Abstract
We studied the function of Arabidopsis (Arabidopsis thaliana) Botrytis Susceptible1 Interactor (BOI) in plant responses to pathogen infection and abiotic stress. BOI physically interacts with and ubiquitinates Arabidopsis BOS1, an R2R3MYB transcription factor previously implicated in stress and pathogen responses. In transgenic plants expressing the BOS1-β-glucuronidase transgene, β-glucuronidase activity could be detected only after inhibition of the proteosome, suggesting that BOS1 is a target of ubiquitin-mediated degradation by the proteosome. Plants with reduced BOI transcript levels generated through RNA interference (BOI RNAi) were more susceptible to the necrotrophic fungus Botrytis cinerea and less tolerant to salt stress. In addition, BOI RNAi plants exhibited increased cell death induced by the phytotoxin α-picolinic acid and by a virulent strain of the bacterial pathogen Pseudomonas syringae, coincident with peak disease symptoms. However, the hypersensitive cell death associated with different race-specific resistance genes was unaffected by changes in the level of BOI transcript. BOI expression was enhanced by B. cinerea and salt stress but repressed by the plant hormone gibberellin, indicating a complex regulation of BOI gene expression. Interestingly, BOI RNAi plants exhibit reduced growth responsiveness to gibberellin. We also present data revealing the function of three Arabidopsis BOI-RELATED GENES (BRGs), which contribute to B. cinerea resistance and the suppression of disease-associated cell death. In sum, BOI and BRGs represent a subclass of RING E3 ligases that contribute to plant disease resistance and abiotic stress tolerance through the suppression of pathogen-induced as well as stress-induced cell death.
Plants recruit diverse cellular regulatory mechanisms to respond to various internal and external signals. Ubiquitin-mediated modification of proteins is a widespread regulatory mechanism that is important for normal cellular functions and plant responses to environmental signals. In Arabidopsis (Arabidopsis thaliana), nearly 6% of the proteome is devoted to ubiquitin-mediated regulatory changes (Vierstra, 2009), suggesting the importance of this pathway for normal cellular processes. Three enzymes, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3), act in consecutive steps to transfer and covalently ligate the 76-amino acid protein ubiquitin to target proteins. E3 ligases provide the specificity by recruiting target proteins for ubiquitination (Smalle and Vierstra, 2004). The Arabidopsis genome contains more than 1,300 E3 ligases, of which at least 469 are predicted RING domain-containing E3 ubiquitin ligases (Stone et al., 2005). The attachment of a polyubiquitin chain of four or more ubiquitins linked via ubiquitin lysyl residue 48 are typically targeted for degradation by the 26S proteosome (Thrower et al., 2000). Modification with a lysyl-63-linked polyubiquitin chain has been implicated in protein activation and stress response (Shi and Kehrl, 2003), whereas monoubiquitination of proteins regulates transcription, receptor internalization, and endosomal sorting independent of proteolysis (Liu et al., 2005). RING E3 ligases studied to date are implicated in hormone signaling, plant development, responses to abiotic and biotic stresses, and the regulation of cell death (Smalle and Vierstra, 2004; Kawasaki et al., 2005; Dong et al., 2006). However, the physiological functions of most E3 ligases have yet to be determined.
Ubiquitination-mediated protein removal or other regulatory changes have emerged as important components of host defense and counterdefense by pathogens (Zeng et al., 2006). Treatment of plants with the proteosome inhibitor MG132 caused increased susceptibility to the necrotrophic fungus Plectosphaerella cucumerina (Llorente et al., 2008). The Arabidopsis RIN2 and RIN3 RING E3 ligases affect the extent of hypersensitive response (HR) mediated by RPM1 and RPS2 (Kawasaki et al., 2005). The tobacco (Nicotiana tabacum) ACRE276 and its Arabidopsis homolog PUB17, encoding U-BOX E3 ubiquitin ligases, are positive regulators of HR (Yang et al., 2006). The loss-of-function mutation in the rice (Oryza sativa) SPOTTED LEAF11 (SPL11) gene encoding an E3 ligase causes lesion-mimic phenotypes and confers resistance to the hemibiotrophic pathogens Magnaporthe grisea and Xanthomonas oryzae pv oryzae, indicating that SPL11 is a suppressor of cell death (Zeng et al., 2004). A membrane-bound E3 ligase that is associated with lipid rafts of the plasma membrane was recently described (Lin et al., 2008). The knockdown of RING1 leads to reduced sensitivity to the fungal toxin fumonisin B1, whereas overexpression of RING1 confers hypersensitivity. The RING E3 ligase HISTONE MONOUBIQUTINATION1 contributes to resistance to necrotrophic fungi independent of proteolysis (Dhawan et al., 2009). Thus, some RING ligases play a role in pathogen defense and the suppression of cell death, although the mechanisms and their targets are not known.
Cell death is a ubiquitous phenomenon during plant-pathogen interactions, occurring in both susceptible and resistant responses. Cell death also occurs in response to abiotic stresses, senescence, and other physiological and developmental processes. The most common form of cell death is the HR, a plant resistance reaction against strains of biotrophic pathogens carrying effector molecules. Plant resistance proteins and pathogen effectors are key determinants of resistance-related HR, although many modulating components are known, including plant hormones and reactive oxygen species. Necrotrophic pathogens cause phytotoxin-mediated host cell death (Wolpert et al., 2002). The distinction between cell death caused by necrotrophic pathogens, HR, and other forms of programmed cell death at the molecular and symptom level is unclear.
In Arabidopsis, many genes regulate cell death associated with disease resistance and susceptibility. A common genetic control for disease susceptibility and resistance-associated cell death does exist. The plant mitogen-activated protein kinase kinase kinase-α is a positive regulator of cell death associated with both plant resistance and susceptibility (del Pozo et al., 2004). The Arabidopsis LSD1 is a zinc finger protein required to limit cell death initiated by various signals (Jabs et al., 1996). Many more mutations define the genetic control of different forms of cell death (Greenberg and Yao, 2004). The contribution of cell death to the plant immune response is dependent on the nutrient acquisition strategy of the invading pathogen. As necrotrophic fungi are adapted to extract nutrients from dead cells, HR cell death enhances pathogen growth and colonization (Govrin and Levine, 2000). Plant mutants with enhanced cell death have increased resistance to biotrophic pathogens but susceptibility to necrotrophic fungi (Kachroo et al., 2001; Veronese et al., 2004).
Previously, we described the Arabidopsis Botrytis Susceptible1 (BOS1) gene encoding an R2R3MYB transcription factor that is required for resistance to pathogens and tolerance to certain abiotic stress factors (Mengiste et al., 2003). Here, we studied the role of Botrytis Susceptible1 Interactor (BOI) and three BOI-RELATED GENES (BRGs) encoding a small subclass of RING E3 ligases. BOI physically interacts with BOS1, has E3 ligase activity, and ubiquitinates BOS1 in vitro. BOI RNA interference (RNAi) plants are more susceptible to Botrytis cinerea and less tolerant to salt stress than the wild-type plants, consistent with the phenotypes of the bos1 mutant allele (Mengiste et al., 2003). BOI is required to restrict the extent of cell death induced by the fungal toxin α-picolinic acid (PA), a known inducer of cell death in animal and plant cells, as well as disease-associated cell death caused by a virulent strain of Pseudomonas syringae. Ectopic expression of BOI reduced toxin-induced cell death but failed to restrict HR cell death mediated by the disease resistance genes RPM1 and RPS2, thus limiting the role of BOI to certain types of cell death. The BOI RNAi plants exhibit reduced growth responses to GA, suggesting a link between reduced growth responsiveness to GA and disease and stress tolerance. Together, our data suggest that Arabidopsis BOI and BOI-related RING E3 ligases contribute to plant stress and disease tolerance through the suppression of pathogen and abiotic stress-induced necrosis.
RESULTS
Identification of BOI RING E3 Ligase as the BOS1-Interacting Protein
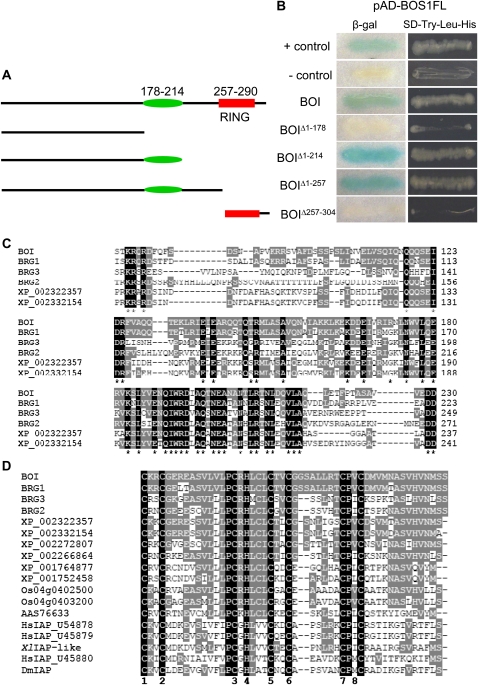
The Arabidopsis R2R3MYB transcription factor BOS1 is required for resistance to B. cinerea and tolerance to abiotic stress factors (Mengiste et al., 2003). To gain insights into the functions of BOS1, we screened for BOS1-interacting proteins from a yeast two-hybrid (Y2H) cDNA library. Full-length BOS1 autoactivated in the absence of an interacting partner in yeast cells. Deletion analysis of the BOS1 cDNA mapped the autoactivation activity to the C-terminal 50-amino acid region of BOS1. The BOS1 cDNA C-terminal deletion construct lacking autoactivation activity was used as bait for screening an Arabidopsis cDNA library constructed from B. cinerea-induced tissue. A cDNA clone corresponding to the Arabidopsis At4g19700 gene encoding a RING E3 ligase was identified as a strong BOI. BOI encodes a protein of 304 amino acids that contains the C-terminal HCa-type RING E3 ligase domain (Fig. 1A). We then swapped the vectors and coexpressed full-length BOS1 cDNA in the prey vector (pAD-BOS1) with BOI in the bait vector (pBD-BOI), which resulted in growth on selective medium and β-galactosidase activity, confirming interaction in yeast (Fig. 1B). In combination with the pAD empty vector, the bait plasmid (pBD-BOI) did not activate the transcription of the β-galactosidase reporter gene and growth on selective medium, confirming the lack of autoactivation by BOI.
Figure 1.
BOI interacts with BOS1 in yeast. A, Overall structure of BOI and deletion constructs used in interaction studies. B, Interactions between BOS1 and BOI proteins in Y2H assays. Yeast strains containing full-length BOS1 in the prey vector (pAD-BOS1FL), full-length BOI, or deletion derivatives of BOI in the bait (pBD-BOI) were assayed for growth on selective medium (−Leu, −Trp, −His; right) and β-galactosidase (β-gal; left) activity. β-Galactosidase activity was assayed from yeast cells grown on synthetic complete medium. Positive (+) and negative (−) controls from the Stratagene Y2H kit were assayed in parallel. C, Sequence comparison between BOI and related plant proteins covering the conserved central WRD domain. D, Comparison of the RING domains of BOI and animal IAP proteins. The black-shaded residues are identical in all the proteins compared, and gray shading indicates residues identical to the BOI protein. In D, the numbers show the conserved metal ligand positions. Dashes denote gaps introduced to maximize alignment. Sequences used in the alignments are from Arabidopsis (At4g19700, At5g45100, At3g12920, At1g79110), poplar (XP_002322357, XP_002332154), Vitis vinifera (XP_002272807, XP_002266864), Physcomitrella patens subsp. patens (XP_001764877, XP_001752458), rice (Os04g0402500, Os04g0403200), Solanum chacoense (AAS76633), human (U54878, U45879, U45880), Xenopus laevis (NP_001082290), and Drosophila melanogaster (AAC46988).
The BOI protein contains three distinct regions: an N-terminal-unique region with limited sequence conservation, a conserved central domain we designated the WRD domain, and a C-terminal RING domain (Fig. 1, A, C, and D; Supplemental Fig. S1). Four deletion constructs of BOI were tested for interaction with full-length BOS1 to determine regions required for interaction (Fig. 1A). The WRD domain covering the amino acid residues from 178 to 214 is conserved in BOI, three Arabidopsis BRGs, and a poplar (Populus trichocarpa; XP_002322357) gene encoding RING E3 ligase (Fig. 1C). The functions of these E3 ligases and the WRD domain are not known. BOI cDNA with the WRD domain interacted with BOS1, but the deletion lacking this region failed to interact, suggesting that this domain represents the interaction module of BOI (Fig. 1B). Consistent with this, the WRD domain is predicted to form the coiled-coil structure (Supplemental Fig. S2) normally required for protein-protein interactions in other proteins. Most RING domains in E3 ligases are associated with various substrate-binding regions including a coiled coil (Stone et al., 2005). The RING domain is dispensable and not sufficient for interaction with BOS1 in Y2H assays.
The RING domain of BOI is closely related to the RING domain of predicted proteins from Arabidopsis, poplar, and rice (Fig. 1D). In addition, BOI shares high sequence conservation with the RING domain of mammalian and Drosophila inhibitor of apoptosis (IAP) proteins. IAPs are regulators of apoptosis and other cellular processes in different organisms, but their function in plants is not known (Deveraux and Reed, 1999; Vaux and Silke, 2005). Based on the nature of the metal ligand residues present and/or the number of amino acids between them (Stone and Walker, 1995), the RING domain of BOI is of the RING HCa type and has three amino acids between the metal ligand positions 4 and 5, a spacing that is also conserved in the animal IAP proteins, as shown in Figure 1D (Stone et al., 2005). The full-length sequence alignment between BOI and BRGs is shown in Supplemental Figure S1A and shows overall identity ranging from 38% to 64% at the amino acid level. Next, the poplar sequence (XP_002322357) is the only other putative plant RING E3 ligase with high overall sequence identity of 47% to BOI. Phylogenetic analysis further confirmed that BOI is related to diverse predicted proteins from Arabidopsis and other plants (Supplemental Fig. S3). The functions of these BOI-related proteins have not been determined in any other plants.
BOS1 and BOI Colocalize and Interact in the Nucleus of the Plant Cell
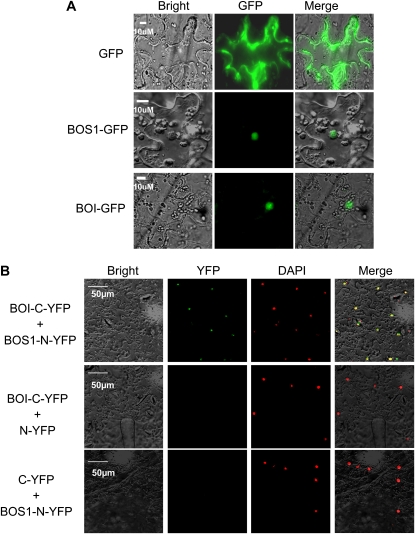
BOS1 and BOI are both localized to the nucleus when expressed individually in Nicotiana benthamiana cells as GFP fusion proteins (Fig. 2A). To confirm in vivo interaction, a bimolecular fluorescence complementation (BiFC) assay was performed. BOS1 was translationally fused with the N-terminal 173-amino acid portion of the yellow fluorescent protein (YFP; pBOS1-nYFP), and BOI was fused with the C-terminal 155-amino acid portion of YFP (pBOI-cYFP). pBOS1-nYFP and pBOI-cYFP were introduced into Agrobacterium tumefaciens and coinfiltrated into N. benthamiana leaves. Microscopic examination revealed YFP fluorescence only when the two constructs were coexpressed (Fig. 2B). Leaves from plants infiltrated with either of the constructs alone or in combination with the empty vector showed no fluorescence. When cells were stained with the fluorescent nuclear stain 4′,6-diamidino-2-phenylindole (DAPI), YFP fluorescence was observed in the nucleus of cells cotransformed with both constructs (Fig. 2B, DAPI column), suggesting an interaction in the nucleus. The interaction of BOS1 and BOI in the nucleus is consistent with the function of BOS1 as a transcription factor. BRG1, an E3 ligase that is closely related to BOI, failed to interact with BOS1 in BiFC and Y2H assays, showing the unique interaction between BOI and BOS1 proteins (Supplemental Fig. S4).
Figure 2.
BOI and BOS1 proteins interact in the nucleus of plant cells. A, BOS1 and BOI are localized to the nucleus when individually expressed as GFP fusion proteins. B, BiFC assay showing interaction between BOI and BOS1 in the nucleus of plant cells. In B, pBOI-cYFP and pBOS1-nYFP were transiently coexpressed or coexpressed with a respective empty vector in N. benthamiana leaf cells. YFP fluorescence was detected when pBOI-cYFP was coexpressed with pBOS1-nYFP. Cells were examined under bright field (Bright), fluorescence (YFP), and as a merged image (Merge) showing either no interaction or interaction in the nucleus.
BOI Is Required for Resistance to B. cinerea
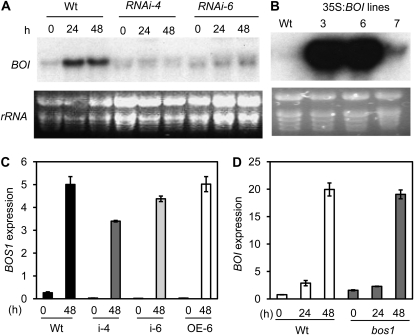
No loss-of-function T-DNA insertion alleles of the BOI gene were available from T-DNA insertion collections to conduct functional studies (Alonso et al., 2003; Rosso et al., 2003). We generated BOI RNAi lines using the 5′ BOI gene-specific region lacking any conserved domain. Two BOI RNAi lines with clearly reduced BOI gene expression were selected (Fig. 3A). We also generated 35S:BOI transgenic Arabidopsis lines that overexpress the BOI transcript relative to the wild-type plants (Fig. 3B). The expression of BOS1 was reduced in BOI RNAi lines after B. cinerea infection but was comparable to the wild-type level in the BOI overexpression lines (Fig. 3C). The BOI gene is expressed at low but detectable levels in healthy leaf tissues but is significantly induced by B. cinerea (Fig. 3, A and D). The BOS1 gene did not regulate BOI gene expression, as the bos1 mutant allele showed no altered BOI transcript level (Fig. 3D). The BOI RNAi plants show no altered growth or any other morphological changes. In addition, BOI RNAi lines have no effect on the BRG1 gene expression (Supplemental Fig. S5).
Figure 3.
BOI RNAi and overexpression lines and their effects on BOS1 gene expression. A and B, RNA blots showing BOI RNAi lines with reduced BOI gene expression (A) and 35S:BOI lines with increased BOI expression (B). C and D, qPCR showing expression of BOS1 in BOI RNAi and 35S:BOI lines (C) and BOI in the bos1 mutant (D) in response to B. cinerea. For RNA blots, total RNA (15 μg) was loaded per lane. qPCR was performed as described in “Materials and Methods.” The qPCR data represent expression levels calculated by the comparative cycle threshold method (Applied Biosystems), with the Arabidopsis Actin2 gene as the endogenous reference for normalization. Experiments were repeated at least two times with similar results. h, Hours after inoculation; Wt, wild type. rRNA was used as a loading control.
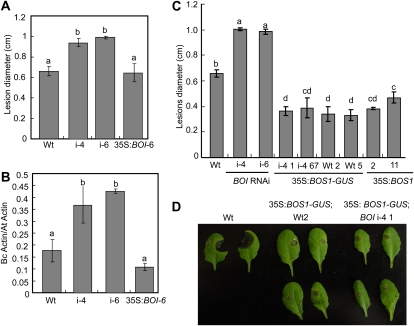
The BOI RNAi lines were significantly more susceptible to B. cinerea infection with a larger disease lesion and increased fungal growth relative to the wild-type plants (Fig. 4, A and B). However, the 35S:BOI plants did not exhibit altered responses to B. cinerea. BOI RNAi and wild-type plants expressing the translation fusion of BOS1 and GUS genes under the control of the cauliflower mosaic virus 35S promoter (35S:BOS1-GUS) were analyzed to determine the functional relationship between BOS1 and BOI. Interestingly, BOI RNAi plants expressing 35S:BOS1-GUS were more resistant to B. cinerea than the wild-type plants, suggesting that BOS1 is a likely direct downstream target of BOI (Fig. 4, C and D). Similarly, the transgenic 35S:BOS1-GUS plants were more tolerant to oxidative stress caused by the herbicide paraquat (Supplemental Fig. S6). These data are consistent with the increased susceptibility or sensitivity of the bos1 T-DNA insertion allele to pathogens and abiotic stress agents (Mengiste et al., 2003).
Figure 4.
BOI and BOS1 contribute to Arabidopsis resistance to B. cinerea. A and B, Mean size of disease lesion (A) and accumulation of B. cinerea ActinA DNA as a measure of fungal growth (B) in BOI RNAi and 35S:BOI plants inoculated with B. cinerea. C and D, Mean size of disease lesion (C) and disease symptoms (D) in 35S:BOS1 and 35S:BOS1-GUS plants inoculated with B. cinerea. In all panels, i-4 and i-6 represent BOI RNAi lines; i-4 1, i-4 67, Wt2, and Wt5 represent BOI1 RNAi or wild-type plants expressing the 35S:BOS1-GUS construct, respectively; and 2 and 11 represent transgenic 35S:BOS1 lines. Plants were inoculated with a single drop (4 μL) of conidial suspension (2.5 × 105 B. cinerea spores mL−1) per leaf. Data represent averages ± se from a minimum of 40 disease lesions. B. cinerea growth was determined by qPCR amplification of B. cinerea ActinA DNA (Bc Actin) relative to the Arabidopsis Actin2 DNA (At Actin) as the endogenous reference for normalization using the comparative cycle threshold method (Applied Biosystems; Bluhm and Woloshuk, 2005). In A to C, analysis of variance was performed to determine the statistical significance of the differences between mean values using SAS software (SAS Institute, 1999). Means with different letters are significantly different from each other (P = 0.05). Disease assays were repeated at least three times.
BOI Reduces Disease- and Toxin-Related Cell Death But Not HR
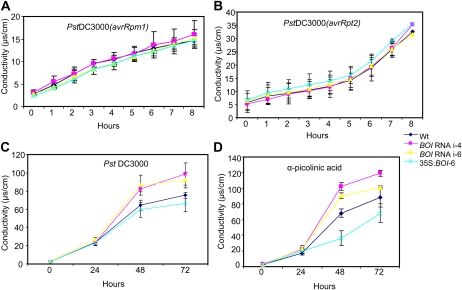
To determine the role of BOI in resistance to bacterial pathogens, the BOI RNAi and 35S:BOI lines were tested for responses to the virulent strains of P. syringae pv tomato (Pst). Bacterial growth and disease symptoms were not changed in BOI RNAi and 35S:BOI lines inoculated with a virulent strain of Pst (Supplemental Fig. S7). We then measured electrolyte leakage as a sensitive assay for detecting changes in cell death (Dellagi et al., 1998; Kawasaki et al., 2005). Higher cell death normally correlates with higher electrolyte leakage. Ion leakage measured after inoculation of BOI RNAi and 35S:BOI plants with avirulent Pst DC3000 (AvrRpm1, AvrRpt2) was not different from that of the wild-type plants, suggesting that BOI has no role in RPM1- and RPS2-mediated HR cell death in Arabidopsis (Fig. 5, A and B). In contrast, plants inoculated with virulent Pst DC3000 showed significantly increased electrolyte leakage in the BOI RNAi plants, indicating increased cell death associated with disease susceptibility (Fig. 5C). The increase in electrolyte leakage was coincident with the expression of disease symptoms caused by virulent P. syringae in Arabidopsis plants, thus marking cell death associated with susceptibility.
Figure 5.
BOI is a suppressor of toxin- and disease-associated cell death but not HR cell death. Electrolyte leakage in BOI RNAi and 35S:BOI plants was induced by treatment or inoculation with Pst DC3000 (avrRpm1; A), Pst DC3000 (avrRpt2; B), Pst DC3000 (C), and PA (D). Plants were inoculated with a bacterial suspension (optical density at 600 nm of 0.01) or infiltrated with PA toxin (1 mg mL−1), and electrolyte leakage was measured as described (Dhawan et al., 2009). Experiments were repeated at least three times. i-4 and i-6 represent BOI RNAi lines 4 and 6, respectively. Wt, Wild type. [See online article for color version of this figure.]
The 35S:BOI and BOI RNAi lines were assayed for their effects on cell death caused by α-picolinic acid (PA), a toxin produced by some fungi that induces apoptosis in animal as well as plant cells (Zhang et al., 2004; Kim et al., 2006). The BOI RNAi plants exhibited increased ion leakage following PA infiltration, whereas the 35S:BOI plants showed reduced ion leakage, suggesting that BOI plays a key role in limiting cell death caused by PA in Arabidopsis (Fig. 5D). Thus, BOI suppresses cell death caused by PA and Pst DC3000 but has no role in the HR cell death mediated by race-specific interactions.
The Arabidopsis LSD1 is a zinc finger protein that suppresses the spread of cell death to uninfected cells surrounding HR sites (Dietrich et al., 1997). The lsd1 mutant is impaired in the control of cell death initiated by various signals. In order to determine whether BOI has an effect on runaway cell death, we constructed lsd1;35S:BOI through genetic crosses between the Arabidopsis lsd1 mutant and 35S:BOI lines. The lsd1/lsd1;35S:BOI plants were comparable to lsd1 in their survival under growth conditions that triggered cell death in lsd1 plants (data not shown), suggesting that BOI failed to substitute for LSD1 in the control of cell death. These data suggest that LSD1 and BOI function in independent pathways.
BOI Is a Functional E3 Ligase and Ubiquitinates BOS1 in Vitro
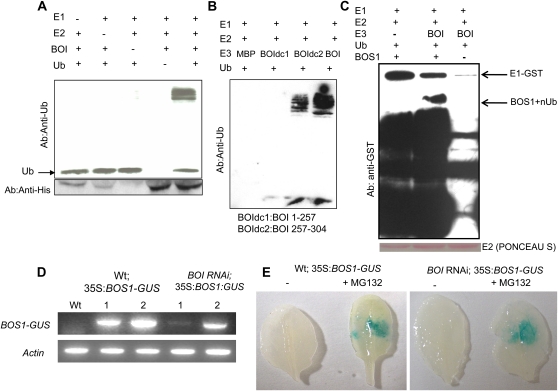
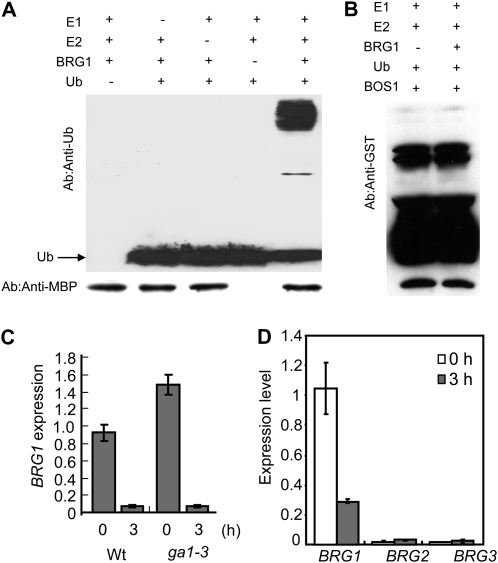
To determine whether the BOI protein has an E3 ligase activity, BOI was expressed in Escherichia coli as a fusion with a His tag. The recombinant His-BOI protein exhibited an E3 ligase activity in vitro and formed polyubiquitinated proteins in the presence of E1, E2, and purified His-BOI. The ubiquitination activity is dependent on the presence of His-BOI as well as E1, E2, and all the components required for ubiquitination, suggesting its involvement in protein ubiquitination and that BOI is a functional E3 ligase (Fig. 6A). To test the requirement of the RING domain of BOI for its ubiquitin ligase activity, the RING domain was deleted and the remaining portion was expressed as a fusion with the maltose-binding protein (MBP). Although the full-length MBP-BOI protein showed ubiquitination activity, the BOI protein lacking the C-terminal RING domain abolished the ubiquitination activity despite being dispensable for interaction with BOS1 (Figs. 1 and 6B). Thus, the RING ligase domain is required for ligase activity but not for its interaction with BOS1. This is consistent with the general notion that substrate binding and the E3 ligase activity are encoded by separate regions of RING E3 ligases.
Figure 6.
Recombinant BOI protein shows E3 ligase activity, and BOS1 accumulates after inhibition of the plant proteosome. A, Recombinant His-BOI protein shows ubiquitin (Ub) E3 ligase activity. B, The RING domain of BOI is essential for its ubiquitin E3 ligase activity. C, Recombinant MBP-BOI ubiquitinates recombinant BOS1 in vitro. D, RT-PCR data showing the expression of BOS1-GUS in 35S:BOS1-GUS transgenic plants. E, Histochemical assay showing GUS activity in transgenic BOS1:GUS plants before and after treatment with the proteosome inhibitor MG132. In E, plants were infiltrated with MG132 or buffer, and leaves were collected and stained with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid for GUS activity 6 h after treatment. Ponceau S is a protein stain used to determine the amount of BOS1 in the reaction. dc, Deletion constructs; Wt, wild type.
The interaction between BOS1 and the E3 ligase BOI suggests that BOS1 is ubiquitinated. Interestingly, the BOS1 protein contains the ubiquitin-interacting motif that is defined by a conserved spacing of a hydrophobic residue, an Ala, and a Ser residue (Miller et al., 2004). A consensus ubiquitin-interacting motif is present in BOS1 at Ile-151, Ala-154, and Ser-158, consistent with what is described (Miller et al., 2004). Recombinant MBP-BOI ubiquitinated glutathione S-transferase (GST)-BOS1 in the presence of all the components required for ubiquitination, consistent with the data on physical interaction (Fig. 6C).
BOS1 Is Regulated by the Plant Proteosome
We reasoned that BOI functions through ubiquitin mediated-modification of the BOS1 protein, leading to proteolysis or other posttranscriptional regulations. Transgenic 35S:BOS1-GUS generated in wild-type and BOI RNAi plants was analyzed for GUS activity to determine the regulatory relationship between BOI and BOS1. The reverse transcription (RT)-PCR data in Figure 6D show a typical example revealing that BOS1-GUS was expressed at constitutive levels in both wild-type and BOI RNAi transgenic plants. Histochemical assays failed to detect any GUS activity in more than 200 BOS1-GUS transgenic plants in the wild type and 96 in the BOI RNAi background. However, after treatment of 35S:BOS1-GUS plants with the proteosome inhibitor MG132, the BOS1-GUS activity was detected at the site of MG132 infiltration in many transgenic plants (Fig. 6E). These data indicate that BOS1:GUS is rapidly degraded by the 26S proteosome, consistent with the ubiquitination of BOS1 by BOI shown in Figure 6C. The levels of BOS1-GUS activity in the BOI RNAi did not differ significantly from the activity in wild-type plants based on the histochemical staining of MG132-infiltrated tissue. This may be due to the presence of residual BOI protein in the RNAi plants, which may be sufficient for ubiquitination of BOS1, or to the functional redundancy with other closely related E3 ligases. Due to the lack of the T-DNA insertion alleles of BOI, it is not possible to validate this first possibility.
Defense Gene Regulation and BOI Gene Expression Patterns
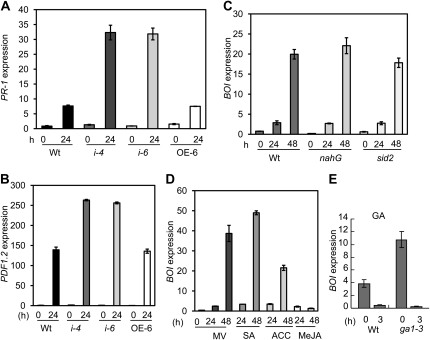
The expression of PR-1 and PDF1.2, two defense marker genes associated with resistance responses to biotrophic and necrotrophic pathogens, respectively, was tested in BOI RNAi and 35S:BOI plants using quantitative RT-PCR (qPCR). In BOI RNAi plants, PR-1 gene expression was higher than in the wild-type plants (Fig. 7A). This may be due to the increased susceptibility and pathogen growth in the BOI RNAi plants or to altered defense signaling. Expression of the PDF1.2 gene was also higher in BOI RNAi but showed no changes in 35S:BOI plants (Fig. 7B). BOI is normally induced by B. cinerea in sid2 and nahG plants, genotypes impaired in salicylic acid (SA) accumulation, suggesting that BOI gene expression is independent of endogenous SA levels (Fig. 7C). The BOI gene is induced by 1-aminocyclopropane-1-carboxylic acid (ACC), the natural precursor of ethylene (ET), and SA but significantly repressed by methyl jasmonate (MeJA; Fig. 7D). Methyl viologen (paraquat), an herbicide that produces reactive oxygen species, slightly induced the BOI gene expression. GA suppressed the expression of BOI by about 8.5-fold relative to mock controls (Fig. 7E). Consistent with the repression of BOI by GA, BOI expression is higher in the GA-deficient mutant ga1-3 (Sun and Kamiya, 1994; Fig. 7E). Exogenous application of GA in the ga1-3 mutant repressed BOI gene expression. In addition, publicly available data (https://www.genevestigator.ethz.ch) reveal that BOI gene expression increases in response to Pst DC3000, drought, uniconazole, and palcobutrazol, suggesting an important function for BOI in mediating responses to biotic and abiotic stresses and plant hormones.
Figure 7.
Basal and induced BOI and defense gene expression. A to C, Expression of PR-1 (A), plant defensin PDF1.2 (B), and BOI (C) during B. cinerea infection. D and E, Expression of BOI in response to ACC, MeJA, methyl viologen (MV), and SA (D) and GA (E). Plants were treated with the various compounds for the durations shown as described in “Materials and Methods.” Experiments were repeated at least two times. qPCR data were calculated as described in the legend for Figure 3. The qPCR data represent means ± se from three replicates. Wt, Wild type.
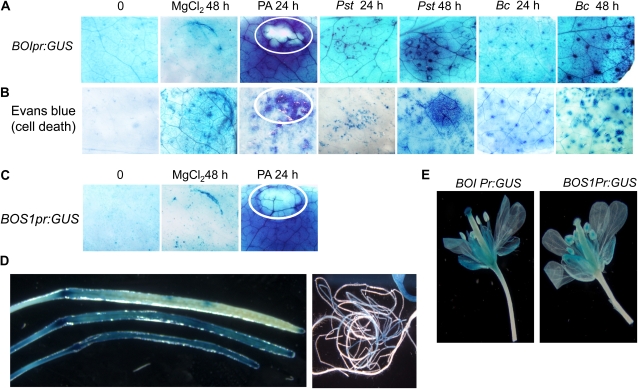
Plant lines expressing the GUS gene driven by the BOI or BOS1 promoter were generated to study the pattern of BOI and BOS1 gene expression relative to infection sites. Expression of GUS in BOI promoter:GUS (BOIPr:GUS) lines localized to PA, Pst DC3000, or B. cinerea-induced necrotic sites (Fig. 8A). PA induced the most intense GUS staining surrounding the site of infiltration. Evans blue staining reveals that PA, Pst DC3000, and B. cinerea induce cell death (Fig. 8B). The necrotic sites revealed by Evans blue staining and the sites of BOI expression appear to coincide. This is particularly evident in PA-treated tissue, where the site of PA infiltration heavily stains with Evans blue, marking cell death (Fig. 8B), but remains free of GUS due to the cell death (Fig. 8, A and C). BOS1Pr:GUS was highly expressed surrounding the site of PA treatment, similar to BOI1 expression (Fig. 8C). In addition, the BOI gene is expressed in siliques, roots, and flowering tissues (Fig. 8, D and E). Intriguingly, comparison of expression patterns in reproductive tissues clearly show that BOI is highly expressed in the stigma tips, while BOS1 is mainly expressed in anthers (Fig. 8E). Thus, both are expressed in distinct parts of the reproductive tissues, suggesting complementary functions.
Figure 8.
BOI gene expression enhanced at disease- and toxin-induced necrotic sites. A and B, BOI gene expression (A) and Evans blue staining for cell death (B) in response to PA, P. syringae, and B. cinerea. C, BOS1 gene expression induced by PA. D, Expression of BOIPr:GUS in siliques (left) and roots (right). E, Expression of BOIPr:GUS (left) and BOS1Pr:GUS (right) in reproductive tissues. In A to C, the circles mark the site of PA infiltration, revealing the lack of GUS activity (A and C) but intense Evans blue staining (B). Plants were infiltrated with buffer (MgCl2), Pst DC3000 (Pst), PA, or spray inoculated with B. cinerea and stained at the times indicated. Evans blue staining was performed as described (Watanabe and Lam, 2006). h, Hours after infection or treatment.
Arabidopsis BRGs Encoding RING E3 Ligases Have Overlapping Functions in the Suppression of Cell Death
BOI is most closely related to Arabidopsis At5g45100, hereafter designated BRG1. BRG1 interacted weakly with BOS1 in a Y2H assay but failed to interact in a BiFC assay in N. benthamiana cells (Supplemental Fig. S4, A and B). The other two BOI-related E3 ligases (BRG2 [At1g79100] and BRG3 [At3g12920]) failed to interact with BOS1 in Y2H assays. The recombinant BRG1 protein had E3 ubiquitin ligase activity but did not ubiquitinate the BOS1 protein, consistent with the lack of in vivo interaction (Fig. 9, A and B). BOI and BRG1 show high sequence conservation, yet only BOI interacted with BOS1 in vivo. BOI and BRG1 sequences carry multiple deletions relative to BRG2 and BRG3 (Supplemental Fig. S1A). At2g12290 was only recently annotated as a putative RING E3 ligase of this family and thus was not studied in detail here. At2g12290 is unique in terms of structural features, with predicted protein size that is only half that of the other BRGs, and devoid of many of the features of BOI and the BRGs. However, it shares the WRD domain and carries a variant RING domain with many of the invariant Cys residues absent (Supplemental Fig. S1, A and B). It is likely that At2g12290 is a pseudogene or is mistakenly annotated.
Figure 9.
E3 ubiquitin (Ub) ligase activity of BRG1 and expression of BRGs in response to GA and salt. A, Recombinant BRG1 protein has ubiquitin ligase activity. B, Recombinant BRG1 protein does not ubiquitinate BOS1. C, Expression of BRG1 in the wild type (Wt) and the GA-deficient ga1-3 mutant. D, Expression of BRG1 to BRG3 in response to GA. qPCR was performed as described previously (Dhawan et al., 2009) and in “Materials and Methods.” Experiments were repeated at least two times with similar results.
GA significantly repressed BRG1, similar to BOI, but increased the expression of BRG2 and BRG3 from a very low basal level of expression (Fig. 9, C and D). Basal BRG1 gene expression is higher in the GA-deficient mutant ga1-3. In addition, the basal and induced BRG gene expression levels are presented in Supplemental Figure S8. BOI was induced by B. cinerea, but BRG1 expression was unchanged during infection. However, BRG1 was induced by SA but repressed by MeJA, similar to the BOI gene. BRG2 and BRG3 are expressed at low basal levels with a slight reduction of gene expression after B. cinerea infection. Expression of BRG3 did not change significantly upon SA, ACC, or MeJA treatment, whereas BRG2 was consistently repressed by SA, ACC, and MeJA. The different plant hormones and B. cinerea differentially regulated BRG expression, with up-regulation of some but down-regulation of others. The main features of the BOI and BRG proteins and their expression in response to various signals are summarized in Table I. Collectively, these data suggest that BOI and the BRGs have overlapping and contrasting expression patterns in response to pathogen infection and plant hormones.
Table I. Summary of the main features of BOI and BRGs.
Bc, B. cinerea; BRG1, At5g45100; BRG2, At1g79110; BRG3, At3g12920; Nt, not tested; +, significant induction; −, significant repression; =, expression unchanged.
| Gene Identifier | Interaction with BOS1 | Open Reading Frame, Molecular Mass (kD) | pI | Identity to BOI | Expression |
||||||
| Bc | PA | GA | MeJA | SA | ACC | NaCl | |||||
| BOI | Strong | 304 amino acids, 33.50 | 5.51 | 100 | + | + | − | − | + | + | + |
| BRG1 | No | 294 amino acids, 32.54 | 5.56 | 64 | = | Nt | − | − | + | = | + |
| BRG2 | No | 358 amino acids, 40.08 | 6.73 | 47 | − | Nt | + | + | − | = | + |
| BRG3 | No | 335 amino acids, 37.70 | 6.22 | 38 | − | Nt | + | + | − | − | + |
| At2g12290 | No | 133 amino acids, 15.44 | 8.93 | 80a | Nt | Nt | Nt | Nt | Nt | Nt | Nt |
The identity is limited to the C-terminal region.
BOI-Related Genes Are Required for Full Resistance to B. cinerea
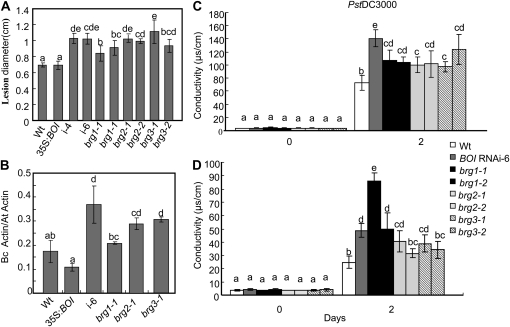
In order to study the functions of BRG genes, two T-DNA insertion alleles for each of the BRGs were isolated from SALK and SAIL T-DNA lines obtained from the Arabidopsis Biological Resource Center (Ohio State University). All of the mutants have a T-DNA insertion in an exon causing loss-of-function mutations. The brg mutant alleles exhibit wild-type morphology, suggesting that the BRGs are dispensable for normal plant development under the growth conditions tested. After challenge with B. cinerea, compared with the wild-type plants, a statistically significant increase in disease lesion size was observed for all the brg mutants (Fig. 10A). Fungal growth also increased in all the brg mutants except for brg1, where increased susceptibility was not accompanied by increased fungal growth (Fig. 10B). Thus, BRGs have overlapping roles in disease resistance but may function through different mechanisms. BOI functions through interaction with BOS1, but BRGs may function independently of BOS1.
Figure 10.
BRGs contribute to B. cinerea resistance and the suppression of PA- and bacteria-induced cell death. A and B, Mean size of disease lesion (A) and fungal growth (B) in B. cinerea-inoculated leaves. C and D, Electrolyte leakage after Pst DC3000 inoculation (C) and PA toxin treatment (D). Disease assays, sample treatment, and electrolyte leakage measurements were performed as described (Dhawan et al., 2009). In B, fungal growth was determined based on the relative PCR amplification of the B. cinerea ActinA and Arabidopsis Actin2 genes. ANOVA and t test (lsd) were performed to determine the statistical significance of the differences between the mean values using SAS software (SAS Institute, 1999). Mean values followed by different letters are significantly different from each other (P = 0.05). Wt, Wild type.
Similarly, the brg1, brg2, and brg3 alleles were tested for disease-related cell death caused by inoculation with Pst DC3000 or treatment with PA. All of the brg mutant alleles showed a significant increase in ion leakage at 2 d postinoculation, with Pst DC3000 indicative of enhanced cell death (Fig. 10C). PA treatment also caused a statistically significant increase in cell death relative to wild-type plants (Fig. 10D). Thus, members of BOI and the BRG gene family share a common role in the control of cell death.
BOI and Its Closely Related E3 Ligase Are Involved in GA and Abiotic Stress Responses
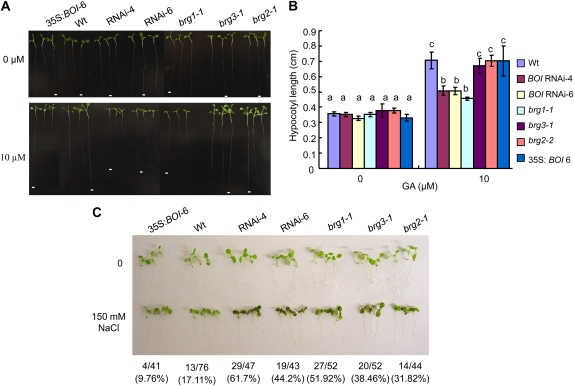
BOI RNAi lines and T-DNA insertion lines of BRGs were assayed for altered responses to stress and defense-mediating plant hormones. Seedling growth or seed germination was not altered in medium containing abscisic acid, ET, MeJA, and SA for BOI RNAi or any of the brg mutants (data not shown). However, in the presence of GA, overall growth and hypocotyl elongation of the BOI RNAi and the brg1 mutant were reduced, relative to the wild-type plants (Fig. 11, A and B). The mean hypocotyl lengths in BOI RNAi and brg1 mutants were significantly reduced relative to wild-type, brg2, and brg3 seedlings. The brg2 and brg3 mutants exhibited wild-type GA responses. GA did not affect the germination of seeds from BOI RNAi and the brg mutant plants.
Figure 11.
BOI is required for full GA-responsive growth and abiotic stress tolerance. A and B, Reduced hypocotyl and overall seedling growth (A) and mean hypocotyl length (B) in BOI RNAi and brg mutants in response to GA. C, Sensitivity of BOI RNAi and brg mutants to salt stress. Seedlings were germinated on plain MS medium and transferred to medium supplemented with GA or NaCl. In B, ANOVA and t test were performed to determine the statistical significance of the differences between the mean values using SAS software (SAS Institute, 1999). Mean values followed by different letters are significantly different from each other (P = 0.05). In C, the total number of seedlings tested and the percentage of seedlings showing salt-induced damage including necrotic or chlorotic symptoms are shown below the image. Wt, Wild type.
Due to the increased sensitivity of the BOS1 mutant alleles to various abiotic stress factors, we evaluated the BOI RNAi and brg mutants for their roles in NaCl-induced stress tolerance. BOI RNAi and brg1 mutant plants have reduced growth in medium containing NaCl relative to wild-type plants (Fig. 11C). BOI RNAi seedlings turned brown and necrotic on medium containing NaCl. The 35S:BOI was less sensitive to salt compared with the wild-type plants. Interestingly, all the mutant lines varied in their sensitivity to salt and differed from the sensitivity of the wild-type plants. The BOI RNAi lines and brg1 mutants exhibited the most severe salt sensitivity symptoms, followed by the brg2 and brg3 mutants. Thus, BOI and BRG1 contribute to survival under abiotic stress conditions.
DISCUSSION
We describe the function of BOI in the suppression of cell death, fungal resistance, and responses to salt stress. BOI physically interacts with BOS1 in yeast and plant cells. Consistent with this, BOI is a functional E3 ligase that ubiquitinates BOS1 in vitro and possibly in vivo, revealing the regulatory relationship between the two proteins. In transgenic plants expressing the BOS1-GUS fusion construct, the GUS activity could be detected only after treatment with the proteosome inhibitor MG132, suggesting that BOS1 has a rapid turnover rate and is a target of ubiquitin-mediated degradation. In addition, the resistance of the BOI RNAi lines that express BOS1-GUS to B. cinerea and tolerance to stress provide strong genetic evidence for the interactions between BOI and BOS1. Furthermore, we provide evidence for the function of BRGs through the phenotypic analysis of T-DNA insertion mutants and gene expression studies. The BOI and BRG genes all contribute to disease resistance and stress tolerance through mostly overlapping and yet some distinct mechanisms involving complex regulation of gene expression and interaction with BOS1. The overall structure and the order of domains are conserved between BOI and the BRGs. The primary sequences in the WRD and RING domains also are highly conserved. However, outside these regions, the level of amino acid conservation is very low. Structurally, many RING E3 ligases have a common C-terminal RING domain but also carry other domains that are thought to contribute to substrate binding. Thus, BOI represents a subclass of RING E3 ligases with overlapping functions and structural features, including a substrate-binding domain predicted to form a coiled-coil structure.
Arabidopsis RNAi lines with reduced BOI levels show increased susceptibility to the necrotrophic fungus B. cinerea and reduced tolerance to salt stress. In addition, the BOI RNAi plants have increased cell death triggered by the fungal toxin PA or inoculation with a virulent P. syringae strain. Interestingly, overexpression of BOI was sufficient to reduce PA-induced cell death and increase salt stress tolerance but failed to improve resistance to B. cinerea, most likely due to the complex virulence strategy of this fungus. Besides physical interaction, the BOS1 and BOI genes are coregulated during B. cinerea infection and by agents that promote cell death, including toxins, confirming their common function in planta. Interestingly, BOI is induced by SA and ACC but is repressed by MeJA and GA, suggesting that a complex regulation is required to maintain normal levels of BOI in wild-type plants. B. cinerea infection is known to increase the accumulation of SA, ET, MeJA, and abscisic acid in wild-type plants. B. cinerea-induced BOI expression is clearly independent of endogenous SA levels. It is also possible that BOI expression during B. cinerea infection is a result of stress rather than an effect of increased hormone levels. BOI and BOS1 are both expressed in reproductive tissues, with BOI expressed more on the stigma while BOS1 is expressed in the anther. These observations suggest that BOS1 and BOI may also have complementary functions in developmental processes.
We also describe the functions of three BRGs encoding RING E3 ligases that display overlapping and distinct gene expression and biological functions with BOI. BRGs also function in the suppression of cell death, a central mechanism of tolerance to necrotrophic fungi. Among the BRGs, BRG1 shares the highest sequence identity and the most overlapping function and coexpression with BOI. The BOI and BRG1 genes are induced by NaCl but repressed by GA and MeJA, although the extent of induction varies significantly between the two genes. Despite their role in resistance to B. cinerea and the suppression of cell death, BRG2 and BRG3 have different levels of basal and induced expression compared with BOI and BRG1. BRG2 and BRG3 have a very low basal level of expression but are further repressed by B. cinerea and SA, whereas BOI is induced. BRG2 and BRG3 are divergent, sharing only 47% and 38% overall identity, respectively, with the BOI protein. All BRGs have a common function in conferring resistance to B. cinerea and suppressing cell death. However, the BRGs do not interact with BOS1, and brg2 and brg3 loss-of-function mutants exhibit no altered responses to GA, suggesting that BRG2 and BRG3 function in disease resistance through mechanisms different from BOI. BRG2 and BRG3 are suppressed by B. cinerea, but their mutants show increased susceptibility rather than increased resistance, as would be predicted. Thus, rather than transcript abundance and regulation, the posttranscriptional regulation may be more important in the case of these two genes.
Recombinant BOI ubiquitinated BOS1 in vitro, and the BOS1-GUS fusion protein could be detected only after treatment of plants with MG132, a proteosome inhibitor, which suggested that BOS1 is normally degraded rapidly. The levels of BOS1-GUS did not vary between BOI RNAi and wild-type plants, as would be expected if BOS1 ubiquitination results in its degradation by the 26S proteosome. This lack of differences may be either due to the requirement for additional factors or to functional redundancy. Also, the BOI RNAi lines may retain BOI protein levels sufficient to ubiquitinate BOS1. Although BOS1 is likely degraded by the proteosome and is rapidly turned over, it is possible that during salt stress, oxidative stress, and B. cinerea infection, BOS1 may become stabilized to confer tolerance to stress. Consistent with this, the 35S:BOS1 and 35S:BOS-GUS plants are more tolerant to oxidative stress and resistant to B. cinerea. Stress-induced stabilization of proteins is a documented regulatory mechanism that promotes stress tolerance. Arabidopsis RGA, a DELLA protein, is stabilized during NaCl stress and promotes plant survival under stress (Achard et al., 2008). The mammalian p53 protein is normally rapidly degraded by the proteosome. Upon DNA damage, p53 levels must increase rapidly; therefore, the ubiquitination of p53 is rapidly terminated to allow the accumulation of p53 (Brooks and Gu, 2003).
Plant responses to pathogens have been studied extensively in connection with plant hormone synthesis, accumulation, and signaling. Altered hormone levels and signaling promote resistance or susceptibility depending on the nature of the pathogens. Hormonal imbalances create a range of plant pathological conditions and are exploited by pathogens (Grant and Jones, 2009). Some plant hormones affect plant defense due to their role in potentiating cell death, while others regulate the activation of protective mechanisms and/or the synthesis of antimicrobial compounds. Consequently, many genes involved in the regulation of cell death and hormone biosynthesis and signaling are also components of the plant immune response and stress tolerance. The roles of ET and jasmonic acid (JA) signaling as positive regulators of resistance to necrotrophic fungi have been firmly established based on genetic data from Arabidopsis and tomato (Solanum lycopersicum) mutants (Thomma et al., 1999; Abuqamar et al., 2008). SA is the major positive regulator of defense against many biotrophic fungi but can suppress resistance to some necrotrophic fungi (Veronese et al., 2006; Spoel et al., 2007). JA and ET act synergistically to promote resistance to necrotrophic fungi, but abscisic acid suppresses or promotes defense depending on the specific plant-pathogen interaction (Mauch-Mani and Mauch, 2005). Recent reports suggest that GAs are suppressors of resistance to necrotrophic fungi. GAs activate a plant growth response pathway that results in the degradation of DELLA proteins, known suppressors of GA-mediated plant growth responses (Harberd et al., 2009). The DELLA triple/quadruple mutant lacking four of the five DELLA genes is susceptible to B. cinerea, likely due to the increased accumulation of reactive oxygen species that promote cell death and susceptibility to B. cinerea (Achard et al., 2008). The quadruple DELLA mutant is also susceptible to Alternaria brassicicola but resistant to P. syringae, attributed to the attenuated JA-dependent defense and enhanced cell death, respectively (Navarro et al., 2008). In the case of BOI RNAi, reduced GA growth responses correlated with susceptibility to B. cinerea independent of changes in the JA-mediated defense response pathway deduced from the normal expression of the JA-regulated PDF1.2 gene.
The BOI RING E3 ligase was recently described as a target of DELLA proteins based on global microarray analysis and chromatin immunoprecipitation experiments (Zentella et al., 2007). However, both the loss-of-function triple DELLA mutant (rga-24gait6ga1-3) and the dominant DELLA mutant protein that stabilizes the DELLA protein RGA express BOI at elevated levels relative to wild-type plants, making it unclear whether BOI is a direct downstream target of DELLAs. Nevertheless, the BOI and BRG1 genes are both significantly repressed by GA, and the BOI RNAi and the brg1 mutant have reduced seedling growth and hypocotyl elongation in response to GA. It is not clear whether the GA-mediated suppression of BOI and BRG1 is through the removal of DELLAs by GA or indirectly. It is possible that GA-mediated removal of the DELLA protein RGA results in the recruitment of other regulatory factors that then suppress BOI and BRG1 gene expression. Plants that carry a dominant mutation that stabilizes DELLA and BOI RNAi have attenuated responses to GA, suggesting that they function in an antagonistic fashion in plant responses to GA. However, BOI and BRG1 may still act as positive factors of GA responses downstream of DELLA, consistent with the suppression of their gene expression by GA as well as the reduced GA responses in the BOI RNAi and the brg1 mutant. Interestingly, our data demonstrate that BOI and BRG1 are required for resistance to B. cinerea and tolerance to salt stress, consistent with the role of DELLAs in defense and stress tolerance (Achard et al., 2006, 2008; Navarro et al., 2008).
BOI and BOS1 define components of host defense that restrict the extent of necrosis and hence are positive regulators of B. cinerea resistance and stress tolerance. The susceptibility of BOI RNAi and the brg mutants is most likely due to the impaired ability to restrict initial pathogen-induced necrosis leading to increased ion leakage. The phenotypes of the bos1 allele are consistent with an impaired control of necrosis during infection and salt-induced stress (Mengiste et al., 2003). Interestingly, the BOI gene has a limited effect on HR cell death mediated by the Arabidopsis disease resistance genes RPM1 and RPS2 and the suppressor of cell death, LSD1, despite the similarities of these different forms of cell death at the molecular level. These data indicate that the genetic control of the various types of cell death may be different. Future progress on the identification and characterization of toxin receptors and their signal transduction components may shed light on the genetic control of phytotoxin and necrotrophic pathogen-mediated cell death. The BOI RNAi plants and brg mutants do not show a lesion-mimic phenotype, and the altered responses are most likely due to the loss of an active mechanism that occurs after pathogen infection. Similarly, in the presence of NaCl, the BOI RNAi and brg2 mutants exhibit increased browning and necrosis of the cotyledons and emerging leaves. This, combined with the significant impact of loss-of-function and gain-of-function alleles of BOI and the T-DNA insertion alleles of brg mutants on toxin- and disease-induced cell death, suggests that this subclass of E3 ligases is crucial in disease and stress tolerance in Arabidopsis.
Plant disease resistance pathways responding to pathogens of different lifestyles function in a synergistic or antagonistic manner. The SA-mediated and JA/ET-mediated defense pathways interact antagonistically (Spoel et al., 2007). Many Arabidopsis mutants show contrasting defense gene expression and plant responses to biotrophic and necrotrophic pathogens (Glazebrook, 2005; Veronese et al., 2006). In the BOI RNAi plants, expression of the PR1 and PDF1.2 genes are both slightly enhanced, suggesting that the altered disease response is not related to defense signal antagonism. Despite the high frequency of Arabidopsis mutations that reveal antagonism between defense pathways, many other genes regulate defense without apparent antagonistic interactions (Berrocal-Lobo et al., 2002; Coego et al., 2005; Dhawan et al., 2009), consistent with the function of BOI. Most such pathways are likely to function independently of hormone-mediated responses.
Overall, we provide strong evidence for the role of BOI and BRGs in plant resistance to B. cinerea and tolerance to salt stress. BOS1 and BOI may integrate plant responses to diverse signals in a manner comparable to DELLA proteins that are also required for stress tolerance as well as resistance to necrotrophic fungi (Achard et al., 2008; Navarro et al., 2008). The reduced GA responses of BOI RNAi and brg1 seedlings as well as the dramatic suppression of BOI and BRG1 genes by GA and their up-regulation by salt suggest that these E3 ligases contribute to plant immunity and salt stress tolerance through a function that involves the GA response pathway. A complex transcriptional and posttranscriptional regulation and additional genetic factors likely govern the functions of BOI, BRG1, and BOS1 in plant defense and stress tolerance. The exact points of action of BOI, BOS1, and the BRG proteins in GA-dependent and independent defense responses need to be defined further.
MATERIALS AND METHODS
Plant Growth Conditions and Disease Assays
Plant growth conditions, growth media, and disease assays were as described (Dhawan et al., 2009). Botrytis cinerea disease assays were performed on soil-grown plants by drop inoculation (4–5 μL) of 2.5 × 105 spores mL−1. As a measure of B. cinerea growth in inoculated plants, the levels of the B. cinerea ActinA DNA were determined by qPCR using the primers shown in Supplemental Table S1. The DNA concentrations were calculated by the comparative cycle threshold method (Applied Biosystems) with the Arabidopsis (Arabidopsis thaliana) Actin2 gene as the endogenous reference for normalization as described (Bluhm and Woloshuk, 2005).
The culture and disease assay for Pseudomonas syringae pv tomato were as described (Zheng et al., 2006). Procedures for measuring electrolyte leakage were as described (Kawasaki et al., 2005; Dhawan et al., 2009). Briefly, a bacterial suspension (optical density at 600 nm of 0.01) or PA (1 mg mL−1) was infiltrated into leaves. Six leaf discs (0.8 cm diameter) were collected from the infiltrated area and washed with water for 50 to 60 min and then placed in a tube containing 8 mL of water. Conductivity of five replicates for each treatment was measured with a conductivity meter (model AB30, Accumet BASIC; Fisher Scientific) following the procedure described (Kawasaki et al., 2005).
Generation of Transgenic Lines and Identification of Mutant Alleles
To generate BOI overexpression lines, full-length BOI cDNA was cloned after the cauliflower mosaic virus 35S promoter into a modified version of binary vector pCAMBIA 1200. The binary vector was then transferred into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis (Clough and Bent, 1998). Transgenic plants were selected on medium containing hygromycin, and the lines overexpressing BOI were identified by RNA blots hybridized to the full-length BOI cDNA. To generate BOI RNAi lines, the first 250-bp fragment of the BOI cDNA starting from the translation initiation codon was amplified using primer pairs (Supplemental Table S1) and cloned into the RNAi vector pGSA1252 (http://www.chromdb.org/rnai/order_vectors.html). BOI RNAi lines with reduced BOI gene expression were selected based on the levels of RNA by hybridizing to the 3′ terminus of the BOI gene. BOI promoter:GUS lines were generated by cloning 1.5-kb fragments of the BOI or BOS1 upstream sequence into the binary vector pCAMBIA 1391 carrying the GUS gene, and transformants were selected on Murashige and Skoog (MS) medium containing hygromycin.
We identified the mutant alleles of BRGs from a segregating population of T-DNA insertion lines obtained from the Arabidopsis Biological Resource Center using T-DNA and gene-specific primers (Supplemental Table S1). The mutant alleles for At5g45100 (brg1-1, Salk_010178; brg1-2, SAIL_1214_E04), At3g12920 (brg2-1, SAIL_302_F07; brg2-2, SAIL_261_G05), and At1g79110 (brg3-1, SAIL_95_F06; brg3-2, SAIL_73_H10) all carry a T-DNA insertion in an exon.
RNA Blots and RT-PCR
For RNA-blot and RT-PCR experiments, total RNA was isolated as described (Lagrimini et al., 1987) or with Trizol reagent according to the manufacturer’s instructions (Invitrogen). For RNA blots, total RNA was separated on 1.2% agarose-formaldehyde gels and blotted to Hybond N+ nylon membranes by a standard protocol. Probes were labeled with 32P by random priming with a commercial kit (Sigma-Aldrich). Hybridization of probe and subsequent washings were performed as described (Church and Gilbert, 1984). For qPCR, total RNA (2 μg) was reverse transcribed with oligo(dT) primers and SuperScript II reverse transcriptase (Invitrogen). The cDNA was then subjected to qPCR with gene-specific primers. The expression levels were calculated by the comparative cycle threshold method (Applied Biosystems) with Arabidopsis Actin2 (At3g18780) as the endogenous reference for normalization. Primer sequences for qPCR expression of BOI and BRGs are provided in Supplemental Table S1. Induced expression was from plants treated by spraying of ACC (0.5 mm), SA (5 mm), MeJA (1 mm), or methyl viologen (50 μm) on soil-grown leaves or in vitro-grown seedlings with 100 μm GA for 3 h or 200 mm NaCl for 6 or 12 h.
RT-PCR for BOS1-GUS transgenic plants was performed after DNase treatment of RNA and first-strand cDNA synthesis. cDNA was synthesized from both control and treated samples with equal amounts of total RNA (2 μg), avian myeloblastosis virus reverse transcriptase (Promega), and oligo(dT15) primers according to standard protocols. The PCR was performed for 28 cycles with 2.5 μL of cDNA as a template and specific primer pairs (94°C for 30 s, 52°C for 30 s, and 72°C for 1 min).
Y2H Screen
Y2H assays were performed with the GAL4 system according to the manufacturer’s instructions (Stratagene). The full-length BOS1 cDNA and various deletion constructs were cloned into pBD-GAL4 to generate a DNA-binding domain bait protein fusion. The partial BOS1 cDNA without the 50-amino acid sequence lacking the transcriptional activation of LacZ was used for screening the Y2H library. We built a cDNA library from B. cinerea-infected Arabidopsis tissue in the HybriZAP-2.1 vector according to the manufacturer’s instructions (Stratagene). At least 106 yeast colonies were screened by transformation into the YRG-2 yeast strain (Stratagene) expressing BOS1 cDNA. Interacting proteins were initially selected for complementation of His auxotrophy on selective medium lacking His, Leu, and Trp. The putative interactors were then tested by assaying for the LacZ reporter gene activation by performing the filter-lift assay as described in the Stratagene protocol. Interactions were retested for His3+, Trp+, and Leu+ auxotrophy and LacZ reporter activity (β-galactosidase assay). The plasmids from the positive clones were then isolated, sequenced, and reintroduced into the original yeast bait and control bait strains to verify interaction.
BiFC Assays
Two vectors, pCAMBIA-N-YFP and pCAMBIA-C-YFP, were generated based on pCAMBIA 1200 for use in the BiFC analysis (Dhawan et al., 2009). The BOI full-length cDNA was inserted into pCAMBIA-C-YFP to generate the C-terminal in-frame fusions with C-YFP (pBOI-cYFP), whereas BOS1 was introduced into pCAMBIA-N-YFP to form the N-terminal in-frame fusion with N-YFP (pBOS1-nYFP). After sequence verification, the constructs were verified by sequencing and the plasmids were introduced into Agrobacterium strain GV3101. The Agrobacterium carrying the appropriate plasmids was expressed in Nicotiana benthamiana leaf tissue by agroinfiltration. In vivo interaction was observed with an epifluorescence microscope (Nikon Eclipse E800).
Recombinant Protein Purification and Ubiquitination Assay
Arabidopsis BOI, BOI257–304, BOI1–257, and BOS1 cDNAs were cloned into pRSET A (Invitrogen), pMAL-c2x (New England Biolabs), and pGEX 4T-1 (GE Healthcare) to produce the fusion protein plasmids named pRSET-BOI, pMAL-BOI, pMAL-BOI257–304, pMAL-BOI1–257, and pGEX-BOS1. The expression and purification of the fusion proteins were performed as described in the product manuals.
In vitro ubiquitination assays were performed in a reaction volume of 30 μL that contained 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 2 mm ATP, 2 mm dithiothreitol, 10 mm phosphocreatine, 2 units of creatine kinase (Sigma-Aldrich), 10 μg of ubiquitin, 200 ng of yeast E1 (Boston Biochem), 400 ng of E2 UbcH5b (Boston Biochem), and approximately 1 μg of HIS-BOI, MBP- BOI, MBP-BOI257–304, MBP-BOI1–257, and/or GST-BOS1. The reactions were incubated at 30°C for 6 h, stopped by the addition of 6× SDS-PAGE sample buffer (125 mm Tris-HCl, pH 6.8, 2% SDS, 20% glycerol, and 0.2% bromphenol blue) at 100°C for 5 min, and analyzed by SDS-PAGE followed by protein gel blotting using anti-ubiquitin (Cell Signaling), anti-His (Amersham Biosciences), anti-GST (Amersham Biosciences), and anti-MBP (New England Biolabs) antibodies.
Hormone Sensitivity Assay
Effects of various plant hormones on seed germination were tested by directly plating seeds on medium containing plant hormones and evaluating changes in the percentage of germination. In addition, seedlings germinated on plain MS medium were transferred to medium containing various concentrations of SA, MeJA, ACC, NaCl, and GA to determine the effect on growth. Surface-sterilized seeds were sown on 1.0% (w/v) agar medium containing MS salts, 2% (w/v) Suc, MES (0.5 g L−1), and different concentrations of MeJA, ACC, 2,4-dichlorophenoxyacetic acid, or indole-3-acetic acid, pH 5.7. The plant hormones were added to the autoclaved medium from filter-sterilized stock solutions. The effects of the plant hormones on overall seedling growth and hypocotyl elongation were evaluated. The hypocotyl length was measured after 7 d of growth in culture. In each case, 25 randomly selected seedlings were measured. The experiments were repeated at least twice on different seed lots.
Sequence data for the genes described in this study can be found in the GenBank/EMBL data libraries under the following accession numbers: BOS1 (At3g06490), BOI (At4g19700) BRG1 (At5g45100) BRG2 (At1g79110), BRG3 (At3g12920), PR1 (At2g14610), and PDF1.2 (At5g44420).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence alignment of BOI and BOI-RELATED proteins.
Supplemental Figure S2. The BOI protein contains a predicted coiled-coil region.
Supplemental Figure S3. Phylogenetic analysis of BOI and BOI-RELATED proteins.
Supplemental Figure S4. Interaction assays between BOS1 and BRG proteins.
Supplemental Figure S5. BRG1 expression in BOI RNAi plants.
Supplemental Figure S6. 35S:BOS1-GUS lines are tolerant to oxidative stress.
Supplemental Figure S7. Bacterial growth in BOI RNAi and 35S:BOI plants.
Supplemental Figure S8. Expression of BRGs.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Tai-ping Sun (Duke University) for Arabidopsis ga1-3 seeds and Dr. Larry Dunkle (Purdue University) for comments on the manuscript.
References
- Abuqamar S, Chai MF, Luo H, Song F, Mengiste T. (2008) Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 20: 1964–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Renou JP, Berthomé R, Harberd NP, Genschik P. (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Bluhm BH, Woloshuk CP. (2005) Amylopectin induces fumonisin B1 production by Fusarium verticillioides during colonization of maize kernels. Mol Plant Microbe Interact 18: 1333–1339 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. (2003) Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol 15: 164–171 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coego A, Ramirez V, Gil MJ, Flors V, Mauch-Mani B, Vera P. (2005) An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17: 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellagi A, Brisset MN, Paulin JP, Expert D. (1998) Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol Plant Microbe Interact 11: 734–742 [DOI] [PubMed] [Google Scholar]
- del Pozo O, Pedley KF, Martin GB. (2004) MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Reed JC. (1999) IAP family proteins: suppressors of apoptosis. Genes Dev 13: 239–252 [DOI] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T. (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88: 685–694 [DOI] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK. (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Govrin EM, Levine A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10: 751–757 [DOI] [PubMed] [Google Scholar]
- Grant MR, Jones JD. (2009) Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Yao N. (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6: 201–211 [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL. (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF. (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci USA 98: 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Nam J, Boyes DC, Holt BF, III, Hubert DA, Wiig A, Dangl JL. (2005) A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. Plant J 44: 258–270 [DOI] [PubMed] [Google Scholar]
- Kim M, Lim JH, Ahn CS, Park K, Kim GT, Kim WT, Pai HS. (2006) Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 18: 2341–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. (1987) Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA 84: 7542–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Martin R, Mongrand S, Vandenabeele S, Chen KC, Jang IC, Chua NH. (2008) RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J 56: 550–561 [DOI] [PubMed] [Google Scholar]
- Liu YC, Penninger J, Karin M. (2005) Immunity by ubiquitylation: a reversible process of modification. Nat Rev Immunol 5: 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, Muskett P, Sánchez-Vallet A, López G, Ramos B, Sánchez-Rodríguez C, Jordá L, Parker J, Molina A. (2008) Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol Plant 1: 496–509 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F. (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol 8: 409–414 [DOI] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron JM, Dietrich RA. (2003) The BOS1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SL, Malotky E, O’Bryan JP. (2004) Analysis of the role of ubiquitin-interacting motifs in ubiquitin binding and ubiquitylation. J Biol Chem 279: 33528–33537 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JD. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- SAS Institute (1999) The SAS System for Windows, Release 8.0. SAS Institute, Cary, NC [Google Scholar]
- Shi CS, Kehrl JH. (2003) Tumor necrosis factor (TNF)-induced germinal center kinase-related (GCKR) and stress-activated protein kinase (SAPK) activation depends upon the E2/E3 complex Ubc13-Uev1A/TNF receptor-associated factor 2 (TRAF2). J Biol Chem 278: 15429–15434 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Walker JC. (1995) Plant protein kinase families and signal transduction. Plant Physiol 108: 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y. (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Tierens KF, Broekaert WF. (1999) Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Silke J. (2005) IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol 6: 287–297 [DOI] [PubMed] [Google Scholar]
- Veronese P, Chen X, Bluhm B, Salmeron J, Dietrich RA, Mengiste T. (2004) The BOS loci of Arabidopsis are required for resistance to Botrytis cinerea infection. Plant J 40: 558–574 [DOI] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. (2006) Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J 45: 884–894 [DOI] [PubMed] [Google Scholar]
- Wolpert TJ, Dunkle LD, Ciuffetti LM. (2002) Host-selective toxins and avirulence determinants: what’s in a name? Annu Rev Phytopathol 40: 251–285 [DOI] [PubMed] [Google Scholar]
- Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JD, Sadanandom A. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Vega-Sánchez ME, Zhu T, Wang GL. (2006) Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res 16: 413–426 [DOI] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HK, Zhang X, Mao BZ, Li Q, He ZH. (2004) Alpha-picolinic acid, a fungal toxin and mammal apoptosis-inducing agent, elicits hypersensitive-like response and enhances disease resistance in rice. Cell Res 14: 27–33 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.