Abstract
Purpose
Pandemic influenza A (H1N1) virus has spread rapidly and prompt diagnosis is needed for successful treatment and prevention of transmission. We investigated clinical predictors, validated the use of previous criteria with laboratory tests, and evaluated the clinical criteria for H1N1 infection in the Korean population.
Materials and Methods
We analyzed clinical and laboratory evaluation data from outpatient clinics at Severance Hospital in Seoul, Korea between November 11 and December 5, 2009.
Results
This analysis included a total of 828 patients. Of these, 372 (44.9%) patients were confirmed with H1N1 infection by real-time reverse transcriptase-polymerase chain reaction (RT-PCR). The most common and predictive symptom was cough (90.3%, OR 8.87, 95% CI 5.89-13.38) and about 40% of H1N1-positive patients were afebrile. The best predictive model of H1N1 infection was cough plus fever or myalgia. The sensitivities, specificities, positive predictive values, and negative predictive values of our suggested criteria were 73.9%, 69.5%, 66.4%, and 76.6%, respectively.
Conclusion
Cough was the most common independent symptom in patients with laboratory-confirmed H1N1 infection, and while not perfect, the combination of cough plus fever or myalgia is suggested as clinical diagnostic criteria. Health care providers in Korea should suspect a cough without fever to be an early symptom of H1N1 infection.
Keywords: Influenza A virus (H1N1 subtype), criteria, diagnosis
INTRODUCTION
A novel influenza A (H1N1) virus spread widely in 2009 and emerged as a global health problem.1 Although most cases were self-limiting, H1N1 influenza spread more rapidly than seasonal influenza2 in 2009 and fatalities occurred in many countries.3 During a pandemic, appropriate and prompt diagnosis are critical for successful treatment and prevention of transmission.4 Although several laboratory tests, including real-time reverse transcriptase-polymerase chain reaction (RT-PCR), are known to be confirmatory tests for novel influenza A (H1N1),5 clinical judgment plays a major role in the timely identification of influenza, and laboratory tests are not always available during an influenza outbreak.6,7 Several organizations have documented their own clinical diagnostic criteria,8 and these criteria vary with regard to sensitivity and predictive values for laboratory confirmed influenza.9 However, most studies have validated the use of clinical diagnostic criteria only for seasonal influenza infection,6,10 and to the best of our knowledge, have never been evaluated for influenza A (H1N1). This study was conducted to evaluate the best clinical predictors for influenza A (H1N1) infection and to compare different clinical diagnostic criteria for influenza A (H1N1) with laboratory diagnostic tests.
MATERIALS AND METHODS
Data collection
This study was approved by the Severance Hospital Ethics Committee. Clinical and laboratory evaluations were analyzed from all patients who were admitted to the flu center [sentinel surveillance outpatient clinic for influenza A (H1N1) infection] at Severance Hospital. The study period was from November 11 to December 5, 2009. Individuals older than 18 years of age with clinical evidence of acute respiratory symptoms over a duration of less than 7 days were included in this study. Acute respiratory symptoms were defined as self-reported influenza-like symptoms including chills/feverishness, cough, sore throat, headache, rhinorrhea, and/or myalgia, with or without a documented fever.11 We also included afebrile individuals (< 37.8℃) who had been taking antipyretics in the previous 12 hours. Fever was defined as a body temperature above 37.8℃
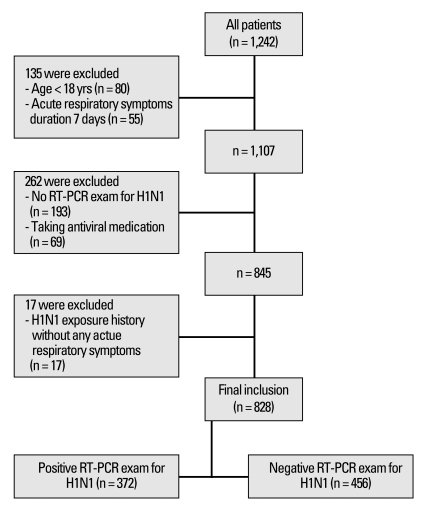
The analysis excluded individuals with a history of contact with a confirmed influenza A (H1N1) patient without any acute respiratory symptoms. We also excluded individuals who had not been confirmed by RT-PCR and individuals who had been taking antiviral medication (Fig. 1). Medical records were analyzed for demographic data, history of contact with influenza A (H1N1) within 7 days, antiviral medication history, and pre-existing medical conditions including pregnancy. Clinical symptoms and lifestyle behaviors such as cigarette smoking, alcohol consumption, and physical activity were also recorded. Body mass index (BMI) was calculated as weight (kg)/height (m2).
Fig. 1.
Study diagram for influenza A (H1N1) clinical criteria comparisons and suggestions. Clinical and laboratory evaluations were analyzed from all patients who were admitted to the flu center at Severance Hospital. The study period was from November 11 to December 5, 2009. Acute respiratory symptoms were defined as self-reported influenza-like symptoms including chills/feverishness, cough, sore throat, headache, rhinorrhea, and/or myalgia, with or without a documented fever. RT-PCR, reverse transcriptase-polymerase chain reaction.
We analyzed several clinical diagnostic criteria of H1N1 influenza. The Centers for Disease Control and Prevention (CDC), in US12 and Korea,13 defined the clinical diagnostic criteria as the presence of fever (greater than 37.8℃ or previous medication with antipyretics) plus one or more of the following: rhinorrhea, nasal congestion, sore throat or cough. The World Health Organization (WHO)14 defined the clinical diagnostic criteria as the sudden onset of fever (> 38℃) and cough or sore throat. Influenza like illness (ILI) criteria is defined by the presence of fever plus two of the following four symptoms: cough, sore throat, myalgia, and headache.10
Laboratory assessment
Specimens were collected using nasopharyngeal swabs by trained physicians. An influenza A (H1N1)-positive case was defined as a positive result on real-time RT-PCR using the Lightcycler 480 (Roche, Mannheim, Germany).15 Conversely, a negative result on RT-PCR was defined as influenza A (H1N1)-negative.
Statistical analysis
Continuous variables are expressed as mean ± standard deviation or medians (with interquartile ranges), and categorical variables are expressed as percentages. Demographic and clinical characteristics were compared between influenza A (H1N1)-positive cases and influenza A (H1N1)-negative cases using the independent t-test for continuous variables and the Chi-square test for categorical variables. The most predictive clinical variables of an illness attributable to influenza A (H1N1) were determined with forward stepwise logistic regression models. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and area under the curve (AUC) were calculated to identify the best clinical diagnostic criteria (suggested criteria) of influenza A (H1N1) infection. We also compared sensitivity, specificity, PPV, NPV, AUC of the influenza like illness (ILI) criteria,7 the Korea CDC criteria,13 and the WHO criteria14 with our suggested criteria. All analyses were conducted using SAS statistical software, version 9.0 (SAS Institute Inc., Cary, NC, USA). All statistical tests were two-sided, and statistical significance was defined as p < 0.05.
RESULTS
Demographic and clinical characteristics
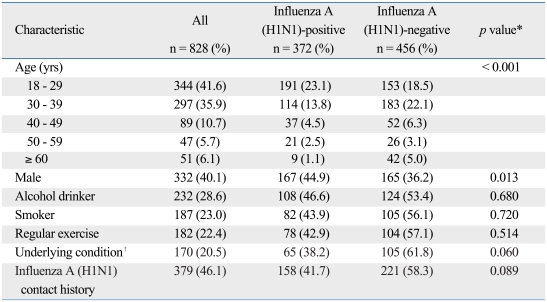
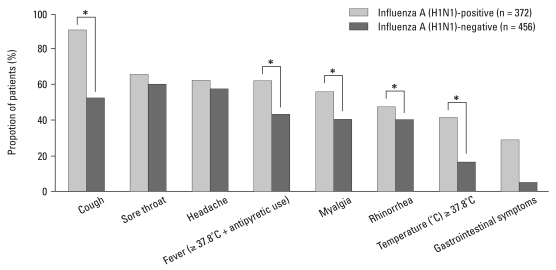
A total of 828 patients were included in the analysis, and 372 (44.9%) patients were confirmed to have influenza A (H1N1) infection by real-time RT-PCR. The median age of all patients was 32 years old (18-81 years old), and younger patients were more susceptible than the elderly [influenza A (H1N1)-positive: 31.6 ± 10.72 years vs. negative: 36.3 ± 13.04 years, p < 0.001]. BMI was not significantly different between the two groups [influenza A (H1N1)-positive: 22.6 ± 3.74 kg/m2 vs. negative: 22.4 ± 3.19 kg/m2, p = 0.442]. Influenza A (H1N1) was more frequently diagnosed in women than in men (p = 0.013), and 38.2% of patients with a confirmed infection had underlying conditions [asthma, 8 (2.2%); chronic pulmonary disease, 4 (1.1%); coronary heart disease, 7 (1.9%); hypertension, 15 (4.1%); DM, 5 (1.4%); thyroid disease, 6 (1.6%); chronic renal failure, 3 (0.8%); cancer, 11 (3.0%); immune deficiency disease, 4 (1.1%); cerebrovascular disease, 1 (0.3%); chronic liver disease, 2 (0.5%); pregnancy 11 (3.0%)] (Table 1). The most frequently reported symptom in the influenza A (H1N1)-positive group was cough (336, 90.3% of patients), followed by sore throat (246, 66.1% of patients), and headache (246, 66.1% of patients). A total of 139 (37.4%) influenza A (H1N1)-positive patients did not have a fever or history of antipyretic use. Cough, myalgia, fever (temperature greater than 37.8℃ or antipyretic use within 12 hours before visit), and rhinorrhea were more frequent in the influenza A (H1N1)-positive group (Fig. 2). The mean duration of the influenza A (H1N1)-positive illness was 2.1 days.
Table 1.
Demographic Characteristics of 828 Patients Admitted to the Flu Center at Severance Hospital with Acute Respiratory Symptoms (November 11 - December 5, 2009)
An alcohol drinker was defined as a person with current alcohol intake more than once a week and a smoker was defined as a current cigarette smoker. Regular exercise was defined as exercise or physical work for more than 30 minutes three times a week.
RT-PCR, reverse transcriptase-polymerase chain reaction.
*Influenza A (H1N1)-positive was defined by a positive result on real-time RT-PCR. Conversely, a negative result on RT-PCR was defined as influenza A (H1N1)-negative. p values between influenza A (H1N1)-positive and negative were calculated with the t-test, Wilcoxon's rank sum test (for continuous variables) or χ2-test, and Fisher exact test (for categorical variables).
†Underlying conditions: asthma, chronic pulmonary disease, coronary heart disease, Hypertension, Diabetes Mellitus, thyroid disease, chronic renal failure, cancer, immune deficiency disease, cerebrovascular disease, chronic liver disease or pregnancy.
Fig. 2.
Clinical symptoms and signs of influenza A (H1N1)-positive cases and influenza A (H1N1)-negative cases. Influenza A (H1N1)-positive was defined as a positive result on real-time RT-PCR. Conversely, a negative result on RT-PCR was defined as influenza A (H1N1)-negative. Cough, myalgia, fever (greater than 37.8℃ or antipyretic use), and rhinorrhea are more frequent in the influenza A (H1N1)-positive group. *p < 0.05, calculated by the χ2-test and Fisher exact test (for categorical variables). RT-PCR, reverse transcriptase-polymerase chain reaction.
Comparison of current clinical diagnostic criteria and our suggested criteria
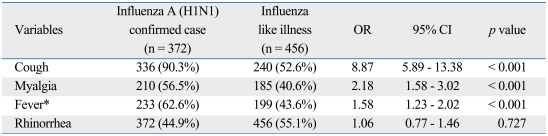
Stepwise logistic regression showed that cough, myalgia, and fever (≥ 37.8℃ or the use of antipyretics within 12 hours before visit) were the only factors significantly associated with a positive PCR test for influenza A (H1N1). The best predictive symptom of influenza A (H1N1) infection was cough. When a history of antipyretics use was not included in the model, there were no statistically significant differences (data not shown) (Table 2).
Table 2.
Clinical Predictors of Influenza A (H1N1) Infection by Multivariate Stepwise Logistic Regression Analysis
Variables included in the stepwise model: age, sex, fever, cough, sore throat, myalgia, headache, rhinorrhea, and gastrointestinal symptoms (nausea, vomiting, diarrhea).
OR, odds ratio; CI, confidence interval.
*Body temperature ≥ 37.8℃ or the use of an antipyretic within 12 hours before visit.
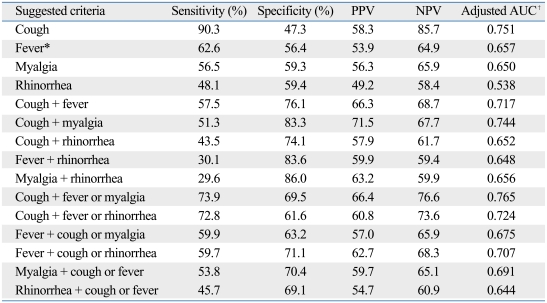
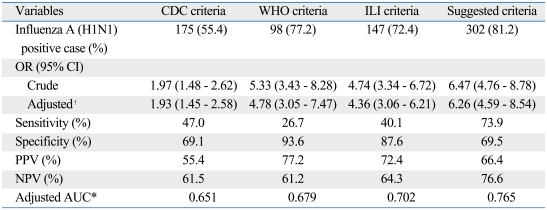
The Korean CDC criteria for 2009 pandemic influenza A (H1N1) were defined by the presence of fever (greater than 37.8℃ or previous medication with antipyretics) plus one or more of the following: rhinorrhea or nasal congestion, sore throat, and cough. However, about 40% of the influenza A (H1N1)-positive patients in this study were afebrile and only 55.4% fulfilled the clinical criteria of the CDC. In order to identify a more appropriate clinical criteria, we examined sensitivity, specificity, PPV, and NPV in combination with cough, fever, myalgia, and rhinorrhea (Table 3). The sensitivity for cough was 90.3%, but the specificity and PPV were relatively lower than others in combination (specificity 47.3%, PPV 58.3%). Considering sensitivity and PPV together, cough plus fever or myalgia was the best predictive model for influenza A (H1N1) infection (sensitivity 73.9%, PPV 66.4%). More than 80% of patients with influenza A (H1N1) met these suggested criteria, and the sensitivity of these suggested criteria (sensitivity 73.9%) was higher than the Korea CDC criteria, WHO criteria, and ILI criteria (Table 4).
Table 3.
Sensitivity, Specificity, Positive Predictive Predictive Value (PPV), and Negative Predictive Value (NPV) of Suggested Clinical Criteria of Influenza A (H1N1)
AUC, area under the curve.
*Body temperature ≥ 37.8℃ or the use of an antipyretic within 12 hours before visit.
†Age, sex adjusted AUC.
Table 4.
Validation of Current Diagnostic Criteria for Influenza A (H1N1) and Comparison with our Suggested Criteria
The CDC criteria are the presence of fever (> 37.8℃ or previous medication with antipyretics) plus one or more of the following: rhinorrhea, nasal congestion, sore throat, or cough. The WHO criteria are the sudden onset of fever (> 38℃) and cough or sore throat. The ILI criteria are the presence of fever (≥ 37.8℃) plus two of the following four symptoms: cough, sore throat, myalgia, and headache. Our suggested criteria are the presence of cough plus fever or myalgia.
OR, odds ratio; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; AUC, area under the curve; ILI, influenza like illness.
*Age, sex adjusted OR, AUC.
DISCUSSION
Since early April 2009, the unique genetic and antigenic features of influenza A (H1N1) have resulted in a more rapid spread than is typical of seasonal influenza,16 and the epidemiologic profile of H1N1 has varied considerably depending on the country and continent being examined.17 During major outbreaks, early and accurate diagnosis of influenza has the ability to ensure prompt and appropriate treatment and can ultimately decrease the economic and public health burden. Several different diagnostic criteria have been used worldwide for surveillance purposes, but the sensitivity and positive predictive value of such criteria vary among studies.7 In addition, there are few data available to assess the diagnostic criteria associated with pandemic influenza A (H1N1) infection.
In Korea, the Korean CDC recommends that antiviral medication be considered for patients with an acute febrile respiratory illness (fever: ≥ 37.8℃ or the use of antipyretics within 12 hours before visit, plus one or more of the following: rhinorrhea or nasal congestion, sore throat, and cough) for the 2009 pandemic influenza A (H1N1). According to our data, diagnostic criteria that focus on fever are inappropriate and may miss about 40% of all influenza infections, because many of the patients in our sample had afebrile influenza A (H1N1) infections. The CDC, WHO, and ILI criteria were specific enough to differentiate suspected cases but were not sensitive enough to detect all cases. When there is a pending influenza A (H1N1) pandemic, the use of clinical diagnostic criteria with high sensitivity is important for early detection and isolation of suspected individuals.15
While fever was a common symptom in previous studies,2 the most frequent symptom in our study was cough (90.3%). Moreover, the most predictive model of influenza A (H1N1) infection was cough plus fever or myalgia. The sensitivity of our suggested criteria was 73.9%, the specificity was 69.5%, the PPV was 66.4%, the NPV was 76.6% and the age and sex adjusted AUC was 0.765. Screening patients with our criteria for ILI would only have missed 19% of cases of pandemic influenza A (H1N1).
We did not analyze other subtypes of influenza viruses, nor did we analyze nonviral pathogens with the ability to cause acute respiratory illness.7 Therefore, these criteria may only be valid for future H1N1 pandemics and may not be relevant to pandemics with other influenza strains and respiratory viruses. The small sample size and the absence of children from the sample are also limitations of the current study. A much larger patient sample that includes children is required to more accurately represent the characteristics of influenza A (H1N1) infection.
In summary, cough was the best independent indicator in laboratory-confirmed influenza A (H1N1), and while not perfect, a combination of cough plus fever or myalgia is suggested as diagnostic criteria. According to our data, this suggested set of criteria demonstrates better sensitivity than of the conventional criteria used to diagnose H1N1. Health care providers in Korea should suspect cough without fever to be early symptoms of influenza A (H1N1) infection.
Footnotes
The authors have no financial conflicts of interest.
References
- 1.Carrat F, Tachet A, Rouzioux C, Housset B, Valleron AJ. Evaluation of clinical case definitions of influenza: detailed investigation of patients during the 1995-1996 epidemic in France. Clin Infect Dis. 1999;28:283–290. doi: 10.1086/515117. [DOI] [PubMed] [Google Scholar]
- 2.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 3.Sym D, Patel PN, El-Chaar GM. Seasonal, avian, and novel H1N1 influenza: prevention and treatment modalities. Ann Pharmacother. 2009;43:2001–2011. doi: 10.1345/aph.1M557. [DOI] [PubMed] [Google Scholar]
- 4.Call SA, Vollenweider MA, Hornung CA, Simel DL, McKinney WP. Does this patient have influenza? JAMA. 2005;293:987–997. doi: 10.1001/jama.293.8.987. [DOI] [PubMed] [Google Scholar]
- 5.Jain R, Goldman RD. Novel influenza A(H1N1): clinical presentation, diagnosis, and management. Pediatr Emerg Care. 2009;25:791–796. doi: 10.1097/PEC.0b013e3181c3c8f8. [DOI] [PubMed] [Google Scholar]
- 6.Monto AS, Gravenstein S, Elliott M, Colopy M, Schweinle J. Clinical signs and symptoms predicting influenza infection. Arch Internal Med. 2000;160:3243–3247. doi: 10.1001/archinte.160.21.3243. [DOI] [PubMed] [Google Scholar]
- 7.Boivin G, Hardy I, Tellier G, Maziade J. Predicting influenza infections during epidemics with use of a clinical case definition. Clin Infect Dis. 2000;31:1166–1169. doi: 10.1086/317425. [DOI] [PubMed] [Google Scholar]
- 8.Gerrard J, Keijzers G, Zhang P, Vossen C, Macbeth D. Clinical diagnostic criteria for isolating patients admitted to hospital with suspected pandemic influenza. Lancet. 2009;374:1673. doi: 10.1016/S0140-6736(09)61983-8. [DOI] [PubMed] [Google Scholar]
- 9.Bellei N, Carraro E, Perosa A, Watanabe A, Arruda E, Granato C. Acute respiratory infection and influenza-like illness viral etiologies in Brazilian adults. J Med Virol. 2008;80:1824–1827. doi: 10.1002/jmv.21295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro-Marí JM, Pérez-Ruiz M, Cantudo-Muñoz P, Petit-Gancedo C, Jiménez-Valera M, Rosa-Fraile M. Influenza Surveillance Network in Andalusia, Spain. Influenza-like illness criteria were poorly related to laboratory-confirmed influenza in a sentinel surveillance study. J Clin Epidemiol. 2005;58:275–279. doi: 10.1016/j.jclinepi.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Ong AK, Chen MI, Lin L, Tan AS, Nwe NW, Barkham T, et al. Improving the clinical diagnosis of influenza--a comparative analysis of new influenza A (H1N1) cases. PloS One. 2009;4:e8453. doi: 10.1371/journal.pone.0008453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Interim guidance for infection control for care of patients with confirmed or suspected novel influenza A (H1N1) virus infection in a healthcare setting. Centers for Disease Control and Prevention. [Accessed November 30, 2009]. http://www.cdc.gov/h1n1flu/guidelines_infection_control.htm.
- 13.Diagnosis and Treatment of Influenza A (H1N1) Korea Centers for Disease Control and Prevention. [Accessed December 2, 2009]. http://flu.cdc.go.kr.
- 14.World Health Organization. Human infection with pandemic (H1N1) 2009 virus: updated interim WHO guidance on global surveillance. 2009. [Google Scholar]
- 15.Lorusso A, Faaberg KS, Killian ML, Koster L, Vincent AL. One-step real-time RT-PCR for pandemic influenza A virus (H1N1) 2009 matrix gene detection in swine samples. J Virol Methods. 2010;164:83–87. doi: 10.1016/j.jviromet.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang LY, Shih SR, Shao PL, Huang DT, Huang LM. Novel swine-origin influenza virus A (H1N1): the first pandemic of the 21st century. J Formos Med Assoc. 2009;108:526–532. doi: 10.1016/S0929-6646(09)60369-7. [DOI] [PubMed] [Google Scholar]
- 17.Vaillant L, La Ruche G, Tarantola A, Barboza P epidemic intelligence team at InVS. Epidemiology of fatal cases associated with pandemic H1N1 influenza 2009. Euro Surveill. 2009;14:pii: 19309. doi: 10.2807/ese.14.33.19309-en. [DOI] [PubMed] [Google Scholar]