Abstract
Intercellular signaling is essential for the coordination of growth and development in higher plants. Although hundreds of putative receptors have been identified in Arabidopsis (Arabidopsis thaliana), only a few families of extracellular signaling molecules have been discovered, and their biological roles are largely unknown. To expand our insight into the developmental processes potentially regulated by ligand-mediated signal transduction pathways, we undertook a systematic expression analysis of the members of the Arabidopsis CLAVATA3/ESR-RELATED (CLE) small signaling polypeptide family. Using reporter constructs, we show that the CLE genes have distinct and specific patterns of promoter activity. We find that each Arabidopsis tissue expresses at least one CLE gene, indicating that CLE-mediated signaling pathways are likely to play roles in many biological processes during the plant life cycle. Some CLE genes that are closely related in sequence have dissimilar expression profiles, yet in many tissues multiple CLE genes have overlapping patterns of promoter-driven reporter activity. This observation, plus the general absence of detectable morphological phenotypes in cle null mutants, suggest that a high degree of functional redundancy exists among CLE gene family members. Our work establishes a community resource of CLE-related biological materials and provides a platform for understanding and ultimately manipulating many different plant signaling systems.
Plant growth and survival are critically dependent on the communication of information between cells. Intercellular signaling pathways convey cell fate information, regulate cell division and differentiation processes, propagate and amplify specific signaling states, and coordinate tissue responses and functions. The importance of cell-to-cell communication is underscored by the classification of 10% of the Arabidopsis (Arabidopsis thaliana) proteome as playing roles in signal transduction (Arabidopsis Genome Initiative, 2000). Yet, whereas more than 400 Arabidopsis genes encode receptor-like kinases (Shiu and Bleecker, 2001) that presumably function as transmembrane sensors for extracellular signals, only a few families of putative signaling molecules have been identified.
The CLE genes encode small polypeptides (less than 15 kD) that share several structural features. Each possesses either an N-terminal signal peptide or membrane anchor sequence, a large variable domain, and a highly conserved 14-amino acid motif called the CLE domain near the C terminus (Cock and McCormick, 2001). Biochemical evidence indicates that the full-length CLAVATA3 (CLV3) polypeptide is proteolytically processed (Ni and Clark, 2006) to a mature active 12- or 13-amino acid arabinosylated glycopeptide consisting of the CLE domain (Kondo et al., 2006; Ohyama et al., 2009). Synthetic peptides corresponding to the CLE motif of other CLE family members also show biological activity in various bioassays (Fiers et al., 2005; Ito et al., 2006), suggesting that such peptides are likely to be the functional CLE gene products.
The biological functions of only a few CLE genes are known. CLV3 is a founding member of the family and plays a key role in the intercellular communication of stem cell fate during Arabidopsis development. CLV3 is specifically expressed in the stem cell population of shoot and floral meristems (Fletcher et al., 1999). Secreted into the extracellular space (Rojo et al., 2002), the CLV3 polypeptide is perceived by CLV1 (Clark et al., 1997; Ogawa et al., 2008) and other transmembrane receptors (Jeong et al., 1999; Müller et al., 2008) in the underlying cells. This signal transduction pathway is a core component of a negative feedback loop linking the stem cell reservoir and the underlying organizing center (Brand et al., 2000). Signaling through the CLV pathway restricts stem cell accumulation by limiting the expression domain of the WUSCHEL RELATED HOMEOBOX (WOX) family transcription factor gene WUSCHEL (Laux et al., 1996), which promotes stem cell activity and CLV3 expression (Schoof et al., 2000).
CLE gene activity also controls stem cell homeostasis in the root meristem. CLE40 transcripts are present at low levels in all Arabidopsis tissues (Hobe et al., 2003), but in roots, CLE40 is specifically expressed in the stele and in differentiating columella cells of the protective root cap (Stahl et al., 2009). CLE40 activity emanating from the columella cells promotes distal root meristem differentiation by acting through the receptor kinase ARABIDOPSIS CRINKLY4 in a negative feedback loop that limits the expression domain of the WUS-related gene WOX5 (Stahl et al., 2009). WOX5 is present in the quiescent center and promotes columella stem cell fate in the distal domain of the root meristem. Thus, CLE/WOX-mediated signaling modules regulate Arabidopsis stem cell fate in both the shoot and the root apical meristems.
Overexpression studies have uncovered other developmental processes that respond to CLE peptide activity. A number of CLE genes when misexpressed in the root apical meristem gradually inhibit root meristem maintenance (Casamitjana-Martínez et al., 2003; Fiers et al., 2004, 2005; Ito et al., 2006; Meng et al., 2010) via a signaling pathway that appears to be distinct from the CLE40 pathway. Plants overexpressing CLE19, CLE21, or CLE25 form miniature rosettes and inflorescences and display anthocyanin overproduction and developmental delays (Strabala et al., 2006). Overexpression of CLE42 or CLE44 results in bushy, dwarfed plants with delayed development and reduced apical dominance, whereas CLE18 or CLE26 overexpression leads to enhanced root elongation (Strabala et al., 2006). Simultaneous overexpression of CLE6 and CLE41 produces stunted, bushy plants with increased hypocotyl vascular cell proliferation, suggesting that CLE peptides can function synergistically (Whitford et al., 2008). These phenotypes indicate roles for CLE family members in regulating many different aspects of development. Yet, although many Arabidopsis CLE genes cause morphological phenotypes when overexpressed, others still remain to be evaluated by this type of study.
Results from overexpression studies as well as in vitro bioassays revealed that multiple CLE peptides have the capacity to generate the same morphological phenotypes. More than a dozen CLE peptides can activate the CLV3 signaling pathway when ectopically expressed in the shoot meristem (Ni and Clark, 2006; Strabala et al., 2006; Jun et al., 2008; Meng et al., 2010). Application of 19 different synthetic CLE peptides in root growth assays can arrest root meristem growth (Ito et al., 2006; Strabala et al., 2006; Whitford et al., 2008), whereas the application of CLE41, CLE42, CLE43, or CLE44 peptides leads to suppressed xylem differentiation (Ito et al., 2006; Whitford et al., 2008). The CLE family has been divided into two functional classes on this basis, with A-type CLE peptides (CLV3, CLE1–CLE27, and CLE40) being capable of inducing root and/or shoot meristematic cell differentiation, whereas B-type CLE peptides (CLE41–CLE44) are not (Whitford et al., 2008). These observations also raise the possibility that another key determinant of CLE peptide specificity, in addition to their primary amino acid sequence (Meng et al., 2010), may be their tissue distribution. Yet, although reverse transcription (RT)-PCR studies show that CLE genes are transcribed in many different tissues (Sharma et al., 2003), their specific expression patterns remain to be characterized.
To obtain a more precise understanding of the extent of overlap between CLE genes and gauge their possible functional redundancy, we used reporter assays to obtain high-resolution expression data for the entire Arabidopsis A-type CLE gene family. We observed highly specific CLE gene promoter activity patterns in roots, shoots, leaves, stems, and flowers, with most tissues expressing multiple CLE genes. We thus uncover a number of new biological processes that may be regulated by small-peptide signal transduction pathways. In addition, we characterized the overexpression phenotypes of previously unstudied CLE genes and identified hypomorphic or null insertion alleles for eight A-type CLE genes. Morphological examination of homozygous cle single mutant plants revealed no detectable growth or development phenotypes. Taken together, our expression and functional data indicate that CLE family members have diverse activities, yet significant functional redundancy exists among them.
RESULTS
Analysis of A-Type CLE Promoter Activity in Vegetative Tissues
Among the Arabidopsis A-type CLE genes, the expression patterns of CLV3, CLE19, and CLE40 have already been reported (Fletcher et al., 1999; Hobe et al., 2003; Fiers et al., 2004). To determine the expression patterns of the other A-type CLE genes, we generated CLE promoter:GUS or GFP fusion constructs using from 974 to 3,398 bp of the 5′ genomic region upstream of each CLE coding sequence. At least 10 independent transgenic lines were analyzed for each gene promoter to monitor consistent reporter activity, with the exception of the CLE11, CLE12, and CLE13 promoters, for which four to eight independent transgenic lines were analyzed. Each CLE promoter drove GUS expression in one consistent pattern except for CLE10 (Supplemental Materials and Methods S1). We observed that all but one A-type CLE gene reporter is expressed in vegetative tissues (Table I; Figs. 1–4).
Table I. Summary of pCLE:GUS activity during vegetative development.
| CLE Gene | Shoot |
Root |
||||||||
| Shoot Apexa | Hypocotyl | Vasculature | Leaf Blade | Otherb | Tipc | Vasculature | Ground Tissues | Epidermis | Otherd | |
| CLE1 | + | + | + | |||||||
| CLE2 | + | |||||||||
| CLE3 | + | + | + | |||||||
| CLE4 | + | + | ||||||||
| CLE5 | + | + | + | |||||||
| CLE6 | + | + | ||||||||
| CLE7 | + | + | ||||||||
| CLE8 | ||||||||||
| CLE9 | + | |||||||||
| CLE10 | + | ±e | ||||||||
| CLE11 | + | + | + | + | ||||||
| CLE12 | + | + | + | |||||||
| CLE13 | + | |||||||||
| CLE14 | + | + | ||||||||
| CLE16 | + | + | + | + | + | + | + | + | ||
| CLE17 | + | + | + | + | + | + | + | + | + | |
| CLE18 | + | + | + | |||||||
| CLE20 | + | + | ||||||||
| CLE21 | + | + | ||||||||
| CLE22 | + | + | + | + | ||||||
| CLE25 | + | + | + | |||||||
| CLE26 | + | + | + | + | ||||||
| CLE27 | + | + | + | + | + | |||||
Shoot apex includes SAM and rosette leaf primordia.
Other includes pith, stipules, stomata, hydathodes, leaf margins, trichomes, and the leaf base.
Root tip includes root cap, root apical meristem, and cell division zone.
Other includes root hair cells and lateral root branch points.
Eight of 15 pCLE10:GUS lines showed root tip expression.
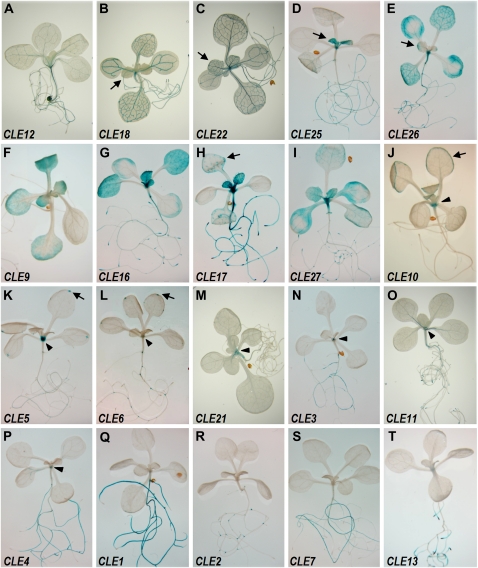
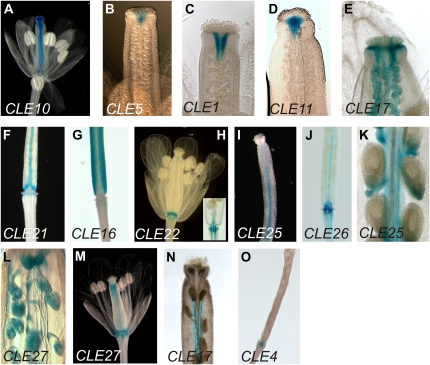
Figure 1.
CLE promoter activity in 11-d-old-seedlings. A, CLE12. B, CLE18. C, CLE22. D, CLE25. E, CLE26. F, CLE9. G, CLE16. H, CLE17. I, CLE27. J, CLE10. K, CLE5. L, CLE6. M, CLE21. N, CLE3. O, CLE11. P, CLE4. Q, CLE1. R, CLE2. S, CLE7. T, CLE13. Arrows indicate GUS activity in the vasculature of young leaves (A–D), leaf margins (H and J), and hydathodes (K and L). Arrowheads (J–P) indicate GUS activity in the shoot apex.
Figure 4.
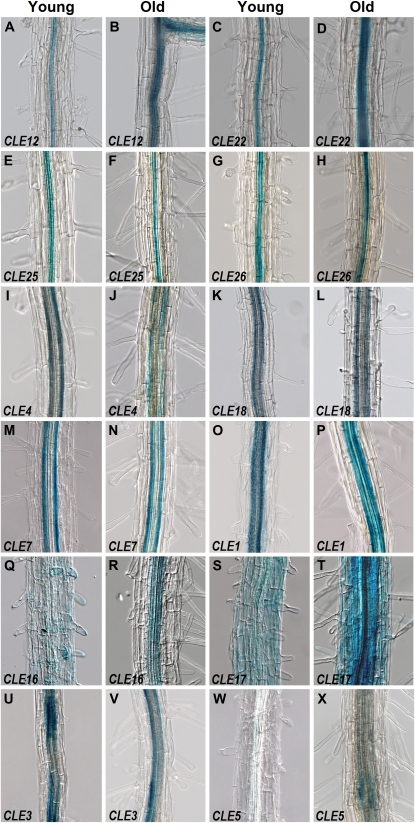
CLE promoter activity in primary root cell files. A to H, CLE12 (A and B), CLE22 (C and D), CLE25 (E and F), and CLE26 (G and H) in the stele. I to L, CLE4 (I and J) and CLE18 (K and L) in the pericycle. M to O, CLE7 (M and N) and CLE1 (O) in the pericycle and endodermis. P, CLE1 in the stele and endodermis. Q, CLE16 in the epidermis. R, CLE16 in the stele. S, CLE17 in the epidermis. T, CLE17 throughout the root. U to X, CLE3 (U and V) and CLE5 (W and X) in patches around the vascular bundle. Images in A, C, E, G, I, K, M, O, Q, S, U, and W were taken from regions of 11-d-old primary or lateral roots where the root hair cells are not fully mature. Images in B, D, F, H, J, L, N, P, R, T, V, and X were taken from the primary root maturation zone with fully differentiated root hair cells.
Seedling Expression Patterns
We first analyzed CLE promoter activity in 11-d-old Arabidopsis seedlings (Fig. 1). From this analysis, the CLE expression patterns could be divided into three groups: (1) those expressed in both shoot and root tissue (15 genes); (2) those expressed only in shoot tissue (two genes); and (3) those expressed only in root tissue (five genes), as listed in Table I. CLE8 was the only CLE gene for which expression was not detected at this stage of development.
Among the CLE genes with promoter activity in aerial tissues, CLE12, CLE18, CLE22, CLE25, and CLE26 all display GUS activity in the vascular tissues (Fig. 1, A–E). Among these, CLE12 staining is relatively weak in secondary and tertiary veins compared with primary veins and is stronger in the root than in the shoot (Figs. 1A and 2C). CLE18 and CLE22 are expressed uniformly in vascular tissue throughout the rosette leaves, although CLE18 promoter activity begins later during leaf growth than CLE22 (Fig. 1, B and C). The patterns of CLE25 and CLE26 promoter activity are complementary during leaf development. CLE25 is strongly expressed in the vascular tissue of young leaf primordia but weaker in mature leaves (Figs. 1D and 2A). CLE26 is initially detected in the leaf tip region where vein patterning initiates and only later expands into the vasculature throughout the entire leaf (Figs. 1E and 2B).
Figure 2.
CLE promoter activity in 11-d-old-seedlings. A to C, CLE25 (A), CLE26 (B), and CLE12 (C) in leaf vascular tissue. D, CLE16 in trichomes and leaf blade. E, CLE17 in trichomes and leaf margin. F to H, CLE5 (F), CLE6 (G), and CLE10 (H) in hydathodes. I, CLE9 in stomata. J, Magnified view of the region boxed in I. K to M, CLE3 (K), CLE11 (L), and CLE21 (M) in stipules. N to P, CLE10 (N), CLE5 (O), and CLE6 (P) in the leaf base. Q, CLE4 in the pith. In A and B, first to fourth rosette leaves are arranged in order from the left. Arrows indicate GUS activity in the vasculature of the basal and apical regions of young leaves (A and B, respectively) and in the midvein (C), trichomes (D and E), hydathodes (F–H), and stipules (K–M). Black and white arrowheads in J indicate a meristemoid cell and a young guard cell, respectively.
The CLE9, CLE16, CLE17, and CLE27 promoters drive GUS activity in rosette leaf blade cells (Fig. 1, F–I). CLE9 is expressed specifically in the stomatal developmental lineage, including meristemoid cells, guard mother cells, and young guard cells, throughout the aerial portions of the plant (Figs. 1F and 2, I and J). CLE16 promoter activity is detected throughout the blade pavement cells, whereas CLE17 and CLE27 promoter activity is broad in young leaves but becomes predominantly marginal as the leaves mature. CLE16 and CLE17 promoter activities are also detected in the trichomes (Figs. 1, G and H, and 2, D and E), as is CLE14 promoter activity (Supplemental Fig. S1, E and G). CLE10 activity is restricted to the rosette leaf margins (Fig. 1J), particularly in the hydathode region (Fig. 2H). The CLE5 and CLE6 promoters also drive specific GUS staining in the hydathode region (Figs. 1, K and L, and 2, F and G).
Other seedling tissues also express multiple CLE gene reporters. Hypocotyls display GUS activity driven by eight CLE promoters (Figs. 1, A, C, E, G, H, I, and M, and 2, L and M; Table I). The shoot apex region exhibits GUS activity driven by the CLE3 to -6, CLE10, CLE11, CLE16, CLE17, CLE21, and CLE27 promoters (Fig. 1, G–P). Among these, CLE3 and CLE11 show stipule-specific expression (Figs. 1, N and O, and 2, K and L). CLE4 is limited to the pith region (Figs. 1P and 2Q), whereas CLE5, CLE6, CLE10, and CLE21 expression is restricted to the base of the rosette leaves and is excluded from the shoot apical meristem (SAM; Figs. 1, J–M, and 2, M–P). CLE16, CLE17, and CLE27 are expressed in initiating leaf primordia (see below). GUS activity driven by the CLE1, CLE2, CLE7, and CLE13 promoters is specific to root tissue (Fig. 1, Q–T).
Root Expression Patterns
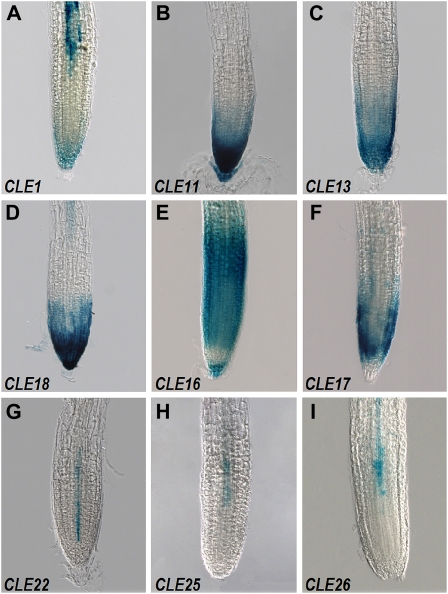
The promoters of many CLE genes are active in the root system of 11-d-old seedlings. GUS activity from four CLE promoters is detected throughout the primary root cap (Fig. 3, A–D). CLE16 activity localizes to the root cap and the elongation zone but is absent from the meristematic division zone (Fig. 3E), whereas CLE17 activity is absent from the root cap but localizes to the meristematic zone and the distal end of the elongation zone (Fig. 3F). CLE22 activity is limited to a single file of newly differentiating vascular cells (Fig. 3G), whereas CLE25 and CLE26 activity is restricted to the vascular parenchyma (Fig. 3, H and I). Vascular tissue in the elongation zone exhibits GUS activity from both the CLE1 and CLE18 promoters (Fig. 3, A and D).
Figure 3.
CLE promoter activity in the primary root tips of 11-d-old-seedlings. A, CLE1 in the root cap and vascular parenchyma. B to D, CLE11 (B), CLE13 (C), and CLE18 (D) in the root cap and apical meristem. E, CLE16 in the root cap and throughout the elongation zone. F, CLE17 in the root apical meristem. G, CLE22 in the newly differentiating vascular tissue. H and I, CLE25 (H) and CLE26 (I) in the vascular parenchyma.
More mature root tissues express multiple CLE gene reporters in overlapping patterns. We examined two different areas of 11-d-old primary and lateral roots: a younger region with immature hair cells as well as an older region with fully differentiated hair cells. We found that the stele in both areas of the root exhibits GUS activity driven by five different CLE promoters (Fig. 4, A–H; Table I). pCLE20:CLE20-GFP fusion protein activity is observed in the protoxylem and metaxylem (Supplemental Fig. S1B), whereas CLE25 and CLE26 promoter activity is restricted to the metaxylem (Supplemental Fig. S2A; data not shown). CLE22 is strongly detected in the vascular parenchyma (Supplemental Fig. S2B). Pericycle cells specifically display GUS activity from the CLE4 and CLE18 promoters (Fig. 4, I–L; Supplemental Fig. S2C), whereas both the pericycle and endodermis express CLE7 (Fig. 4, M and N; Supplemental Fig. S2D). CLE1 promoter activity is detected throughout the endodermis and the stele (Fig. 4, O and P; Supplemental Fig. S2E). In contrast, pCLE14:GFP activity is restricted to the root epidermis (Supplemental Fig. S1F).
Several CLE promoters are active in a spatially and temporally restricted fashion in the primary root. In the less mature region of the root, pCLE16:GUS and pCLE17:GUS activity are found exclusively in the epidermis (Fig. 4, Q and S), whereas in the older region, CLE16 promoter activity localizes to the stele (Fig. 4R) and CLE17 promoter activity expands throughout the root (Fig. 4T). CLE3 promoter activity is patchy in the stele in the less mature root region (Fig. 4U) but becomes limited to the pericycle and endodermis in older tissue (Fig. 4V). Finally, CLE5 promoter activity is not detected until the primary root is fully differentiated, when very weak activity is observed in the stele (Fig. 4, W and X).
CLE promoter activity is highly dynamic during lateral root formation. CLE27 promoter-driven GUS activity is detected in the cortex of the primary root at lateral root inception (Supplemental Fig. S3A). As the lateral root cells begin to grow out, the CLE27 reporter is expressed throughout the dome but most strongly in a ring around the base (Supplemental Fig. S3, B and C). As outgrowth continues, CLE27 promoter activity is lost from the lateral root tip but remains detectable in the more basal region (Supplemental Fig. S3, D and E). In mature lateral roots, CLE27 promoter activity is found at the base of the lateral root where it joins the primary root (Supplemental Fig. S3F) as well as in the meristematic zone (Supplemental Fig. S3G). CLE2 and CLE20 reporter expression is likewise detected at the base of initiating lateral roots (Supplemental Figs. S1, C and D, and S3, H and I), although at a slightly later stage than CLE27. As root outgrowth progresses, CLE2 promoter activity becomes confined to the interior cells at the junction between the primary and lateral roots (Supplemental Fig. S3J). Similarly, CLE11 promoter-driven GUS activity is observed in a small group of cells surrounding the initiating lateral roots (Supplemental Fig. S3K) as well as at the root tip and is sustained at the junction between the primary and lateral roots (Supplemental Fig. S3L). CLE5 and CLE6 promoter activity is also seen specifically in the interior cells at the junction between the primary root and the mature lateral roots (Supplemental Fig. S3, M and N).
CLE promoter activity is also dynamic in the lateral root vasculature. CLE22 is expressed earliest during lateral root formation (Supplemental Fig. S3, O and P), followed by CLE25 (Supplemental Fig. S3Q). pCLE25:GUS and pCLE26:GUS activity both localize to the vasculature of outgrowing lateral roots (Supplemental Fig. S3, Q and R), whereas CLE4 (Supplemental Fig. S3S) and CLE12 (Supplemental Fig. S3T) activity is restricted to the mature lateral root vasculature. CLE7 promoter-driven activity is likewise limited to the mature lateral root vasculature, but unlike the others, its expression is absent from the junction between the primary and lateral roots (Supplemental Fig. S3U). Thus, the expression of half a dozen CLE reporters is induced at various stages during the development of the lateral root vasculature.
Lateral root tips initiate the expression of different CLE reporters at various stages during their formation. GUS activity driven by the promoters of both CLE16 and CLE17 is detected throughout the initiating lateral roots, before becoming confined to the tips and meristematic zones as the roots grow out (Supplemental Fig. S4, A–H). CLE11 (Supplemental Fig. S4I) and CLE13 (Supplemental Fig. S4K) activity initiates at the lateral root tip at a slightly later point during outgrowth, followed by CLE18 promoter activity (Supplemental Fig. S4M) and CLE1 activity (Supplemental Fig. S4O). The promoters of all six of these CLE genes are active in the root caps of mature lateral roots (Supplemental Fig. S4, D, H, J, L, N, and P). Yet, unlike the other five, which are localized in all root cap cells, CLE1 promoter activity in lateral roots is restricted to the interior layers of the root cap. CLE18 reporter signal is strong in the root cap and weaker in cells at the very distal end of the elongation zone. In addition, initiating vascular tissues display CLE22 promoter activity (Supplemental Fig. S4Q) and more mature vascular tissues display CLE25 and CLE26 promoter activity (Supplemental Fig. S4, R and S). Lateral roots, therefore, express different combinations of CLE gene reporters in overlapping patterns throughout their development.
Eight CLE genes are represented on the ATH1 microarray used to generate a spatiotemporal map of gene expression in the roots of 5- to 7-d-old seedlings (Brady et al., 2007). Comparison of our results with this data set using the eFP Browser (Cartwright et al., 2009) showed congruent profiles for CLE3, CLE12, and CLE17. However, we did not detect activity of CLE2 in the phloem, CLE6 in phloem companion cells, CLE21 in epidermal cells of the elongation region, or CLE27 in primary root cortex and lateral root cap cells (Brady et al., 2007). Furthermore, CLE26 is reported in the metaphloem, whereas we detect it in the metaxylem. These discrepancies likely reflect the increased sensitivity of transcriptional profiling compared with GUS reporter analysis, the use of insufficient CLE regulatory sequence for some reporter constructs, and/or the difference in developmental age between the samples evaluated.
Analysis of A-Type CLE Promoter Activity in Reproductive Tissues
With a few exceptions, such as those for CLE2 and CLE8, most A-type CLE gene reporters are expressed during reproductive growth (Table II; Figs. 5–7). As in seedlings, each reproductive tissue expresses more than one CLE gene reporter. Conversely, most CLE gene reporters are expressed in more than one reproductive tissue, yet their individual expression patterns are highly specific and restricted.
Table II. Summary of pCLE:GUS activity during reproductive development.
| CLE Gene | Stem/Pedicels | Branching Points | Cauline Leaves | Sepals/Petals | Stamens |
Gynoecium |
Flower Basea | ||||
| Anthers | Filament | Stigma | Style | Ovaryb | Ovules | ||||||
| CLE1 | + | + | |||||||||
| CLE2 | |||||||||||
| CLE3 | + | ||||||||||
| CLE4 | + | + | +c | ||||||||
| CLE5 | + | + | + | ||||||||
| CLE6 | + | + | + | ||||||||
| CLE7 | + | ||||||||||
| CLE8 | |||||||||||
| CLE9 | + | + | +d | ||||||||
| CLE10 | + | + | + | + | + | + | |||||
| CLE11 | + | + | |||||||||
| CLE12 | +e | + | + | + | |||||||
| CLE13 | +e | + | + | ||||||||
| CLE14 | +d | ||||||||||
| CLE16 | + | +e | + | + | + | ||||||
| CLE17 | + | + | + | + | + | + | +c | ||||
| CLE18 | + | +de | + | ||||||||
| CLE20 | +e | ||||||||||
| CLE21 | + | + | + | ||||||||
| CLE22 | +e | + | +e | +e | + | ||||||
| CLE25 | +e | + | +e | + | |||||||
| CLE26 | +e | + | +de | + | +e | + | |||||
| CLE27 | + | + | + | ||||||||
Includes receptacle and abscission zone.
Includes valves, replum, and septum.
Siliques only.
Sepals only.
Vasculature.
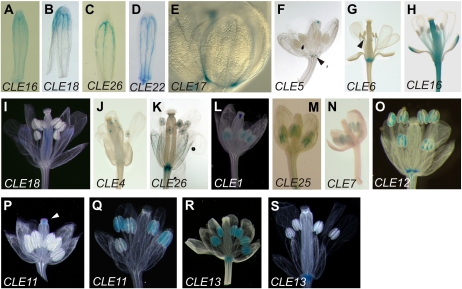
Figure 5.
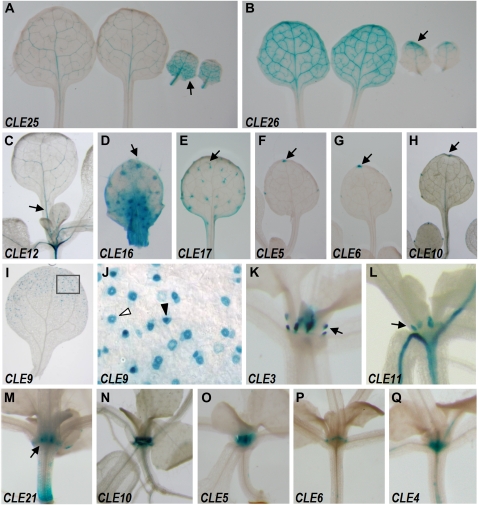
CLE promoter activity in inflorescences and cauline leaves. A, CLE16 in the epidermis. B to F, CLE12 (B), CLE13 (C), CLE22 (D), CLE25 (E), and CLE26 (F) in the vasculature. The CLE22 stem is overstained to visualize GUS activity in the cauline leaf. G to M, CLE4 (G), CLE5 (H), CLE6 (I), CLE10 (J), CLE17 (K), CLE21 (L), and CLE27 (M) in the inflorescence branching points. N, CLE9 in the stomata. O, CLE18 in the cauline leaf vasculature and marginal cells. P, CLE16 in the epidermis. Q and R, CLE22 (Q) and CLE26 (R) in the vasculature. S, CLE27 throughout the pedicels.
Figure 7.
CLE promoter activity in the gynoecium. A, CLE10 in the stigma, style, and central tissues. B to D, CLE5 (B), CLE1 (C), and CLE11 (D) in the style. E, CLE17 at the stigma/style boundary. F, CLE21 in the valve margins. G, CLE16 in the valves. H to J, CLE22 (H), CLE25 (I), and CLE26 (J) in the vasculature. The inset in H shows CLE22 in the basal vasculature and abscission zone. K, CLE25 in the funiculi and ovules. L and M, CLE27 in the ovules (L) and abscission zone (M). N, CLE17 in the silique valve margins. O, CLE4 in the silique receptacle.
Inflorescence Expression Patterns
The promoters of many CLE genes are active in inflorescence tissues. GUS activity from the CLE16 promoter is found specifically in the epidermis (Fig. 5A). The CLE12, CLE13, CLE20, CLE22, CLE25, and CLE26 reporters are expressed in the stem vasculature (Fig. 5, B–F; data not shown), in each case more strongly in the primary inflorescence branching points than elsewhere in the stem. Activity from seven CLE promoters is largely restricted to the primary branching points of the stem (Fig. 5, G–M; Table II), although CLE17 promoter activity is also present in the stem trichomes. Consistent with its expression in vegetative tissues, signal from the CLE9 reporter is detected in the stomata of stem epidermal cells (Fig. 5N). Pedicels display promoter-driven GUS activity from CLE16 in the epidermis (Fig. 5P), from CLE22 and CLE26 in the vasculature (Fig. 5, Q and R), and from CLE27 throughout (Fig. 5S).
Eight CLE promoters are active in the cauline leaves. CLE12 and CLE22 promoter activity is strong in the proximal vasculature and at the very tip of the cauline leaf (Fig. 5, B and D). The CLE26 promoter drives patchy vascular expression in the blade (Fig. 5F). CLE18-driven expression is detected in the vasculature and at the leaf margins (Fig. 5O), whereas CLE10 and CLE17 promoter-driven GUS activity is limited to the leaf margins (Fig. 5, J and K). CLE9 expression is only detected in stomatal cells (Fig. 5N), and CLE3 expression is restricted to stipules (data not shown). Promoter activity from all eight genes is also detected in rosette leaves.
Flower Expression Patterns
Floral tissues display highly complex CLE promoter activity patterns (Fig. 6). Sepals and petals express nine different CLE gene reporters. CLE9 promoter activity is detected in the sepal stomatal cells, consistent with what is observed in vegetative tissues (Fig. 2J). The CLE14 reporter is expressed in sepal trichomes (Supplemental Fig. S1H). CLE16 promoter activity is detected in sepal and petal vasculature throughout flower development, although it is stronger at the distal ends in young flowers (Fig. 6A) and becomes confined to these regions in fully mature flowers (Fig. 6H). CLE18 and CLE26 reporters are expressed in sepal veins, more strongly in their distal portion (Fig. 6, B and C). CLE22 promoter activity is observed uniformly in the vasculature of both sepals and petals, although it becomes more pronounced in the sepal veins in fully developed flowers (Fig. 6D). CLE17 promoter activity is detected along the margins of sepals and petals in young flower buds (Fig. 6E). CLE5 and CLE6 reporters are expressed at the base of each flower organ type, above the abscission zone (Fig. 6, F and G).
Figure 6.
CLE promoter activity in flowers. A to D, CLE16 (A), CLE18 (B), CLE26 (C), and CLE22 (D) in sepal vasculature. E, CLE17 in sepal and petal margins. F and G, CLE5 (F) and CLE6 (G) at the base of the flower. H to K, CLE16 (H), CLE18 (I), CLE4 (J), and CLE26 (K) in the stamen filaments. L to N, CLE1 (L), CLE25 (M), and CLE7 (N) in the anthers. O to S, CLE12 (O), CLE11 (P and Q), and CLE13 (R and S) in the pollen grains.
The male reproductive structures express a variety of CLE gene reporters (Fig. 6). Each stamen-expressed reporter is present in either the anther or the filament but not in both. Five CLE reporters are expressed in the filament (Table II). CLE16 and CLE18 are uniformly detected throughout the entire filament (Fig. 6, H and I). In contrast, CLE6 promoter activity is detected very weakly in the distal region of the filament (Fig. 6G), whereas CLE4 and CLE26 promoter activity is limited to the distal tip of the filament, where it connects to the anther (Fig. 6, J and K). CLE4 is the only one of these that is expressed exclusively in the filament and not in other floral tissues.
Anthers express six different CLE gene reporters (Table II). CLE1 promoter activity is found in both pollen grains and tapetum cells of anthers from their emergence in stage 6 floral buds until maturation (Fig. 6L). The CLE25 reporter is expressed throughout the anthers in young developing flowers (Fig. 6M). In mature flowers, CLE25 expression become more restricted and overlaps with that of CLE7 in the central region of the anther sacs, along the boundaries with the connective tissue (Fig. 6N). CLE11, CLE12, and CLE13 promoter activity is evident solely in pollen grains. The CLE12 reporter is expressed throughout all stages of pollen development (Fig. 6O). However, CLE11 and CLE13 reporters show a dynamic and complementary expression pattern: the CLE11 reporter is expressed only in fully mature pollen grains (Fig. 6, P and Q), whereas the CLE13 reporter is expressed only in young developing anthers (Fig. 6, R and S).
The female reproductive structure exhibits CLE promoter activity in a variety of complex patterns (Fig. 7). The CLE10 reporter is the most broadly expressed, its activity being detected in the stigma, style, transmitting tract, septum, and ovules (Fig. 7A). The style also displays promoter activity from CLE1, CLE5, and CLE11. Among these, CLE5 and CLE1 reporters are expressed in the transmitting tract of the style (Fig. 7, B and C), whereas CLE11 reporter expression is observed in a central region that appears to correspond to the transmitting tract or vascular fans (Fig. 7D). Uniquely, CLE17 promoter activity is detected in a ring at the margin between the stigmatic tissue and the style, in the transmitting tract, and in the septum (Fig. 7E). CLE21 promoter activity is observed in the valve margins (Fig. 7F), whereas CLE16 promoter activity occurs in the valves (Fig. 7G). The vasculature of the gynoecium exhibits CLE22, CLE25, and CLE26 promoter activity (Fig. 7, H–J). CLE25 reporter expression is additionally detected in the septum, funiculi, and at the proximal end of the ovules (Fig. 7K), whereas the CLE27 reporter is expressed throughout the funiculi and ovules of fully mature flowers (Fig. 7L). Finally, the abscission zone exhibits promoter activity from CLE10 (Fig. 5J), CLE12 (Fig. 6O), CLE13 (Fig. 6, R and S), CLE16 (Fig. 6H), CLE21 (Fig. 7F), CLE22 (Fig. 7H), CLE26 (Fig. 7J), and CLE27 (Fig. 7M). These promoter activity patterns are consistent throughout gynoecium and silique development, with two exceptions. First, CLE17 reporter expression shifts from the stigma/style region in carpels to the valve margins in siliques (Fig. 7N). Second, CLE4 reporter expression becomes detectable specifically in the silique receptacle (Fig. 7O). In sum, our observations indicate that different CLE promoters drive GUS activity in complex, overlapping spatial and temporal patterns, particularly in the reproductive tissues.
Detailed Analysis of CLE Promoter Activity in Shoot Apices
Promoter activity from 10 different CLE genes is detected around the shoot apex region of seedlings (Figs. 1 and 2). We examined their expression patterns in greater detail by sectioning 10-d-old GUS-stained seedlings. We found that CLE21 promoter activity is restricted to the stipules (Supplemental Fig. S5A), whereas CLE10 promoter activity is detected in stipules and in a proximal, adaxial domain of leaf primordia (Supplemental Fig. S5B). CLE4 and CLE26 promoter activity is located in the pith and the ground cells of the hypocotyl, respectively (Supplemental Fig. S5, C and D).
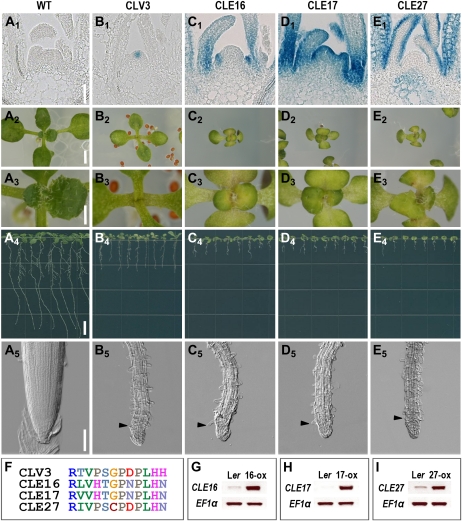
In contrast, the CLE16, CLE17, and CLE27 reporter constructs show consistent activity in or adjacent to the SAM in multiple independent transgenic lines. Compared with untransformed wild-type plants (Fig. 8A1) and pCLV3:GUS plants (Fig. 8B1), CLE16 promoter activity is detected throughout initiating leaf primordia on the SAM flanks (Fig. 8C1). As the leaf primordia develop, CLE16 GUS activity is stronger in the proximal than the distal region and in the L1 cells (Fig. 8C1; Supplemental Fig. S6, A and B). A similar pattern is observed in pCLE17:GUS leaf primordia (Fig. 8D1). However, CLE17 promoter activity is also detected in the SAM (Fig. 8D1). Transgenic plants with weak CLE17 promoter activity show GUS staining in the outer layers of the central zone and the peripheral zone (PZ; Supplemental Fig. S6C). Transgenic plants with strong CLE17 promoter activity exhibit GUS staining throughout the SAM and initiating leaf primordia, most strongly in the PZ and the outer cell layers (Supplemental Fig. S6D). CLE27 promoter activity is exclusively detected in the epidermal layer of developing leaf primordia and young rosette leaves and in the pith region beneath the rib meristem (Fig. 8E1; Supplemental Fig. S6, E and F). However, CLE27 reporter expression is excluded from the SAM and the initiating leaf primordia on the meristem flanks. These results are consistent with Arabidopsis SAM transcription profiling data showing that CLE27 mRNA is absent from the SAM but that CLE17 is expressed at low levels in the central zone and organizing center and at moderate levels in the PZ (Yadav et al., 2009). CLE16 is not represented in the profiling data set.
Figure 8.
CLE16, CLE17, and CLE27 overexpression phenotypes. A1 to E1, Longitudinal sections of 10-d-old wild-type (WT), pCLV3:GUS, pCLE16:GUS, pCLE17:GUS, and pCLE27:GUS plant shoot apices, respectively. A2 to A5, Thirteen-day-old Landsberg erecta (Ler) seedlings. B2 to B5, Thirteen-day-old p35S:CLV3 seedlings. C2 to C5, Thirteen-day-old p35S:CLE16 seedlings. D2 to D5, Thirteen-day-old p35S:CLE17 seedlings. E2 to E5, Thirteen-day-old p35S:CLE27 seedlings. F, Comparison of CLE peptide sequences. Alignment was performed using MUSCLE (Edgar, 2004). Each color represents a different amino acid residue. G to I, RT-PCR analysis of CLE expression in transgenic plants. EF1α is used as a control. A3 to E3 are magnified views of shoot apex in A2 to E2. Nomarski images of the roots in A5 to E5 show root hair differentiation (arrowheads) close to the root tip. Bars = 100 μm in A1 to E1 and A5 to E5, 2.5 mm in A2 to E2, 1 mm in A3 to E3, and 5 mm in A4 to E4.
Because CLE17 reporter expression in the SAM overlapped with the CLV3 expression domain, we tested whether CLE17 could activate the CLV signaling pathway by analyzing the phenotypes of p35S:CLE17 transgenic plants. Ectopic expression of CLV3 causes SAM arrest early during vegetative development (Fig. 8, B2 and B3) as well as premature floral meristem termination (Brand et al., 2000). In contrast, ectopic expression of CLE17 does not confer a SAM termination phenotype (Fig. 8, D2 and D3), nor is floral organ formation affected. These data indicate that CLE17 cannot activate the CLV3 signaling pathway in either shoot or floral meristems. Instead, the rosette leaves of CLE17-overexpressing plants show a delayed growth rate and smaller and epinastic morphology compared with wild-type rosette leaves (Fig. 8, D2 and D3). Developmental timing is also delayed, and apical dominance appears to be reduced. In addition, CLE17 overexpression (Fig. 8H) causes root apical meristem termination (Fig. 8, D4 and D5), as has been observed in plants that have been treated with exogenous CLE peptides or that overexpress other CLE genes, such as CLV3 (Fig. 8, B4 and B5). Transgenic plants overexpressing either CLE16 or CLE27 (Fig. 8, G and I) display phenotypes that closely resemble those of p35S:CLE17 plants (Fig. 8, C2–C5 and E2–E5).
Alignment of the CLE16, CLE17, and CLE27 peptides with that of CLV3 reveals altered amino acids at key residues in the CLE domain. Compared with CLV3, all three peptides contain an Asn instead of a His at position 12, CLE16 and CLE17 contain a His instead of a Pro at position 4 as well as an Asn instead of an Asp at position 8, and CLE27 contains a Cys instead of the highly conserved Gly at position 6 (Fig. 8F). These observations suggest that the failure of these three proteins to activate the CLV3 signaling pathway when overexpressed in the SAM may be due to differences in the composition of their CLE motifs.
CLE Overexpression Phenotypes in Shoots and Roots
Despite extensive overexpression studies that uncovered other developmental processes responding to CLE peptide activity, the overexpression phenotypes of a few CLE genes remain undetermined. To fill in this gap, we generated transgenic plants expressing the coding region of CLE8, CLE12, or CLE22 under the control of the cauliflower mosaic virus 35S promoter and scored them for shoot and root meristem arrest phenotypes (Supplemental Table S1). We found that p35S:CLE8 plants showed neither shoot nor root meristem defects (Supplemental Fig. S7B), whereas both p35S:CLE12 and p35S:CLE22 plants displayed SAM termination (Supplemental Fig. S7, C and D). Reduced root growth and root apical meristem arrest were also observed in CLE12- and CLE22-overexpressing plants (Supplemental Fig. S7, E and F). Our data are consistent with previous work except in the case of CLE8, which has been reported to trigger root apical meristem consumption when overexpressed (Ito et al., 2006). Combined with earlier studies, our results indicate that 16 of 26 A-type CLE genes, including CLV3, can induce SAM termination when overexpressed and that 18 (or 19) A-type CLE genes can induce root apical meristem termination.
Mutational Analysis of A-Type CLE Loci
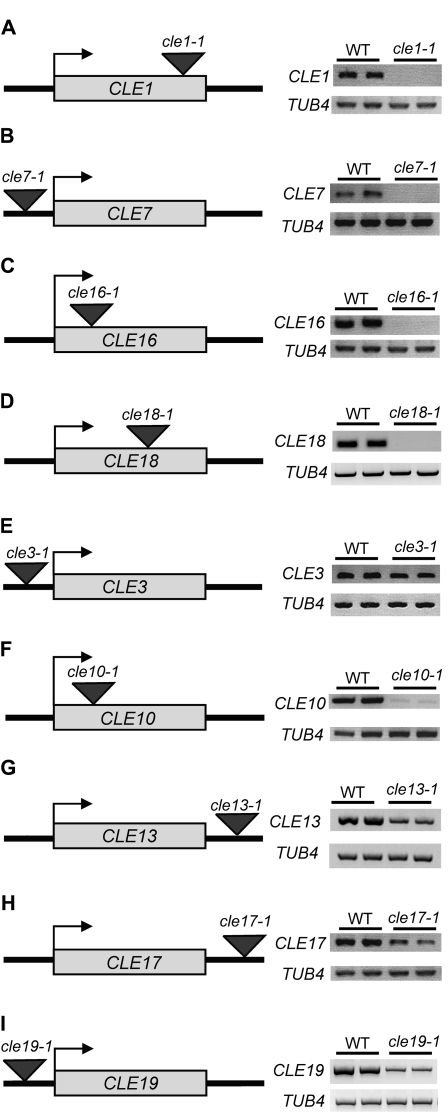
To begin to determine the functions of additional A-type CLE genes, we obtained cle T-DNA insertion alleles from the publicly available collections. We identified one allele each with an insertion within the CLE1, CLE10, CLE16, and CLE18 coding regions, one allele each with an insertion in the CLE3, CLE7, CLE13, and CLE19 5′ untranslated regions, and one allele with an insertion in the CLE17 3′ untranslated region (Fig. 9). We performed RT-PCR to examine CLE mRNA transcript levels in plants homozygous for each of these cle alleles. No transcripts were detected in cle1-1, cle7-1, cle16-1, and cle18-1 plants (Fig. 9, A–D), indicating that they represent null alleles. Reduced transcript levels were detected in cle3-1, cle10-1, cle13-1, cle17-1, and cle19-1 plants (Fig. 9, E–I), showing that these are hypomorphic alleles. For cle3-1, we observed a minor but reproducible decrease in CLE3 mRNA levels (Fig. 9E). In addition, we identified T-DNA insertion alleles within 700 bp upstream or downstream of the CLE2, CLE4, CLE6, CLE9, CLE17, and CLE21 coding regions, but RT-PCR experiments detected no significant reduction in transcript levels (Supplemental Table S2). Detailed inspection of seedling, inflorescence, flower, and root development revealed no detectable morphological defects in plants homozygous for any of these cle alleles, suggesting that substantial functional redundancy occurs among A-type CLE family members.
Figure 9.
Characterization of CLE insertion alleles. Location of the insertion allele relative to each CLE coding region (gray box) and CLE mRNA transcript levels in two individual wild-type (WT) and homozygous cle mutant plants are shown. TUBULIN4 (TUB4) was used as a control.
DISCUSSION
Dozens of potential intercellular signaling molecules as well as hundreds of putative receptors have been cataloged in the Arabidopsis genome, yet relatively little is known about their individual expression patterns or functions. Several members of the A-type CLE family of small secreted polypeptides act to maintain cell fate in shoot and root apical meristems; however, the dearth of mutations in the other small CLE coding sequences has limited our insight into their biological activities. In addition, only a single study of A-type CLE mRNA transcription profiles exists (Sharma et al., 2003), and few CLE genes are represented on microarrays, another source of detailed expression data. To address these deficiencies, we performed a comprehensive characterization of A-type CLE promoter expression during Arabidopsis vegetative and reproductive development and identified null or hypomorphic alleles of seven CLE genes.
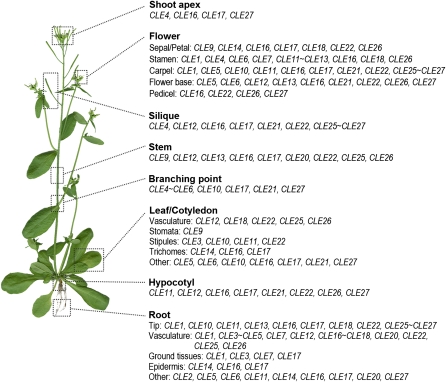
One important finding from our work is that most Arabidopsis tissues express one or more CLE gene reporters (Fig. 10). This includes highly specialized cell types such as stomata, trichomes, and stipules. Primary roots display CLE promoter-driven expression in the root cap, the root apical meristem, and each radial cell layer (Figs. 3 and 4), and lateral root formation is associated with the dynamic activity of multiple CLE promoters (Supplemental Fig. S3). The vasculature is characterized by the expression of 14 CLE reporters, half of which are specific to either the root or the shoot vasculature. Multiple CLE gene reporters also are expressed in different tissues of the inflorescence stem (Fig. 5) as well as in each floral organ. In particular, the reproductive organs express a variety of CLE gene reporters in specific spatial and temporal patterns (Figs. 6 and 7). Seven different CLE reporters are expressed at the base of the flower, where the cognate genes may be involved in the signaling process(es) that controls floral organ abscission. These observations indicate that, beyond their functions in shoot and root apical meristems, CLE-mediated signal transduction pathways are likely to play roles in a wide variety of different biological processes.
Figure 10.
Summary of A-type CLE promoter activity in Arabidopsis. Shown is a list of the CLE genes expressed in the various tissues of a mature Arabidopsis plant. [See online article for color version of this figure.]
Another intriguing finding is that the A-type CLE gene promoters drive highly distinct and specific patterns of expression. For example, the promoters of CLE3, CLE5, CLE16, and CLE17 are active in a spatially and temporally restricted fashion in the primary root (Fig. 4), whereas the CLE1, CLE5, CLE11, CLE16, CLE17, and CLE21 promoters are active in unique subdomains of the developing gynoecium (Fig. 7). No two CLE promoters drive expression in identical patterns throughout the plant, and indeed, we observe that even CLE genes with very similar sequences have divergent reporter expression patterns. This is exemplified by CLE3 and CLE4, which have an identical CLE motif and pair together in the published phylogenies (Ito et al., 2006; Strabala et al., 2006; Jun et al., 2008; Mitchum et al., 2008). We detect CLE3 promoter activity in leaf stipules (Fig. 2K) and the pericycle and endodermis of mature roots (Fig. 4, U and V), whereas CLE4 promoter activity coincides with CLE3 in the root pericycle (Fig. 4, I and J) but is also found in the hypocotyl pith (Supplemental Fig. S5C), inflorescence branch points (Fig. 5G), stamen filaments (Fig. 6J), and receptacle (Fig. 7O). Thus, while these two genes may function interchangeably in the pericycle, they would appear to have unique activities in the other tissues. The importance of the location of gene expression in conferring functional specificity has also been shown with closely related members of the MADS domain transcription factor family (Pinyopich et al., 2003).
Although many CLE proteins act interchangeably when ectopically expressed in shoots or roots, indicating that tissue distribution is important for their functional specificity, the location of gene expression is not the only determinant of CLE function. Studies have shown that the CLE motif itself determines much of the functional specificity of the proteins in different plant tissues (Fiers et al., 2005, 2006; Ito et al., 2006; Kondo et al., 2006; Ni and Clark, 2006; Meng et al., 2010). We found that the CLE17 promoter is active in the SAM in a domain that overlaps with CLV3, yet its overexpression fails to induce a CLV3 overexpression SAM termination phenotype (Fig. 8). The CLE peptide of the SAM-expressed CLE17 gene differs from that of CLV3 at several key residues, including the C-terminal residue (His-12) that plays an essential role in CLV3 peptide function and binding to the CLV1 receptor kinase (Kondo et al., 2008). These data provide an additional piece of evidence that the CLE motif plays a critical role in determining CLE activity and receptor-binding specificity in planta. Other factors contributing to CLE signaling specificity are likely to include the tissue distribution of their cognate receptors as well as of the enzymes that process the CLE proteins to the active arabinosylated peptides (Ohyama et al., 2009).
To date, assessing the biological roles of small signaling peptide gene families has proven to be a significant challenge, primarily due to a lack of hypomorphic or null alleles. These are not available for most CLE family members because the small size of the genes reduces the target size for T-DNA insertion and because the mature molecule consists of only a short stretch of amino acids. We have identified null mutations in the CLE1, CLE7, CLE16, and CLE18 genes, but in each case the homozygous plants lack detectable morphological phenotypes, indicating that their function may be conditioned by environmental factors and/or masked by redundancy with other CLE peptides. In the future, alternative strategies will be required to specifically target CLE genes for down-regulation. Possibilities include artificial microRNAs (Schwab et al., 2006) and increased efficiency homologous recombination (Osakabe et al., 2010; Zhang et al., 2010).
Although each CLE promoter is active in a unique spatial and temporal pattern during Arabidopsis development, we observe that multiple CLE promoters are active in overlapping patterns within a given tissue. These overlapping CLE genes are most likely to have redundant functions; thus, our complete gene family promoter expression analysis serves as a guide to identify potential redundant CLE peptides within specific tissues. However, it should be noted that overlapping expression patterns do not necessarily guarantee redundant activities and that CLE genes expressed in the same cell types could potentially send opposite signals, as has been observed for members of the EPIDERMAL PATTERNING FACTOR family of small peptide ligands during stomatal development (Abrash and Bergmann, 2010).
Finally, our elucidation of CLE promoter activity throughout Arabidopsis development provides a resource for predicting candidate receptors based on their overlapping or neighboring expression patterns. For instance, the PHLOEM INTERCALATED WITH XYLEM (PXY) gene encodes an LRR receptor kinase that belongs to the same clade of LRR-RLK subclass XI proteins as CLV1 (Hirakawa et al., 2008). PXY is expressed in dividing vascular procambium cells (Fisher and Turner, 2007) and interacts with the B-type CLE peptide CLE41 (Etchells and Turner, 2010). We find that 14 different CLE promoters drive GUS activity in the vasculature (Table I). Thus, the products of one or more of these CLE genes could act as ligands for PXY and/or for VASCULAR HIGHWAY1, another LRR-RLK expressed in procambial cells throughout the plant (Clay and Nelson, 2002). An LRR-RLK encoded by the EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS (EMS1/EXS) gene is expressed in the sporogenous and parietal cells of the developing anther, where it controls microsporocyte differentiation and tapetal identity (Canales et al., 2002; Zhao et al., 2002). The CLE1, CLE7, CLE12, CLE13, and CLE25 promoters are all active during early anther formation, making these genes candidates to encode EMS1/EXS ligands. EMS1/EXS is also expressed in developing leaf primordia, inflorescence meristems, and young flower buds (Canales et al., 2002). Yet, among the CLE promoters active in young anthers, only those of CLE12, CLE13, and CLE25 are also active in leaf primordia and none is active in or adjacent to the inflorescence meristem. Thus, broadly expressed receptors such as EMS1/EXS may potentially bind different CLE ligands deriving from different cell types. In conclusion, our systematic analysis of the CLE gene family illustrates the complex expression dynamics of these signaling molecules throughout the Arabidopsis life cycle and provides a foundation for identifying and characterizing many ligand-receptor-mediated signaling pathways during plant development.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotype Columbia and Landsberg erecta plants were used in this study. Seeds were imbibed at 4°C for 3 d before sowing and were grown in a greenhouse under long days (16 h of light and 8 h of dark) with a day/night temperature cycle of 22°C/18°C. Seeds were surface sterilized for 10 min in 5% NaOCl and 0.1% Triton X-100, rinsed in distilled water, and plated on plates containing Murashige and Skoog medium with 0.8% type M agar (Life Technologies), 0.5 mm MES, pH 5.7, 0.5% Suc, and 1 mL/L Gamborg's vitamin solution (Sigma).
Construction of Transgenic Plants
To generate CLE promoter:GUS fusion constructs, the 5′ upstream region (974–3,398 bp) of each CLE gene was PCR amplified from Columbia genomic DNA and cloned into a binary vector for transformation. For each construct, the binary vector in the Agrobacterium tumefaciens strain GV3101 or ASE was introduced into plants by the floral dip method (Clough and Bent, 1998). Primer sequences are listed in Supplemental Table S3.
Histochemical Assays
GUS staining of transgenic plants was performed as described (Jefferson et al., 1987), with the modification that 0.5 mm potassium ferrocyanide and 2.5 mm potassium ferricyanide were used. Incubation times ranged from 2 to 16 h following vacuum infiltration. Subsequent tissue embedding and sectioning were performed as described (Sieburth and Meyerowitz, 1997). For roots and some flower samples, whole-mount clearing was performed in Hoyer’s medium (Liu and Meinke, 1998), and the samples were visualized using a Zeiss Axiophot microscope equipped with Nomarski optics. Whole seedlings, inflorescences, and flower specimens were imaged using a Zeiss Stemi 2000-c or a Zeiss Stemi SV11 microscope. GFP fluorescence was visualized using a Leica DM LB fluorescence microscope or a Zeiss LSM 510 confocal microscope with 488-nm/530-nm excitation/emission light.
mRNA Transcript Analysis
cDNA was synthesized from 1 to 5 μg of total RNA using an oligo(dT18) primer and SuperScript III reverse transcriptase (Invitrogen). For RT-PCR, 1 μL of the first-strand cDNA reaction was used as a template. The annealing temperature for RT-PCR was 55°C to 60°C for all primer pairs, and the number of PCR cycles was as follows: EF1α, 23 cycles; TUB4, 26 cycles; CLE2, CLE16, CLE17, and CLE27, 30 cycles; CLE1, CLE3, CLE4, CLE6, CLE7, and CLE19, 35 cycles; CLE18, 40 cycles. Primer sequences are listed in Supplemental Table S3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. CLE14 and CLE20 promoter activity in vegetative and reproductive tissues.
Supplemental Figure S2. Examples of CLE promoter activity in primary root vasculature.
Supplemental Figure S3. CLE promoter activity during lateral root development.
Supplemental Figure S4. CLE promoter activity in lateral root tips of 11-d-old-seedlings.
Supplemental Figure S5. CLE promoter activity in the shoot apex region.
Supplemental Figure S6. CLE16, CLE17, and CLE27 promoter activity in the shoot apex.
Supplemental Figure S7. CLE8, CLE12, and CLE22 overexpression phenotypes.
Supplemental Table S1. CLE overexpression meristem phenotypes.
Supplemental Table S2. CLE insertion alleles.
Supplemental Table S3. Oligonucleotides used in this study.
Supplementary Material
Acknowledgments
We thank Niki Kubat, Vicky Chen, and Liz Fong for assistance handling plants and generating constructs; the RIKEN, SAIL, GABI-KAT, and SALK collections for supplying indexed insertion mutant lines; and Sheila McCormick and Barbara Baker for helpful discussions.
References
- Abrash EB, Bergmann DC. (2010) Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson HG. (2002) EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol 12: 1718–1727 [DOI] [PubMed] [Google Scholar]
- Cartwright DA, Brady SM, Orlando DA, Sturmfels B, Benfey PN. (2009) Reconstructing spatiotemporal gene expression data from partial observations. Bioinformatics 25: 2581–2587 [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martínez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B. (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Clay NK, Nelson T. (2002) VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell 14: 2707–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cock JM, McCormick S. (2001) A large family of genes that share homology with CLAVATA3. Plant Physiol 126: 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Turner SR. (2010) The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137: 767–774 [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, van der Schors R, van der Geest L, Li KW, Stiekema WJ, Liu CM. (2006) The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol 141: 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM. (2005) The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Hause G, Boutilier K, Casamitjana-Martínez E, Weijers D, Offringa R, van der Geest L, van Lookeren Campagne M, Liu C-M. (2004) Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327: 37–49 [DOI] [PubMed] [Google Scholar]
- Fisher K, Turner S. (2007) PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. (2008) Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobe M, Müller R, Grünewald M, Brand U, Simon R. (2003) Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol 213: 371–381 [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JH, Fiume E, Fletcher JC. (2008) The CLE family of plant polypeptide signaling molecules. Cell Mol Life Sci 65: 743–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Nakamura T, Yokomine K, Sakagami Y. (2008) Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides. Biochem Biophys Res Commun 377: 312–316 [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KFX, Berger J, Jürgens G. (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Liu CM, Meinke DW. (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16: 21–31 [DOI] [PubMed] [Google Scholar]
- Meng L, Ruth KC, Fletcher JC, Feldman L. (2010) The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Mol Plant 3: 760–772 [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Wang X, Davis EL. (2008) Diverse and conserved roles of CLE peptides. Curr Opin Plant Biol 11: 75–81 [DOI] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R. (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Clark SE. (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol 140: 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Osakabe K, Osakabe Y, Toki S. (2010) Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc Natl Acad Sci USA 107: 12034–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC. (2002) CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T. (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Ramirez J, Fletcher JC. (2003) The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol 51: 415–425 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. (2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Strabala TJ, O’Donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HCC, Hudson KR. (2006) Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol 140: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P. (2008) Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci USA 105: 18625–18630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. (2009) Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci USA 107: 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H. (2002) The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.