end-to-end variability in prostate shape by capturing smaller slice-to-slice incremental variability. However, there is a flip side of this step: it will also propagate the error to the adjacent slices if the contour on the current slice is incorrect. Also, canny edge detection may not be reliable in some circumstances, and could be replaced by more robust and scale-independent feature extraction filters like Gabor filter bank [9] or dyadic wavelet transform.
4. CONCLUSIONS AND FUTURE WORK
We presented an algorithm for outlining the prostate in MRI volumes acquired with potentially different endorectal coils and imaging parameters. The algorithm requires a single mouse click input of prostate center from the user at initialization. The algorithm has performed sufficiently fast for intra-operative use. Performance on clinical data indicates that minimal user correction is required, mostly in the apex and also in the base, which has been of no particular concern in actual practice. Importantly, the algorithm has been robust to variations of signal quality due to the use of different endorectal coils, field strengths, imaging sequences, and patient poses.
Various exciting future directions emerge from here and are being evaluated. One possible direction is use of 3D shape models, not compromising on time performance. A multi-segment 3D prostate shape model consisting of base, midsection and apex as segments that can vary independently, is under consideration to more effectively capture the shape variability of prostate. Another possibility is incorporating edge-profile information inside the 3D shape model, as in our case, we have variable edge profile in mid-section, base and apex regions. This also opens up a new challenge of finding metrics for model-to-image registration/segmentation.
We presently are processing a more extensive set of MRI studies with multiple expert segmentations for ground truth. Upon completing this data preparation step, inter-observer disagreements could be calculated and multiple observer studies will support more conclusive statements about curacy claims in regions where no apparent ground truth exists. In this regard, Warfield's definitive work on validation of image segmentation and expert quality should be followed [18].
Another, highly relevant study is currently underway to quantify the actual effects of miscontouring on the clinical targeting accuracy when contour-based data fusion is used. The overall objective here is to derive upper limits for the targeting inaccuracy in the presence of contour errors in the pre-operative and intra-operative datasets.
Acknowledgments
SUPPORT: NIH 1R01EB002963, NIH 1R01CA111288
REFERENCES
- [1].Adusumilli S, Pretorius ES. Magnetic resonance imaging of prostate cancer. Semin Urol Oncol. 2002;20:192–210. doi: 10.1053/suro.2002.35333. [DOI] [PubMed] [Google Scholar]
- [2].d'Aische AD, De Craene M, Haker S, Weisenfeld N, Tempany C, Macq B, Warfield SK. Lecture Notes in Computer Science. Vol. 3216. Springer-Verlag; Berlin Heidelberg New York: 2004. Improved non-rigid registration of prostate MRI; pp. 845–852. [Google Scholar]
- [3].Alterovitz R, Goldberg K, Pouliot J, Hsu IC, Kim Y, Norworolski SM, Kurhanewicz J. Registration of MR prostate images with biomechanical modelling and nonlinear parameter estimation. Med Phy. 2006;33:446–454. doi: 10.1118/1.2163391. [DOI] [PubMed] [Google Scholar]
- [4].Pathak SD, Chalana V, Haynor DR, Kim Y. Edge-guided boundary delineation in prostate ultrasound images. IEEE Trans. Med. Imag. 2000;19:1211–1219. doi: 10.1109/42.897813. [DOI] [PubMed] [Google Scholar]
- [5].Abolmaesumi P, Sirouspour M. An interacting multiple model probabilistic data association filter for cavity boundary extraction from ultrasound images. IEEE Trans. Med. Imag. 2004;23:772–784. doi: 10.1109/tmi.2004.826954. [DOI] [PubMed] [Google Scholar]
- [6].Badiei S, Salcudean SE, Varah J, Morris WJ. Lecture Notes in Computer Science. Vol. 4191. Springer-Verlag; Berlin Heidelberg New York: 2006. Prostate segmentation in 2D ultrasound images using image warping and ellipse fitting; pp. 17–24. [DOI] [PubMed] [Google Scholar]
- [7].Hu N, Downey DB, Fenster A, Ladak HM. Prostate boundary segmentation from 3D ultrasound images. Med Phy. 2003;30:1648–1659. doi: 10.1118/1.1586267. [DOI] [PubMed] [Google Scholar]
- [8].Wang Y, Cardinal HN, Downey DB, Fenster A. Semi-automatic 3D segmentation of prostate using 2D US images. Med Phy. 2003;30:887–897. doi: 10.1118/1.1568975. [DOI] [PubMed] [Google Scholar]
- [9].Shen D, Davastikoz C. Segmentation of prostate boundaries from ultrasound images using statistical shape model. IEEE Trans. Med. Imag. 2003;22:539–551. doi: 10.1109/TMI.2003.809057. [DOI] [PubMed] [Google Scholar]
- [10].Gong L, Pathak SD, Haynor DR, Cho PS, Kim Y. Parametric shape modeling using deformable super-ellipses for prostate segmentation. IEEE Trans. Med. Imag. 2004;23:340–349. doi: 10.1109/TMI.2004.824237. [DOI] [PubMed] [Google Scholar]
- [11].Gong L, Ng L, Pathak SD, Tutar I, Cho PS, Haynor DR, Kim Y. Prostate ultrasound image segmentation using level set bases region flow with shape guidance. Proc. of SPIE. 2005;5747:1648–1657. [Google Scholar]
- [12].Tsai A, Yezzi A, Wells W, Tempany C, Tucker D, Fan A, Grimson WE, Willsky A. A shape-based approach to the segmentation of medical imagery using level sets. IEEE Trans. Med. Imag. 2003;22:137–154. doi: 10.1109/TMI.2002.808355. [DOI] [PubMed] [Google Scholar]
- [13].Zwiggelaar R, Zhu Y, Williams S. Lecture Notes in Computer Science. Vol. 2652. Springer-Verlag; Berlin Heidelberg New York: 2003. Semi-automatic segmentation of the prostate; pp. 1108–1116. [Google Scholar]
- [14].Zhu Y, Williams S, Zwiggelaar R. Segmentation of volumetric prostate MRI data using hybrid 2D+3D shape modeling. Proc. of Medical Image Understanding and Analysis. 2004:61–64. [Google Scholar]
- [15].Allen PD, Graham J, Williamson DC, Hutchinson CE. Differential segmentation of the prostate in MR images using combined 3D shape modeling and voxel classification. ISBI IEEE. 2006:410–413. [Google Scholar]
- [16].Chalana V, Kim Y. A methodology for evaluation of boundary detection algorithms on medical images. IEEE Trans. Med. Imag. 1997;16:642–652. doi: 10.1109/42.640755. [DOI] [PubMed] [Google Scholar]
- [17].Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC. Morphometric analysis of white matter lesions in MR images: Method and validation. IEEE Trans. Med. Imag. 1994;13:716–724. doi: 10.1109/42.363096. [DOI] [PubMed] [Google Scholar]
- [18].Warfield SK, Zou KH, Wells WM. Lecture Notes in Computer Science. Vol. 2652. Springer-Verlag; Berlin Heidelberg New York: 2002. Validation of image segmentation and expert quality with an Expectation-Maximization algorithm; pp. 298–306. [Google Scholar]