Abstract
Glutathione, a nonribosomal thiol tripeptide, has been shown to be critical for many processes in plants. Much less is known about the roles of glutathione in cyanobacteria, oxygenic photosynthetic prokaryotes that are the evolutionary precursor of the chloroplast. An understanding of glutathione metabolism in cyanobacteria is expected to provide novel insight into the evolution of the elaborate and extensive pathways that utilize glutathione in photosynthetic organisms. To investigate the function of glutathione in cyanobacteria, we generated deletion mutants of glutamate-cysteine ligase (gshA) and glutathione synthetase (gshB) in Synechocystis sp. PCC 6803. Complete segregation of the ΔgshA mutation was not achieved, suggesting that GshA activity is essential for growth. In contrast, fully segregated ΔgshB mutants were isolated and characterized. The ΔgshB strain lacks reduced glutathione (GSH) but instead accumulates the precursor compound γ-glutamylcysteine (γ-EC). The ΔgshB strain grows slower than the wild-type strain under favorable conditions and exhibits extremely reduced growth or death when subjected to conditions promoting oxidative stress. Furthermore, we analyzed thiol contents in the wild type and the ΔgshB mutant after subjecting the strains to multiple environmental and redox perturbations. We found that conditions promoting growth stimulate glutathione biosynthesis. We also determined that cellular GSH and γ-EC content decline following exposure to dark and blue light and during photoheterotrophic growth. Moreover, a rapid depletion of GSH and γ-EC is observed in the wild type and the ΔgshB strain, respectively, when cells are starved for nitrate or sulfate.
Photosynthetic organisms are constantly faced with the threat of reactive oxygen species (ROS) generated as a by-product of photosynthesis and cellular metabolism (Asada, 1999). To overcome these challenges, photosynthetic organisms have developed robust antioxidant and redox buffering systems composed of enzymatic and small molecule components (Latifi et al., 2009). Glutathione is a small, ubiquitous molecule that is involved in a plethora of cellular processes in addition to its role as an antioxidant and in the maintenance of cellular redox homeostasis (Schafer and Buettner, 2001). Compared with heterotrophic organisms such as yeast (Penninckx, 2000) and Escherichia coli (Masip et al., 2006), less is known regarding the roles of glutathione in photosynthetic organisms, despite an array of studies performed in plants (Meyer, 2008; Rouhier et al., 2008; Foyer and Noctor, 2009). The disparity is likely due to the extensive diversity of pathways involving glutathione metabolism in photoautotrophs compared with heterotrophs (Meyer and Hell, 2005). Surprisingly, even less is known about the functions of glutathione in cyanobacteria. This is especially significant given that glutathione metabolism likely evolved with the advent of oxygenic photosynthesis in cyanobacterial ancestors (Copley and Dhillon, 2002). There are many similarities between processes involving glutathione in plants and cyanobacteria. Cyanobacteria have smaller gene families involving glutathione metabolism compared with plants (Rouhier et al., 2008), making them excellent candidates for the study of glutathione metabolism in photosynthetic organisms. In this study, we investigated the role of glutathione in the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803), a model photosynthetic organism.
Glutathione (l-γ-glutamyl-l-cysteinyl-Gly) is a low-Mr thiol tripeptide that is synthesized through two sequential ATP-dependent steps catalyzed by Glu-Cys ligase (GshA) and glutathione synthetase (GshB; Fig. 1A). GshA catalyzes the ligation of Cys with the γ-carboxyl group of Glu to form γ-glutamyl-Cys (γ-EC). GshB ligates Gly to the Cys residue of γ-EC to form glutathione. The Glu-Cys ligase (Gsh1) in Arabidopsis (Arabidopsis thaliana) has been extensively characterized and shown to be redox regulated (Jez et al., 2004; Hicks et al., 2007). GshA from the cyanobacteria Anabaena sp. PCC 7120 has been biochemically characterized, but there is no evidence that its activity is redox modulated (Ashida et al., 2005). The kinetic mechanism of glutathione synthetase (Gsh2) from Arabidopsis has been described (Jez and Cahoon, 2004), and enzymatic activity has been demonstrated for the GshB protein in the cyanobacterium Synechococcus sp. PCC 7942 (Okumura et al., 1997).
Figure 1.
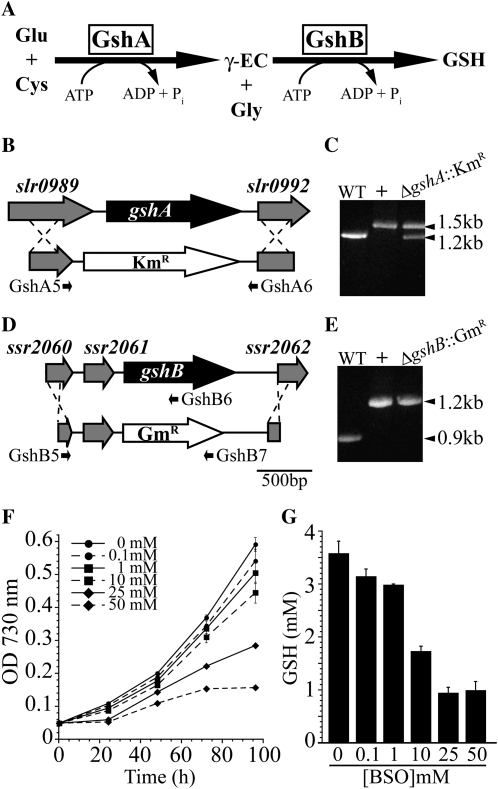
Disruption of glutathione biosynthesis in Synechocystis 6803. A, Diagram of the glutathione biosynthetic pathway. B, The entire open reading frame of the gshA gene was replaced with a kanamycin resistance cassette (KmR) to generate the ΔgshA::KmR strain. C, Segregation of ΔgshA::KmR was tested by PCR using primers GshA5 and GshA6 shown in A. Lanes show wild-type genomic DNA (WT), pSL2083 (+), and ΔgshA::KmR genomic DNA. D, The gshB gene was replaced with a gentamicin resistance cassette (GmR). E, Segregation of ΔgshB::GmR was confirmed by PCR using primers GshB5, GshB6, and GshB7 shown in D. Lanes show wild-type genomic DNA (WT), pSL2085 (+), and ΔgshB::GmR genomic DNA. F, Growth of wild-type cells in the presence of the GshA inhibitor BSO. G, Cellular GSH concentration after 96 h of growth in the presence of BSO. Primer sequences used in cloning and segregation analysis are shown in Table I.
Glutathione accumulates to millimolar levels within the cell, primarily in the reduced form (GSH). GSH can undergo intermolecular oxidation to form glutathione disulfide (GSSG), a process that can be reversed enzymatically by glutathione reductase through NADPH-dependent reduction of the disulfide (Serrano et al., 1984). The reduction potential of the 2GSH/GSSG redox couple is dependent on the absolute concentration and the ratio of reduced to oxidized glutathione, and it has been proposed that this redox couple contributes significantly to the cellular redox environment (Schafer and Buettner, 2001). Changes to the glutathione redox state have been proposed to be involved in cellular signaling pathways in plants (Meyer, 2008). However, analysis of the cellular redox environment is complicated by the fact that even within a single cell, different cellular organelles may maintain the glutathione couple at different reducing potentials (Meyer et al., 2007; Wolf et al., 2008). The comparative simplicity of the cyanobacterial cell makes biochemical interpretations more clear, potentially providing insight into the functions of glutathione within the plant chloroplast.
In plants, glutathione is critical for many cellular functions (Mullineaux and Rausch, 2005; Rouhier et al., 2008; Foyer and Noctor, 2009). Genetic perturbation of glutathione biosynthesis in Arabidopsis has dramatic consequences for cellular development, hampering genetic studies of glutathione deficiency in fully developed plants. Weak alleles of gsh1 (Glu-Cys ligase) in Arabidopsis result in cadmium sensitivity due to the role of GSH in phytocheletin synthesis and heavy metal detoxification (Howden et al., 1995). Strong alleles of gsh1 result in seedlings lacking a root meristem, and null alleles are embryo lethal (Vernoux et al., 2000; Cairns et al., 2006). Null mutations in gsh2 result in a seedling-lethal phenotype in Arabidopsis. Homozygous gsh2 mutant ovules accumulate high levels of the glutathione precursor γ-EC, which is presumably exported from the chloroplast into the cytoplasm (Pasternak et al., 2008). In plants and algae, ascorbate and glutathione function in a glutathione-ascorbate cycle and constitute the major soluble antioxidant network for the degradation of hydrogen peroxide (H2O2; Asada, 1999) in combination with other cellular components, including glutathione reductase and catalase (Mhamdi et al., 2010). However, ascorbate concentrations in cyanobacteria are about 250 times lower than that reported for plant chloroplasts, and while ascorbate peroxidase activities have been reported for Nostoc muscorum 7119 and Synechococcus 6311 and 7942 (Tel-Or et al., 1985; Mittler and Tel-Or, 1991), recent genome analysis has not identified a gene with similarity to a plant-like ascorbate peroxidase in Synechocystis 6803 (Stork et al., 2005). Therefore, glutathione appears to be the major water-soluble antioxidant in cyanobacteria.
It has been known for some time that cyanobacteria contain high levels of glutathione (Fahey et al., 1978; Tel-Or et al., 1985). Furthermore, it has been demonstrated that addition of the precursor amino acid Cys to Synechocystis 6803 results in increased glutathione accumulation (Suginaka et al., 1998) and increased heat tolerance (Suginaka et al., 1999). Recently, immunocytochemical methods were used to identify the subcellular localization of glutathione and Cys in Synechocystis 6803 (Zechmann et al., 2010). This report also claimed that glutathione and Cys levels are reduced when cells are grown in sulfate-deplete medium, a phenomenon also observed in yeast (Elskens et al., 1991) that likely involves degradation by γ-glutamyltranspeptidase. To date, there has been a single report of the genetic deletion of a glutathione biosynthesis gene in cyanobacteria. Okumura et al. (1997) isolated a gshB mutant strain from a screen for pigment biosynthesis mutants in Synechococcus 7942 and found that the mutant appeared more yellow in color compared with the wild type but had similar growth under photoautotrophic conditions. However, this report did not extensively characterize the mutant phenotypes under adverse conditions. Recently, there have been some exciting studies showing the importance of glutaredoxins in Synechocystis 6803, leading to the discovery of a novel pathway for selenate tolerance (Marteyn et al., 2008), insight into their potential role in iron-sulfur cluster delivery (Picciocchi et al., 2007; Iwema et al., 2009), and in arsenate reduction and detoxification (López-Maury et al., 2009). While these studies have shed light on some aspects of the roles of glutathione, it is not known how glutathione levels change in response to cellular perturbations and what role glutathione plays during oxidative stress in cyanobacteria.
In this work, we have investigated the role of glutathione in the model cyanobacterium Synechocystis 6803 by generating deletion mutations of gshA and gshB. We found that glutathione is beneficial to Synechocystis 6803 during acclimation to both environmental and redox perturbations and is essential during extreme oxidative stress. We found that many diverse conditions commonly utilized to probe cellular physiology have dramatic effects on glutathione levels. Furthermore, we found a strong connection between glutathione metabolism and photosynthetic electron transport, in particular, PSII activity, emphasizing the importance of glutathione in oxygenic photosynthetic organisms.
RESULTS
Disruption of the Glutathione Biosynthetic Pathway
The slr1238 gene product is annotated as glutathione synthetase (GshB), whereas the gene encoding GshA in Synechocystis 6803 is not annotated in Kyoto Encyclopedia of Genes and Genomes pathways (Ogata et al., 1999) or on Cyanobase (http://genome.kazusa.or.jp/cyanobase/). Identification of a putative gshA gene (slr0990) is based on 64% sequence identity to the Anabaena sp. PCC 7120 gene (alr3351), the product of which has been biochemically characterized as having Glu-Cys ligase activity (Ashida et al., 2005). The slr1238 gene exhibits 64.4% sequence identity to the functionally characterized gshB gene (Synpcc7942_2324) in Synechococcus elongatus PCC 7942 (Okumura et al., 1997). To study the function of glutathione in Synechocystis 6803, we generated deletion mutations of slr0990 (ΔgshA::KmR) and slr1238 (ΔgshB::GmR; Fig. 1). Because Synechocystis 6803 maintains approximately 10 copies of its chromosome under normal conditions (Labarre et al., 1989), it is necessary to passage the culture through several generations to allow for full segregation of genomic insertions. After multiple passages on kanamycin-containing plates, we were unable to obtain fully segregated colonies of ΔgshA::KmR but instead obtained colonies maintaining approximately 50% wild-type DNA at the gshA locus as determined by PCR (Fig. 1C), suggesting that this gene is essential for survival. Further attempts to segregate the mutant on medium containing kanamycin (40 μg mL−1) and 1 mm GSH were also unsuccessful (data not shown). This strain contains approximately 30% less GSH compared with the wild type under normal growth conditions, which further supports the notion that slr0990 encodes GshA. Complete segregation of the ΔgshB::GmR mutant was confirmed by PCR (Fig. 1E). The ΔgshA::KmR/ΔgshB::GmR double mutant was also constructed. As in each single mutant, full segregation of ΔgshB::GmR but not of ΔgshA::KmR was observed (data not shown). Primer sequences used in construction and analysis of mutants are shown in Table I. Because we were unable to obtain fully segregated ΔgshA::KmR mutants, we also utilized a pharmacological approach to disrupt glutathione biosynthesis by applying the specific GshA inhibitor dl-buthionine-S,R-sulfoximine (BSO), the l-form being active (Griffith and Meister, 1979) to cell cultures. We found a concentration-dependent growth reduction (Fig. 1E) and decrease in GSH levels (Fig. 1F) in the presence of BSO. This finding supports our hypothesis that GshA activity is required for growth. The unusually high concentrations required for growth reduction likely result from the decreased sensitivity of cyanobacterial GshA to BSO (Ki = 29.3 mm; Ashida et al., 2005) compared with the Arabidopsis protein (Ki = 1.2 mm; Jez et al., 2004). Due to the inherent problems in analyzing the partially segregated ΔgshA::KmR mutant, our subsequent analysis mainly focused on the fully segregated ΔgshB::GmR mutant strain.
Table I. Primers used in this study.
Forward (F) and reverse (R) primers were used to amplify upstream (up.) and downstream (dn.) regions of the indicated genes for the generation of transformation plasmids shown in Table II or for segregation analysis (Seg.). Boldface indicates restriction sites used for cloning. ORF, Open reading frame.
| Primer | Description | Sequence (5′-3′) |
| GshA1 | F: up. gshA (pSL2083) | TCTGGATCCGGAAACCGGTTGTGTCAGTT |
| GshA2 | R: up. gshA (pSL2083) | TCTCTGCAGGCACTGCACTCGCCTCTTTA |
| GshA3 | F: dn. gshA (pSL2083) | TCTCTGCAGCTGCTTACCCCTGAGCAAAG |
| GshA4 | R: dn. gshA (pSL2083) | TCTAAGCTTAATCCGTGGATTGGTAGGTG |
| GshA5 | F: Seg. of ΔgshA::KmR | CCACTGTTTCCTGGTGGTCT |
| GshA6 | R: Seg. of ΔgshA::KmR | CCCTCGGCAAAAGTTTATGA |
| GshB1 | F: up. gshB (pSL2085) | TCTGGATCCGACTTTCGTGGCGAAATGGT |
| GshB2 | R: up. gshB (pSL2085) | TCTCTGCAGCAGGGTCAATATCCTTGGGA |
| GshB3 | F: dn. gshB (pSL2085) | TCTCTGCAGCCTGGGATCAAGACAGCTCAA |
| GshB4 | R: dn. gshB (pSL2085) | TCTAAGCTTGTCCCTGTACTGGCACATTG |
| GshB5 | F: Seg. of ΔgshB::GmR | GGAAACCATGGGTCTGCTTA |
| GshB6 | R: Seg. of ΔgshB::GmR | TTTTCATTGGCTTCCCGTAG |
| GshB7 | R: Seg. of ΔgshB::GmR | TCACCGTAATCTGCTTGCAC |
| GshB8 | F: gshB ORF (pSL2086) | TCTCATATGAAACTGGCTTTTATTATCGAT |
| GshB9 | R: gshB ORF (pSL2086) | TCTGTTAACCTAAAATTGTTTTTCCAACCA |
| GshB10 | R: Seg. of T2086 | CTCGCTTCTTGGATGACCTC |
| GshB11 | R: Seg. of T2086 | ATATAAGCGGCCCACTTCCT |
Characterization and Genetic Complementation of ΔgshB::GmR
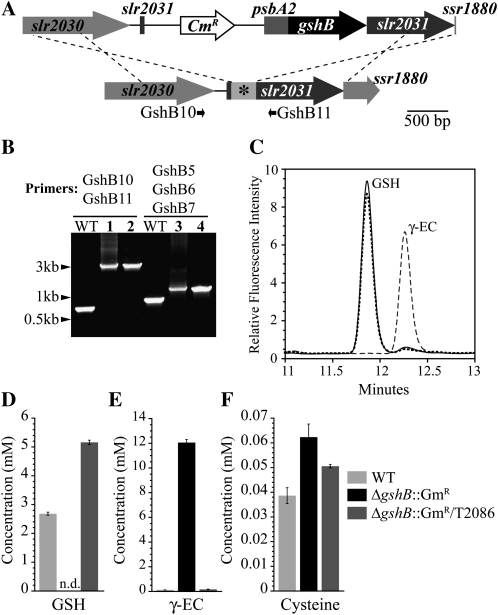
To verify the function of GshB, we generated a vector designed to express the gshB gene (pSL2086) in the ΔgshB::GmR strain using the strategy shown in Figure 2A. Integration of pSL2086 containing the gshB gene into the ΔgshB::GmR mutant strain was confirmed by PCR (Fig. 2B). Cellular thiol content was analyzed in the resulting strain, ΔgshB::GmR/T2086, and compared with the wild type and ΔgshB::GmR (Fig. 2, C–F). GSH is the predominant species in the wild type (2.7 ± 0.05 mm) and ΔgshB::GmR/T2086 (5.1 ± 0.07 mm). In contrast, the ΔgshB::GmR mutant does not contain detectable amounts of glutathione but instead accumulates high levels of γ-EC (12 ± 0.25 mm). γ-EC levels in the ΔgshB::GmR mutant are significantly higher than those in the wild type (0.17 ± 0.005 mm) and ΔgshB::GmR/T2086 (0.21 ± 0.012 mm). In fact, these levels are over 4-fold higher than that of GSH in the wild type, a phenomenon observed in bacteria and yeast gshB mutants (Grant et al., 1997; Harrison et al., 2005). In contrast, the seedling-lethal Arabidopsis gsh2 mutant ovules accumulate γ-EC over 10-fold higher than GSH levels in wild-type plants (Pasternak et al., 2008). Cys levels are also increased in ΔgshB::GmR (62 ± 5 μm) compared with the wild type (39 ± 3 μm) and ΔgshB::GmR/T2086 (51 ± 0.7 μm). Similar accumulation of Cys is also observed in embryos of the Arabidopsis gsh2 mutant (Pasternak et al., 2008). While we did not focus on the oxidation state of the thiols in this work, preliminary data indicate that in wild-type cells, GSSG does not represent more than 10% of the total glutathione pool under normal conditions and is typically maintained under 5%.
Figure 2.
Genetic complementation of the ΔgshB::GmR strain and quantification of cellular thiols. A, The gshB gene was cloned into the pTCP2031V (Satoh et al., 2001; Muramatsu et al., 2009) vector under the control of the psbA2 promoter (top) and targeted to the slr2031 site (asterisk) in the ΔgshB::GmR mutant; the resulting strain is ΔgshB::GmR/T2086. B, Segregation of ΔgshB::GmR/T2086 was confirmed by PCR using primers shown in A and Figure 1D. Lanes show wild-type genomic DNA (WT), pSL2086 (lane 1), ΔgshB::GmR/T2086 genomic DNA (lanes 2 and 4), and pSL2085 (lane 3). C, HPLC elution profile of monobromobimane-derivatized thiols extracted from wild-type (solid line), ΔgshB::GmR (dashed line), and ΔgshB::GmR/T2086 (dotted line) cells. D to F, Quantification of cellular GSH (D), γ-EC (E), and Cys (F) levels. Data are means of three independent cultures ± se. Intracellular concentrations are based on an estimated cellular volume of Synechocystis 6803 equal to 4.4 × 10−15 L; for details, see “Materials and Methods.” n.d., Not detected.
ΔgshB::GmR Is Sensitive to Redox Perturbations
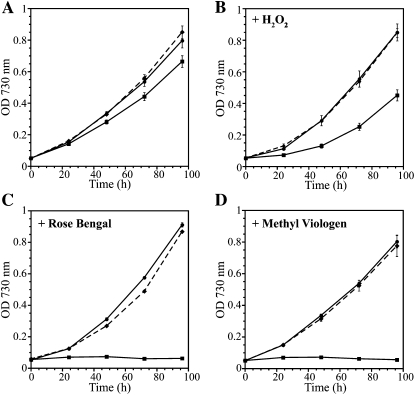
Glutathione is known to function as a redox buffer and cellular antioxidant. Therefore, we tested the growth of the ΔgshB::GmR strain, which does not contain glutathione, in conditions predicted to induce oxidative stress. The ΔgshB::GmR mutant is able to grow photoautotrophically; however, growth is slower compared with wild-type and ΔgshB::GmR/T2086 strains (Fig. 3A). At all light intensities tested (20–1,000 μmol photons m−2 s−1) as well as under light/dark regimes, the ΔgshB::GmR strain was able to grow, albeit at a slower rate than the wild type. After extended growth in batch culture, the ΔgshB::GmR mutant consistently bleached and died before the wild type, suggesting a role for GSH in the maintenance of cellular viability during the stationary phase in Synechocystis 6803. In E. coli, glutathione content increases during transition to the stationary phase (Fahey et al., 1978), and the requirement of antioxidant enzymes during the stationary phase has been observed in yeast (Longo et al., 1996). Growth in the ΔgshB::GmR strain was also examined in conditions promoting extreme oxidative stress. H2O2 is known to oxidize cellular thiols (Gutscher et al., 2009) and to elicit the expression of genes involved in oxidative stress in Synechocystis 6803 (Li et al., 2004; Singh et al., 2004). The ΔgshB::GmR strain exhibited severe growth retardation in the presence of H2O2 at concentrations that did not affect the wild type or the complemented strain (Fig. 3B). To determine whether this response was specific to H2O2 or if it was a general sensitivity to ROS, we grew the mutant in the presence of the type II photosensitizer Rose Bengal (RB) and the herbicide methyl viologen (MV). RB absorbs visible light and transfers excitation energy to molecular oxygen to generate singlet oxygen (Fischer et al., 2004). On the other hand, MV accepts electrons from the reducing side of PSI and donates the electrons to molecular oxygen to generate the superoxide anion radical. The ΔgshB::GmR mutant was extremely sensitive to RB and MV at concentrations that did not significantly reduce the growth of the wild type (Fig. 3, B and C). When started at low culture densities (optical density at 730 nm [OD730] = 0.05; 2.4 × 107 cells mL−1), there was no growth of this mutant strain in the presence of 1 μm MV. Even at higher cell densities (greater than 108 cells mL−1), this concentration of MV led to a reduction of cellular chlorophyll and resulted in cell death (data not shown). Titration of MV in the growth medium indicated that the ΔgshB::GmR mutant cannot grow at levels 0.5 μm or greater, whereas the wild type could grow in the presence of 2.0 μm MV.
Figure 3.
Growth of wild-type, ΔgshB::GmR, and ΔgshB::GmR/T2086 strains. Treatments are as follows: photoautotrophic (A), 1.5 mm H2O2 (B), 5 μm RB (C), 1 μm MV (D). Strains are wild type (circles, solid line), ΔgshB::GmR (squares, solid line), and ΔgshB::GmR/T2086 (diamonds, dashed line). Growth was monitored as turbidity at 730 nm. Error bars represent se of three independent cultures.
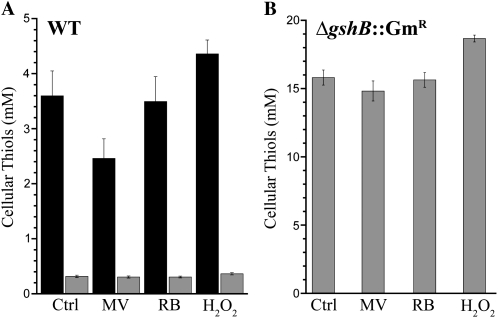
To further examine the cellular response to MV, RB, and H2O2, we measured GSH and γ-EC levels after 3 h of exposure to each of the compounds at the indicated concentrations (Fig. 4). After exposure to 1 μm MV, GSH but not γ-EC levels decreased by approximately 30%. A decrease in foliar GSH levels following MV treatment has also been observed in plants (Iturbe-Ormaetxe et al., 1998). Treatment with 1 mm H2O2 resulted in an approximately 10% increase of GSH and γ-EC levels. Stimulation of glutathione biosynthesis by oxidants such as H2O2 is a well-studied phenomenon in plants (May and Leaver, 1993; Queval et al., 2009; Mhamdi et al., 2010). No increase in GSH or γ-EC was observed after treatment with 5 μm RB; however, a role for GSH in response to RB is not unlikely given that increased glutathione peroxidase transcript abundance has been observed in Chlamydomonas following RB treatment (Fischer et al., 2004). These results highlight the importance of GSH as an antioxidant in cyanobacteria during oxidative stress. During MV treatment, the decrease in GSH levels might indicate that it was being utilized to protect the cells, whereas γ-EC levels did not change to a great extent. Because we only measured reduced thiols in this assay, the decrease might also reflect the oxidation of GSH to GSSG. Additionally, many factors, including concentration of the compound, duration of exposure, light intensity, and cell density, will likely influence the outcome of the experiment. In summary, these results indicate that while γ-EC appears to functionally replace GSH under favorable conditions, it is not sufficient during severe redox perturbations. Moreover, GSH is critical for protection against diverse ROS species, including H2O2, singlet oxygen, and superoxide anion radical.
Figure 4.
Changes in cellular thiol content after redox perturbations. GSH (black bars) and γ-EC (gray bars) were measured in untreated wild-type (WT; A) and ΔgshB::GmR (B) cells (Ctrl) or cells exposed to 1 μm MV, 5 μm RB, or 1 mm H2O2 for 3 h under continuous illumination at 30 μmol photons m−2 s−1. Each bar represents the mean of three independent cultures, and error bars represent se.
Light Intensity and Quality Affect Glutathione Metabolism
Photosynthetic organisms depend on energy obtained from light to drive cellular metabolism and carbon fixation. Therefore, it is crucial that cyanobacteria are able to balance light harvesting and energy transduction with demands at the metabolic level. Our laboratory recently found through transcriptional profiling that changes in light intensity (Aurora et al., 2007; Singh et al., 2008) and quality (Singh et al., 2009) have major effects on cellular physiology and primary metabolism, especially carbon, nitrogen, and sulfur metabolism.
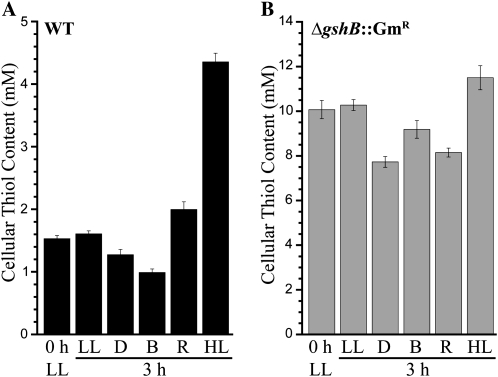
We examined the primary thiol concentration in wild-type (GSH) and ΔgshB::GmR (γ-EC) cells after exposure to different light conditions (Fig. 5). Singh et al. (2009) found that gshA and gshB genes are differentially expressed in response to preferential illumination of PSI (blue light) or PSII (orange-red light). The differential illumination of the photosystems is primarily due to the large pigment-protein antennae complex, the phycobilisome, thought to be mostly associated with PSII (Mullineaux, 2008). Blue light excites chlorophyll in PSI. When cells are exposed to PSII light, gshB expression is up-regulated compared with PSII light. In contrast, gshA expression is decreased in PSII versus PSI light. In concordance with these results, we found that illumination with orange-red or blue light affected GSH and γ-EC levels in the opposite direction. Blue light led to a 25% decrease in GSH levels and only a slight decline in γ-EC levels, whereas red light led to a 30% increase in wild-type GSH levels and a 20% decline in γ-EC levels. In both wild-type and ΔgshB::GmR cells, high light led to increased thiol concentration; however, GSH levels increased by 200% while γ-EC levels increased by only 15%. Exposure to dark led to an approximately 20% decline in both GSH and γ-EC levels. It is important to note that initial γ-EC levels (approximately 10 mm) in the ΔgshB::GmR mutant low-light control were about 6-fold higher than the GSH levels in the wild type (approximately 1.5 mm).
Figure 5.
Influence of light quality and intensity on glutathione metabolism. The major cellular thiol was measured in the wild type (WT; GSH; black bars; A) and ΔgshB::GmR (γ-EC; gray bars; B) after exposure to different light conditions. Low light (LL; 20 μmol photons m−2 s−1)-grown cells were transferred to low light, dark (D), blue (B), orange-red (R), or high light (HL; 150 μmol photons m−2 s−1) for 3 h. Thiols were then analyzed by HPLC. Each bar represents the mean of three independent cultures, and error bars represent se. For further details, see “Materials and Methods.”
These results demonstrate the interplay between photosynthetic electron transfer on glutathione metabolism. Linear photosynthetic electron transport does not occur during illumination with blue light or in the dark, and these conditions both led to a substantial decrease in GSH and γ-EC levels. Additionally, these conditions do not promote growth in Synechocystis 6803 (Singh et al., 2009). Exposure to orange-red light and high light promotes growth, oxygen evolution, and linear photosynthetic electron transport and resulted in significant increases in GSH levels. Furthermore, transcriptional activation of gshB is coordinated with conditions promoting the production of reductant and ATP needed for the biosynthesis of GSH and precursors, including Glu and Cys. These results emphasize a role for GSH during increased growth and metabolism and conditions promoting oxidative stress. However, the ability of the ΔgshB::GmR strain to grow at all light intensities tested suggests that many factors in addition to glutathione are responsible for acclimation to high-light conditions as concluded from transcriptomics studies (Hihara et al., 2001; Singh et al., 2008).
Effect of Glc on Glutathione Metabolism
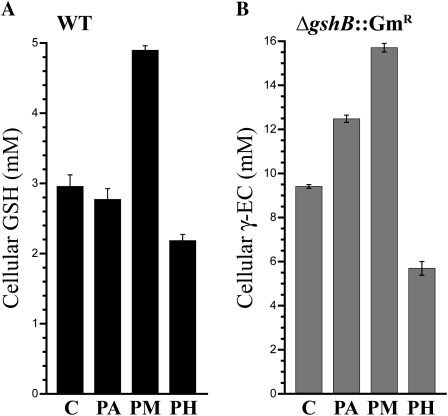
Synechocystis 6803 is able to grow photoautotrophically (PA), photomixotrophically (PM) in the presence of Glc, and photoheterotrophically (PH) in the presence of Glc and the PSII inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). During PM growth, Glc is catabolized via glycolysis and the oxidative pentose phosphate pathway, and high rates of photosynthetic and respiratory electron transport are observed (Takahashi et al., 2008). Increased electron transfer reactions during PM growth could result in increased ROS production compared with PA or PH growth. During PH growth, the linear photosynthetic electron transport chain is blocked and Glc catabolism is utilized for energy production. We analyzed the primary thiol component in wild-type (GSH) and ΔgshB::GmR (γ-EC) cells after 24 h in PA, PM, and PH conditions (Fig. 6) and compared them with the levels in control cultures prior to transfer to experimental conditions. We found that PM growth led to increased thiol levels, while PH growth led to deceased thiol levels in both wild-type and ΔgshB::GmR cells. PM conditions also resulted in increased growth compared with the other conditions. These results agree with the data presented above, which indicate that increased photosynthetic electron transport and conditions promoting rapid growth are correlated with high cellular glutathione content.
Figure 6.
Cellular thiol content after addition of Glc. The major cellular thiol was measured in the wild type (WT; GSH; black bars; A) and ΔgshB::GmR (γ-EC; gray bars; B) grown photoautotrophically (PA), photomixotrophically (PM) in the presence of 5 mm Glc, or photoheterotrophically (PH) in the presence of 5 mm Glc and 10 μm DCMU for 24 h. Values for preculture cells used as the inoculum for the experiment are also shown (C). Error bars represent se of two separate measurements each from two independent cultures.
Glutathione Is involved in Acclimation to Nutritional Perturbations
Cyanobacteria are continually faced with environmental challenges such as nutrient limitation. To overcome these challenges, they must adjust their physiology and metabolism (Schwarz and Forchhammer, 2005). While many responses appear to be specific to a particular stress, there are also many general stress responses elicited during a perturbation. Using integrated analysis of large-scale transcriptome data sets, our laboratory recently determined that oxidative stress is a general phenomenon underlying numerous perturbations in Synechocystis 6803 (Singh et al., 2010). In plants, biotic and abiotic perturbations led to oxidative stress and signaling through the production of ROS and changes in the glutathione redox potential (Mittler, 2002; Meyer, 2008; Foyer and Noctor, 2009).
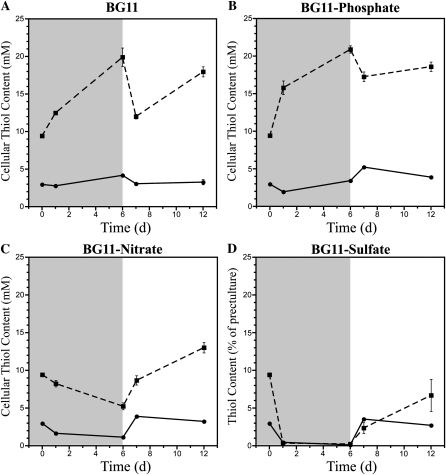
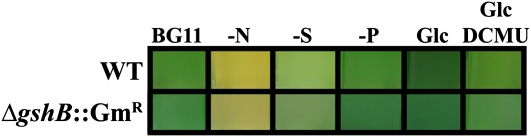
To probe the role of glutathione during physiologically important perturbations, we deprived wild-type and ΔgshB::GmR cells of nitrate, sulfate, or phosphate. These three nutrients play critical roles in growth and photosynthesis in cyanobacteria and plants; therefore, limitation of any one of these seriously impacts cellular physiology (Richaud et al., 2001; Schwarz and Forchhammer, 2005; Schachtman and Shin, 2007; Adams et al., 2008). Furthermore, GSH represents a large portion of reduced sulfur within the cell and contains Glu, a central player in nitrogen metabolism. Thus, there must be strict coordination between nutrient assimilation and glutathione biosynthesis (Kopriva and Rennenberg, 2004). To test the affect of nutrient availability on the glutathione pool, we measured cellular thiols during nutrient depletion and repletion in the wild type and the ΔgshB::GmR strain (Fig. 7). For depletion, cells were transferred to BG11 medium lacking nitrate, sulfate, or phosphate and grown for 6 d. For repletion, cells were transferred from deplete conditions to BG11 and allowed to recover for 6 d. GSH and γ-EC levels were measured in wild-type and ΔgshB::GmR strains, respectively. After 6 d of growth in BG11, GSH and γ-EC levels increased (Fig. 7A). Upon dilution into fresh BG11, GSH and γ-EC concentrations decreased. After 6 d of growth in the replete condition, γ-EC levels increased significantly in the ΔgshB::GmR strain. During phosphate depletion, γ-EC levels in the ΔgshB::GmR mutant strain increased, while GSH in the wild type transiently decreased after 24 h and then increased slightly during the duration of the nutrient depletion. Following phosphate repletion, γ-EC levels in the ΔgshB::GmR mutant transiently decreased during the first 24 h and then increased, while GSH levels in the wild type increased slightly after 24 h and then decreased for the duration of the time course (Fig. 7B). Nitrate limitation led to a 50% decrease of GSH in the wild type within 24 h. γ-EC levels were also reduced by approximately 50% in ΔgshB::GmR after 6 d of growth. After 24 h in replete medium, levels of both GSH and γ-EC increased and continued to rise in the ΔgshB::GmR strain (Fig. 7C). Sulfate depletion led to a dramatic reduction of both GSH and γ-EC within 24 h that was exaggerated after 6 d (Fig. 7D). Other experiments have shown that the depletion occurs in less than 12 h after transfer to sulfate-deplete medium (data not shown). After transfer to complete medium, wild-type GSH levels surpassed control values within 24 h and then returned to near control levels, while γ-EC levels in the ΔgshB::GmR mutant did not fully recover to control levels during this time course. Cultures of both wild-type and ΔgshB::GmR strains visually appeared similar after 24 h of depletion (Fig. 8). The large and dynamic changes in glutathione and γ-EC levels during sulfate and nitrate depletion and repletion suggest that glutathione and γ-EC can be catabolized as a source of sulfur and nitrogen during adverse conditions in cyanobacteria.
Figure 7.
Effect of nutrient depletion on glutathione metabolism. The major primary thiol was measured in the wild type (GSH; solid line, circles) and ΔgshB::GmR (γ-EC; dashed line, squares) over the course of 12 d in deplete (shaded) and replete (white) conditions. Cells were precultured in BG11 medium (0 d) prior to transfer to BG11 (A) or BG11 lacking nitrate (B), sulfate (C), or phosphate (D) for 6 d. After 6 d of growth in deplete conditions, cells were transferred to fresh BG11 medium and grown for an additional 6 d. Error bars represent se of two measurements each from two independent cultures.
Figure 8.
Visual comparison of wild-type (WT) and ΔgshB::GmR cultures. Photographs show cultures after 24 h of growth in BG11 medium lacking nitrate (−N), sulfate (−S), or phosphate (−P) or with the addition of 5 mm Glc with or without 10 μm DCMU.
Reduced Growth and Fitness of the ΔgshB::GmR Strain during Sulfate Starvation and Recovery
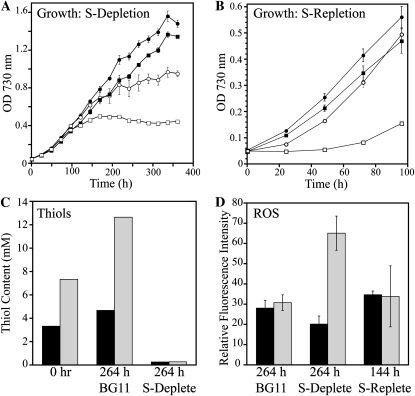
When wild-type and ΔgshB::GmR cells were transferred to medium lacking sulfate, growth stopped within 24 h and both GSH and γ-EC levels were depleted. In order to more thoroughly investigate the role of GSH in acclimation to sulfate deprivation, we grew cells in BG11 (control) or in medium containing one-tenth the sulfate concentration of BG11 (30.3 versus 303 μm; Fig. 9A). The growth rates of all cultures remained similar for the first 100 h. After 100 h, ΔgshB::GmR cells entered a stationary phase in sulfate-deplete medium, followed by the wild type. However, the wild type maintained a higher cell density throughout the experiment. After 264 h of growth, cells were transferred to fresh BG11 medium and growth was monitored (Fig. 9B). Wild-type cells from sulfate-deplete medium grew similar to the BG11-grown controls, but sulfur-deprived ΔgshB::GmR cells recovered very slowly. Sulfur-starved cells exhibited reduced pigment content and appeared lighter in color compared with cells grown in BG11 (Fig. 8). After transfer from sulfate-deplete medium to sulfur-replete medium, wild-type cells quickly recovered pigments and grew, while ΔgshB::GmR cells stayed light green for several days (Supplemental Fig. S1). After several days of growth, ΔgshB::GmR cells were usually able to recover, although sometimes the cells progressively bleached to white and died. During the time course, GSH levels and γ-EC levels were measured for wild-type and ΔgshB::GmR strains, respectively (Fig. 9C), and ROS production was measured using the cell-permeable fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA; Fig. 9D). Increased DCF fluorescence during sulfate starvation in ΔgshB::GmR cells compared with the wild type indicates oxidative stress in the mutant cells during sulfate depletion. The oxidation state of GSH and γ-EC was also determined during growth in BG11 or after 24 h of sulfate starvation. In the wild type, the GSSG levels increased from approximately 2.5% (GSH:GSSG ratio = 38) to 4% (GSH:GSSG ratio = 26) of the glutathione pool during sulfate starvation. In the ΔgshB::GmR strain, bis-γ-glutamylcystine levels rose from about 2% [γ-EC:(γ-EC)2 ratio = 43] to 7% [γ-EC:(γ-EC)2 ratio = 14] of the γ-EC pool during sulfate starvation. Together, these data suggest that during sulfate starvation, the ΔgshB::GmR mutant is experiencing more oxidative stress than the wild type and provides evidence that GSH is important in acclimation to nutrient-deplete conditions. Consistent with our previous results, the ΔgshB::GmR strain is able to survive under normal conditions, but not when faced with extreme environmental and redox perturbations.
Figure 9.
Characterization of sulfate (S) depletion in ΔgshB::GmR cells. A, Growth of the wild type (circles) and ΔgshB::GmR (squares) in BG11 (black symbols) or BG11 containing 10% sulfate (30.3 μm; white symbols). Values represent means ± se (n = 3). B, Growth of the wild type (circles) and ΔgshB::GmR (squares) in BG11 after 264 h of growth in sulfate-deplete (white symbols) or BG11 (black symbols) medium. Values are means ± se of three cultures. C, The major cellular thiol in the wild type (GSH; black bars) and ΔgshB::GmR (γ-EC; gray bars) was quantified by HPLC prior to sulfate depletion (0 h) after 264 h of growth in BG11 or in sulfate-deplete medium. Each value represents a single measurement and is consistent with the results of at least three independent experiments. D, ROS accumulation after 264 h of growth in BG11 or sulfate-deplete medium and after 144 h of recovery in BG11 medium. Error bars represent se of three measurements each from three independent cultures.
DISCUSSION
In this study, we have utilized ΔgshA and ΔgshB deletion mutants in Synechocystis 6803 to investigate the functions of glutathione in cyanobacteria. Furthermore, we analyzed thiol compounds in the wild type and the ΔgshB strain during exposure to environmental and redox perturbations commonly utilized to study cyanobacterial physiology. Our findings demonstrate that glutathione metabolism is invoked under multiple conditions, suggesting a role in the acclimation response to many diverse perturbations. Furthermore, we found that γ-EC is able to functionally replace GSH under favorable conditions but not during conditions promoting extreme oxidative stress.
Our findings suggest that GshA activity is essential in Synechocystis 6803, because we were unable to isolate fully segregated ΔgshA::KmR mutants. In Arabidopsis, gsh1 mutant plants are embryo lethal (Cairns et al., 2006), and gshA mutants in yeast require either dithiothreitol or GSH supplemented in the medium for growth (Spector et al., 2001). However, in E. coli, glutathione is dispensable, because redundancy exists between the glutathione and thioredoxin systems (Prinz et al., 1997). This indicates more specialized roles of GSH in photosynthetic microbes compared with heterotrophic bacteria. In fact, glutathione may be involved in the regulation of many processes in cyanobacteria through glutathionylation (Li et al., 2007).
We were able to generate a fully segregated ΔgshB::GmR mutant that lacks glutathione and instead accumulates γ-EC. In Arabidopsis, mutations in gsh2 result in a seedling-lethal phenotype that can be rescued by expressing gsh2 in the cytoplasm alone (Pasternak et al., 2008). In some organisms, such as halobacteria, γ-EC is the naturally occurring primary thiol due to its increased stability in high salt concentrations (Sundquist and Fahey, 1989). The ΔgshB::GmR strain accumulates γ-EC to levels over 4-fold higher than GSH in the wild type. The redox state of the 2GSH/GSSG couple is dependent on the ratio of reduced and oxidized glutathione in addition to the reducing capacity (absolute concentration; Schafer and Buettner, 2001). This means that changes in the total GSH pool can have dramatic consequences on the cellular redox state. Furthermore, high levels of glutathione increase the buffering capacity of the GSH/GSSG redox couple. The high levels of γ-EC in ΔgshB::GmR may be required to maintain redox poise in the absence of GSH or could represent a decreased efficiency of γ-EC in the feedback regulation of GshA compared with GSH. Feedback inhibition of GshA by GSH has been reported for purified Anabaena 7120 (Ashida et al., 2005) and Arabidopsis (Jez et al., 2004) enzymes. γ-EC also inhibits Arabidopsis GshA, but to a lower degree than GSH: at 10 mm γ-EC, GshA activity was reduced to only 34% of the control compared with 21% with GSH (Pasternak et al., 2008).
Changes in light quality and intensity are thought to modulate the cellular redox environment in photosynthetic organisms. Our results show that increased photosynthetic electron transfer directly impacts cellular glutathione levels. While increased GSH biosynthesis has been observed in response to high-light treatment in plants (Ogawa et al., 2004), our results suggest that PSII illumination, and not PSI illumination, leads to increased GSH biosynthesis. In Synechocystis 6803, thiols increased during high-light and orange-red light treatments and during photomixotrophic growth (Figs. 5 and 6). Furthermore, glutathione levels decrease in conditions where linear photosynthetic electron transport is inactive. This is observed during photoheterotrophic growth (Fig. 6) in the dark and during illumination with blue light (Fig. 5). Singh et al. (2009) concluded that preferential illumination of PSI with blue light leads to cyclic electron transfer for the generation of ATP and stimulation of respiration for the generation of reducing equivalents. While it is possible that increased electron flow surrounding PSI results in increased ROS, thereby oxidizing the glutathione pool, it has been demonstrated that photoreduction of O2 to water in Synechocystis 6803 is mediated by two A-type flavoproteins and does not generate substantial ROS (Helman et al., 2003). This is also supported by the finding that only 1% of photosynthetic electron transport results in the production of H2O2 in Synechocystis 6803 (Tichy and Vermaas, 1999). All of the conditions promoting increased cellular glutathione content also result in increased PSII-mediated oxygen evolution and therefore provide evidence for the role of GSH in the detoxification of ROS.
Photosynthetic electron transport also shuttles electrons into regulatory networks such as those controlled by thioredoxin (Schürmann and Buchanan, 2008). The glutathione system also plays critical roles in the coordination of cellular processes with photosynthetic activity (Foyer and Noctor, 2009). High light is known to promote oxidative stress and to result in many changes at the physiological and transcriptional levels in Synechocystis 6803, including decreasing photosystem content and phycobilisomes and inducing genes involved in cellular protection (Hihara et al., 2001; Singh et al., 2008). It is surprising that ΔgshB::GmR is able to grow at all light intensities tested. While growth of ΔgshB::GmR is consistently reduced compared with the wild type, high light resulted in a proportional decrease in both wild-type and ΔgshB::GmR growth. Multiple protective mechanisms are responsible for acclimation to high light in Synechocystis 6803. Induction of peroxiredoxin genes (slr1198 and sll1621) as well as the NADPH-dependent glutathione peroxidase-like gene (slr1992) are observed following high-light treatment (Singh et al., 2008). In Synechocystis 6803, production of H2O2 under high light intensity is considerably lower than that observed in isolated chloroplasts (Tichy and Vermaas, 1999).
Cyanobacteria contain several enzymatic antioxidant systems that metabolize ROS, including superoxide dismutase (SodB), catalase (KatG), and multiple peroxiredoxins (Pérez-Pérez et al., 2009). While ΔkatG is not sensitive to MV or H2O2 (Tichy and Vermaas, 1999), ΔsodB mutants are extremely sensitive to MV treatment (Thomas et al., 1998). Additional protection from ROS is provided by the membrane-soluble tocopherol, which has been shown to be important during mixotrophic growth in cyanobacteria (Sakuragi et al., 2006). Our results show that GSH plays a critical role in the protection from multiple ROS species in cyanobacteria, despite the presence of other specialized systems. While glutathione appears to play a critical role in protection from ROS, many systems for the detoxification of ROS exist within cyanobacteria (Perelman et al., 2003), and each system may function under a particular condition.
We observed large changes in cellular thiol pools after nitrate, sulfate, and phosphate depletion in the wild type and the ΔgshB::GmR mutant strain. During nitrate and sulfur starvation, GSH and γ-EC are depleted and could be catabolized for amino acids, as seen in yeast (Elskens et al., 1991; Mehdi and Penninckx, 1997) during nutrient deficiency. In plants, a significant shift in cellular metabolism is observed following sulfur deprivation, leading to decreased glutathione, protein, and chlorophyll contents (Nikiforova et al., 2005). We hypothesize that the increased sensitivity of the ΔgshB::GmR mutant to sulfate starvation and recovery reflects increased oxidative stress in the mutant during these conditions. It is possible that recovery of ΔgshB::GmR could reflect defects in sulfate uptake or assimilation. However, γ-EC biosynthesis rates following transfer to sulfate-replete conditions are similar to those of the wild type, and eventually γ-EC levels exceed the levels of GSH in the wild type. In plants, GSH serves as a reductant to convert adenosine 5′ phosphosulfate (APS) into sulfite by APS reductase, but γ-EC can replace GSH in this process (Bick et al., 1998). In Synechocystis 6803, APS is phosphorylated to 3′-phospho-5′-adenylylsufate by APS kinase and subsequently reduced to sulfite by 3′-phospho-5′-adenylylsufate reductase (Schmidt, 1977), an enzyme thought to be redox regulated by the glutathione/glutaredoxin system in E. coli (Lillig et al., 2003). There are still many outstanding questions regarding the regulatory components involved in sulfate assimilation and integration with carbon and nitrogen metabolism, nutrients critical for glutathione metabolism.
CONCLUSION
In summary, our analysis of glutathione metabolism in cyanobacteria reveals that cellular glutathione content is highly responsive to changes in light quality and quantity and nutrient availability. Dynamic changes in cellular thiol content require significant amounts of energy and resources; therefore, these changes must be beneficial for the acclimation to changing environmental conditions. Furthermore, our data indicate that GshA activity is essential in cyanobacteria, because we were unable to obtain fully segregated ΔgshA::KmR mutants. In addition, we find that while the glutathione precursor γ-EC can function during normal growth, GSH is essential for protection against redox stress.
MATERIALS AND METHODS
Culture Conditions
Synechocystis sp. PCC 6803 strains were grown in liquid BG11 (Allen, 1968)medium at 30°C under continuous illumination by cool-white fluorescent lights under 30 μmol photons m−2 s−1, unless otherwise indicated. Mutant strains were maintained on solid BG11 agar plates supplemented with 40 μg mL−1 kanamycin, 5 μg mL−1 gentamicin, or a combination of 40 μg mL−1 kanamycin and 5 μg mL−1 gentamicin for ΔgshA::KmR, ΔgshB::GmR, and ΔgshA::KmR/ΔgshB::GmR strains, respectively; 5 μg mL−1 gentamicin plus 10 μg mL−1 chloramphenicol was used for the ΔgshB::GmR/T2086 strain. All experiments were performed using medium without antibiotics added, as the strains lacking glutathione (ΔgshB::GmR and ΔgshA::KmR/ΔgshB::GmR) were sensitive to low concentrations of aminoglycoside antibiotics present in liquid medium, despite the expression of a functional resistance gene (data not shown). For growth assays, cells were grown to mid log phase and harvested by centrifugation. The cells were washed in fresh BG11 and centrifuged to pellet, and the cell pellets were resuspended in the appropriate medium. The cells were diluted to OD730 = 0.05 in BG11 without antibiotics and grown with shaking (200 rpm). The OD730 was measured every 24 h on a μQuant Microplate spectrophotometer (Biotek Instruments). Where indicated, BSO, H2O2, RB, or MV was added at the concentrations specified in “Results.” For nutrient deprivation, cells were washed and resuspended in appropriate deplete medium (at OD730 ≈ 0.04), transferred to 400-mL square flasks, and bubbled with air under continuous illumination under 40 μmol photons m−2 s−1. After 6 d of growth, deplete cells were harvested by centrifugation, resuspended in BG11 to OD730 = 0.05, and transferred to 250-mL shaker flasks for an additional 6 d. Glc (5 mm) and DCMU (10 μm) were added when specified. For growth in orange-red and blue light, cells grown in shaker flasks at 40 μmol photons m−2 s−1 were transferred to 3-cm-diameter test tubes in a water bath maintained at 30°C and bubbled with air. Illumination was provided by a custom light-emitting diode panel at approximately 10 μmol photons m−2 s−1 as described previously (Singh et al., 2009).
Construction of Mutant Strains
The open reading frame of gshA (slr0990) was replaced with a modified kanamycin gene lacking a transcriptional terminator sequence. Primer pairs GshA1 + GshA2 and GshA3 + GshA4 were used to amplify a 498-bp region upstream and a 499-bp region downstream of gshA, respectively. Upstream and downstream fragments were cloned into pUC18 flanking a kanamycin resistance cassette (KmR), and the resulting plasmid is pSL2083. The gshB (slr1238) reading frame was replaced with a gentamicin resistance cassette (GmR). Primer pairs GshB1 + GshB2 and GshB3 + GshB4 were used to amplify a 529-bp region upstream and a 505-bp region downstream of gshB, respectively. Upstream and downstream PCR products were cloned into the pUC18 plasmid on either side of a gentamicin resistance gene to create pSL2085. Wild-type Synechocystis 6803 was transformed with pSL2083 and pSL2085 to make ΔgshA::KmR and ΔgshB::GmR, respectively. The ΔgshA::KmR/ΔgshB::GmR double mutant was obtained by transforming ΔgshA::KmR with pSL2085. To genetically complement ΔgshB::GmR, primers GshB8 + GshB9 containing NdeI and HpaI restriction sites, respectively, were used to amplify the coding region of gshB. The product of this reaction was cloned into plasmid pTCP2031V (Satoh et al., 2001; Muramatsu et al., 2009), and the resulting plasmid is pSL2086 (Fig. 2A; Table II). This plasmid was transformed into ΔgshB::GmR, and the resulting strain is designated ΔgshB::GmR/T2086. Segregation of mutant alleles was determined by PCR as shown in Figures 1 and 2. All primers used in study are listed in Table I.
Table II. Plasmids and strains.
Plasmids were constructed as described in “Materials and Methods” and used to generate the strains used in this study. ORF, Open reading frame.
| Plasmid | Description (Strain) | Reference |
| pSL2083 | For generation of ΔgshA::KmR | This study |
| pSL2085 | For generation of ΔgshB::GmR | This study |
| pTCP2031v | Contains CmR, psbA2 promoter, and slr2031 targeting sequences | Satoh et al. (2001); Muramatsu et al. (2009) |
| pSL2086 | gshB ORF cloned into pTCP2031v for generation of ΔgshB::GmR/T2086 | This study |
Measurement of Cellular Thiols
Thiols were extracted from Synechocystis 6803, derivatized with monobromobimane, and analyzed essentially as described (Newton and Fahey, 1995). Thiols were separated using a ZORBAX XDB-C18 (4.6 × 250 mm, 5 μm) column on an Agilent 1200 series HPLC apparatus (Agilent Technologies). Fluorescent bimane-thiol conjugates were detected on an Agilent 1200 series fluorescence detector (380 nm excitation, 480 nm emission). HPLC run conditions were as follows: solvent A is 0.1% trifluoroacetic acid in deionized water, and solvent B is 100% methanol. Flow rates were maintained at 1.2 mL min−1, and linear gradients were used for separation (0 min, 10% B; 2 min, 15% B; 8 min, 20% B; 14 min, 40% B; 16 min, 100% B; 18 min, 100% B; 20 min, 10% B; 30 min, 10% B, reinjection). Comparison of peak areas with those of authentic standards was used to quantify thiols. Measurement of reduced and oxidized forms of glutathione and γ-EC was essentially as described (Fey et al., 2005) with the following modifications. In the extraction buffer, we utilized 50% acetonitrile instead of 50% methanol, and we buffered with 20 mm HEPES, pH 8.0, instead of 100 mm phosphate, pH 7.1. Separation of thiols was conducted as above. GSH concentration after growth in BSO (Fig. 1) was measured by the glutathione reductase-5,5′-dithiobis(2-nitrobenzoic acid) recycling assay as described (Queval and Noctor, 2007) after extraction of thiols from cells in 0.2 n HCl. Intracellular concentrations were estimated using an average Synechocystis 6803 cellular volume of 4.4 × 10−15 L. The cellular volume was calculated based on an average cell diameter of 2.0 ± 0.2 μm (n = 300) and assuming a spherical cell. Cell diameter was determined using light microscopy (Nikon Eclipse 80i) and analyzed using MetaVue software (version 6.3).
DCFH-DA Assay for Estimation of ROS
Cells grown in BG11, sulfate-deplete, or sulfate-replete medium were transferred to opaque black 96-well plates (Costar). DCFH-DA (2 mm in 100% ethanol) was added directly to the cells at a final concentration of 10 μm. Cells were incubated in the dark with intermittent shaking for 1 h at room temperature. Fluorescence was measured at 525 nm after excitation at 488 nm every 20 min for the duration of the incubation on a Synergy Mx fluorescence plate reader (Biotek Instruments). Three sample replicates were measured for each of the three biological replicates. Fluorescence intensity was normalized to OD730 and is presented as relative fluorescence.
Chemicals
All chemicals and thiol standards were purchased from Sigma.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Wild-type and ΔgshB::GmR strains during recovery from sulfate depletion.
Supplementary Material
Acknowledgments
We thank Drs. Abhay Singh and Joseph Jez and members of the Pakrasi laboratory for collegial discussions.
References
- Adams MM, Gómez-García MR, Grossman AR, Bhaya D. (2008) Phosphorus deprivation responses and phosphonate utilization in a thermophilic Synechococcus sp. from microbial mats. J Bacteriol 190: 8171–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MM. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 [DOI] [PubMed] [Google Scholar]
- Asada K. (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Ashida H, Sawa Y, Shibata H. (2005) Cloning, biochemical and phylogenetic characterizations of gamma-glutamylcysteine synthetase from Anabaena sp. PCC 7120. Plant Cell Physiol 46: 557–562 [DOI] [PubMed] [Google Scholar]
- Aurora R, Hihara Y, Singh AK, Pakrasi HB. (2007) A network of genes regulated by light in cyanobacteria. OMICS 11: 166–185 [DOI] [PubMed] [Google Scholar]
- Bick JA, Aslund F, Chen Y, Leustek T. (1998) Glutaredoxin function for the carboxyl-terminal domain of the plant-type 5′-adenylylsulfate reductase. Proc Natl Acad Sci USA 95: 8404–8409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NG, Pasternak M, Wachter A, Cobbett CS, Meyer AJ. (2006) Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol 141: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley SD, Dhillon JK. (2002) Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol 3: 25.1–25.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elskens MT, Jaspers CJ, Penninckx MJ. (1991) Glutathione as an endogenous sulphur source in the yeast Saccharomyces cerevisiae. J Gen Microbiol 137: 637–644 [DOI] [PubMed] [Google Scholar]
- Fahey RC, Brown WC, Adams WB, Worsham MB. (1978) Occurrence of glutathione in bacteria. J Bacteriol 133: 1126–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey V, Wagner R, Braütigam K, Wirtz M, Hell R, Dietzmann A, Leister D, Oelmüller R, Pfannschmidt T. (2005) Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem 280: 5318–5328 [DOI] [PubMed] [Google Scholar]
- Fischer BB, Krieger-Liszkay A, Eggen RL. (2004) Photosensitizers neutral red (type I) and rose bengal (type II) cause light-dependent toxicity in Chlamydomonas reinhardtii and induce the Gpxh gene via increased singlet oxygen formation. Environ Sci Technol 38: 6307–6313 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal 11: 861–905 [DOI] [PubMed] [Google Scholar]
- Grant CM, MacIver FH, Dawes IW. (1997) Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to an accumulation of the dipeptide gamma-glutamylcysteine. Mol Biol Cell 8: 1699–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW, Meister A. (1979) Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S-n-butyl homocysteine sulfoximine). J Biol Chem 254: 7558–7560 [PubMed] [Google Scholar]
- Gutscher M, Sobotta MC, Wabnitz GH, Ballikaya S, Meyer AJ, Samstag Y, Dick TP. (2009) Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J Biol Chem 284: 31532–31540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J, Jamet A, Muglia CI, Van de Sype G, Aguilar OM, Puppo A, Frendo P. (2005) Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. J Bacteriol 187: 168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman Y, Tchernov D, Reinhold L, Shibata M, Ogawa T, Schwarz R, Ohad I, Kaplan A. (2003) Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr Biol 13: 230–235 [DOI] [PubMed] [Google Scholar]
- Hicks LM, Cahoon RE, Bonner ER, Rivard RS, Sheffield J, Jez JM. (2007) Thiol-based regulation of redox-active glutamate-cysteine ligase from Arabidopsis thaliana. Plant Cell 19: 2653–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M. (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R, Andersen CR, Goldsbrough PB, Cobbett CS. (1995) A cadmium-sensitive, glutathione-deficient mutant of Arabidopsis thaliana. Plant Physiol 107: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Escuredo P, Arrese-Igor C, Becana M. (1998) Oxidative damage in pea plants exposed to water deficit or paraquat. Plant Physiol 116: 173–181 [Google Scholar]
- Iwema T, Picciocchi A, Traore DA, Ferrer JL, Chauvat F, Jacquamet L. (2009) Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry 48: 6041–6043 [DOI] [PubMed] [Google Scholar]
- Jez JM, Cahoon RE. (2004) Kinetic mechanism of glutathione synthetase from Arabidopsis thaliana. J Biol Chem 279: 42726–42731 [DOI] [PubMed] [Google Scholar]
- Jez JM, Cahoon RE, Chen S. (2004) Arabidopsis thaliana glutamate-cysteine ligase: functional properties, kinetic mechanism, and regulation of activity. J Biol Chem 279: 33463–33470 [DOI] [PubMed] [Google Scholar]
- Kopriva S, Rennenberg H. (2004) Control of sulphate assimilation and glutathione synthesis: interaction with N and C metabolism. J Exp Bot 55: 1831–1842 [DOI] [PubMed] [Google Scholar]
- Labarre J, Chauvat F, Thuriaux P. (1989) Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC 6803. J Bacteriol 171: 3449–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A, Ruiz M, Zhang CC. (2009) Oxidative stress in cyanobacteria. FEMS Microbiol Rev 33: 258–278 [DOI] [PubMed] [Google Scholar]
- Li H, Singh AK, McIntyre LM, Sherman LA. (2004) Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J Bacteriol 186: 3331–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yang Q, Zhang L, Li H, Cui Y, Wu Q. (2007) Identification of novel targets of cyanobacterial glutaredoxin. Arch Biochem Biophys 458: 220–228 [DOI] [PubMed] [Google Scholar]
- Lillig CH, Potamitou A, Schwenn JD, Vlamis-Gardikas A, Holmgren A. (2003) Redox regulation of 3′-phosphoadenylylsulfate reductase from Escherichia coli by glutathione and glutaredoxins. J Biol Chem 278: 22325–22330 [DOI] [PubMed] [Google Scholar]
- Longo VD, Gralla EB, Valentine JS. (1996) Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae: mitochondrial production of toxic oxygen species in vivo. J Biol Chem 271: 12275–12280 [DOI] [PubMed] [Google Scholar]
- López-Maury L, Sánchez-Riego AM, Reyes JC, Florencio FJ. (2009) The glutathione/glutaredoxin system is essential for arsenate reduction in Synechocystis sp. strain PCC 6803. J Bacteriol 191: 3534–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteyn B, Domain F, Legrain P, Chauvat F, Cassier-Chauvat C. (2008) The thioredoxin reductase-glutaredoxins-ferredoxin crossroad pathway for selenate tolerance in Synechocystis PCC6803. Mol Microbiol 71: 520–532 [DOI] [PubMed] [Google Scholar]
- Masip L, Veeravalli K, Georgiou G. (2006) The many faces of glutathione in bacteria. Antioxid Redox Signal 8: 753–762 [DOI] [PubMed] [Google Scholar]
- May MJ, Leaver CJ. (1993) Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol 103: 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi K, Penninckx MJ. (1997) An important role for glutathione and gamma-glutamyltranspeptidase in the supply of growth requirements during nitrogen starvation of the yeast Saccharomyces cerevisiae. Microbiology 143: 1885–1889 [DOI] [PubMed] [Google Scholar]
- Meyer AJ. (2008) The integration of glutathione homeostasis and redox signaling. J Plant Physiol 165: 1390–1403 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R. (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Hell R. (2005) Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res 86: 435–457 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Hager J, Chaouch S, Queval G, Han Y, Taconnat L, Saindrenan P, Gouia H, Issakidis-Bourguet E, Renou J-P, et al. (2010) Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol 153: 1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Tel-Or E. (1991) Oxidative stress responses in the unicellular cyanobacterium Synechococcus PCC 7942. Free Radic Res Commun 12-13: 845–850 [PubMed] [Google Scholar]
- Mullineaux CW. (2008) Phycobilisome-reaction centre interaction in cyanobacteria. Photosynth Res 95: 175–182 [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Rausch T. (2005) Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth Res 86: 459–474 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sonoike K, Hihara Y. (2009) Mechanism of downregulation of photosystem I content under high-light conditions in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 155: 989–996 [DOI] [PubMed] [Google Scholar]
- Newton GL, Fahey RC. (1995) Determination of biothiols by bromobimane labeling and high-performance liquid chromatography. Methods Enzymol 251: 148–166 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R. (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138: 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27: 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Hatano-Iwasaki A, Yanagida M, Iwabuchi M. (2004) Level of glutathione is regulated by ATP-dependent ligation of glutamate and cysteine through photosynthesis in Arabidopsis thaliana: mechanism of strong interaction of light intensity with flowering. Plant Cell Physiol 45: 1–8 [DOI] [PubMed] [Google Scholar]
- Okumura N, Masamoto K, Wada H. (1997) The gshB gene in the cyanobacterium Synechococcus sp. PCC 7942 encodes a functional glutathione synthetase. Microbiology 143: 2883–2890 [DOI] [PubMed] [Google Scholar]
- Pasternak M, Lim B, Wirtz M, Hell R, Cobbett CS, Meyer AJ. (2008) Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J 53: 999–1012 [DOI] [PubMed] [Google Scholar]
- Penninckx M. (2000) A short review on the role of glutathione in the response of yeasts to nutritional, environmental, and oxidative stresses. Enzyme Microb Technol 26: 737–742 [DOI] [PubMed] [Google Scholar]
- Perelman A, Uzan A, Hacohen D, Schwarz R. (2003) Oxidative stress in Synechococcus sp. strain PCC 7942: various mechanisms for H2O2 detoxification with different physiological roles. J Bacteriol 185: 3654–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Mata-Cabana A, Sánchez-Riego AM, Lindahl M, Florencio FJ. (2009) A comprehensive analysis of the peroxiredoxin reduction system in the cyanobacterium Synechocystis sp. strain PCC 6803 reveals that all five peroxiredoxins are thioredoxin dependent. J Bacteriol 191: 7477–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F. (2007) CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry 46: 15018–15026 [DOI] [PubMed] [Google Scholar]
- Prinz WA, Aslund F, Holmgren A, Beckwith J. (1997) The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem 272: 15661–15667 [DOI] [PubMed] [Google Scholar]
- Queval G, Noctor G. (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Queval G, Thominet D, Vanacker H, Miginiac-Maslow M, Gakière B, Noctor G. (2009) H2O2-activated up-regulation of glutathione in Arabidopsis involves induction of genes encoding enzymes involved in cysteine synthesis in the chloroplast. Mol Plant 2: 344–356 [DOI] [PubMed] [Google Scholar]
- Richaud C, Zabulon G, Joder A, Thomas JC. (2001) Nitrogen or sulfur starvation differentially affects phycobilisome degradation and expression of the nblA gene in Synechocystis strain PCC 6803. J Bacteriol 183: 2989–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Lemaire SD, Jacquot JP. (2008) The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu Rev Plant Biol 59: 143–166 [DOI] [PubMed] [Google Scholar]
- Sakuragi Y, Maeda H, Dellapenna D, Bryant DA. (2006) α-Tocopherol plays a role in photosynthesis and macronutrient homeostasis of the cyanobacterium Synechocystis sp. PCC 6803 that is independent of its antioxidant function. Plant Physiol 141: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh S, Ikeuchi M, Mimuro M, Tanaka A. (2001) Chlorophyll b expressed in cyanobacteria functions as a light-harvesting antenna in photosystem I through flexibility of the proteins. J Biol Chem 276: 4293–4297 [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Shin R. (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58: 47–69 [DOI] [PubMed] [Google Scholar]
- Schafer FQ, Buettner GR. (2001) Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med 30: 1191–1212 [DOI] [PubMed] [Google Scholar]
- Schmidt A. (1977) Assimilatory sulfate reduction via 3′-phosphoadenosine-5′-phosphosulfate (PAPS) and adenosine-5′-phosphosulfate (APS) in blue-green algae. FEMS Microbiol Lett 1: 137–140 [Google Scholar]
- Schürmann P, Buchanan BB. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10: 1235–1274 [DOI] [PubMed] [Google Scholar]
- Schwarz R, Forchhammer K. (2005) Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology 151: 2503–2514 [DOI] [PubMed] [Google Scholar]
- Serrano A, Rivas J, Losada M. (1984) Purification and properties of glutathione reductase from the cyanobacterium Anabaena sp. strain 7119. J Bacteriol 158: 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Bhattacharyya-Pakrasi M, Elvitigala T, Ghosh B, Aurora R, Pakrasi HB. (2009) A systems-level analysis of the effects of light quality on the metabolism of a cyanobacterium. Plant Physiol 151: 1596–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Elvitigala T, Bhattacharyya-Pakrasi M, Aurora R, Ghosh B, Pakrasi HB. (2008) Integration of carbon and nitrogen metabolism with energy production is crucial to light acclimation in the cyanobacterium Synechocystis. Plant Physiol 148: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Elvitigala T, Cameron JC, Ghosh BK, Bhattacharyya-Pakrasi M, Pakrasi HB. (2010) Integrative analysis of large scale expression profiles reveals core transcriptional response and coordination between multiple cellular processes in a cyanobacterium. BMC Syst Biol 4: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Li H, Sherman LA. (2004) Microarray analysis and redox control of gene expression in the cyanobacterium Synechocystis sp. PCC 6803. Physiol Plant 120: 27–35 [DOI] [PubMed] [Google Scholar]
- Spector D, Labarre J, Toledano MB. (2001) A genetic investigation of the essential role of glutathione: mutations in the proline biosynthesis pathway are the only suppressors of glutathione auxotrophy in yeast. J Biol Chem 276: 7011–7016 [DOI] [PubMed] [Google Scholar]
- Stork T, Michel KP, Pistorius EK, Dietz KJ. (2005) Bioinformatic analysis of the genomes of the cyanobacteria Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942 for the presence of peroxiredoxins and their transcript regulation under stress. J Exp Bot 56: 3193–3206 [DOI] [PubMed] [Google Scholar]
- Suginaka K, Yamamoto K, Ashida H, Sawa Y. (1999) Effect of intracellular glutathione on heat-induced cell death in the cyanobacterium, Synechocystis PCC 6803. Biosci Biotechnol Biochem 63: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Suginaka K, Yamamoto K, Ashida H, Kono Y. (1998) Cysteine uptake for accumulation of glutathione by the cyanobacterium Synechocystis PCC 6803. Biosci Biotechnol Biochem 62: 424–428 [DOI] [PubMed] [Google Scholar]
- Sundquist AR, Fahey RC. (1989) The function of gamma-glutamylcysteine and bis-gamma-glutamylcystine reductase in Halobacterium halobium. J Biol Chem 264: 719–725 [PubMed] [Google Scholar]
- Takahashi H, Uchimiya H, Hihara Y. (2008) Difference in metabolite levels between photoautotrophic and photomixotrophic cultures of Synechocystis sp. PCC 6803 examined by capillary electrophoresis electrospray ionization mass spectrometry. J Exp Bot 59: 3009–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tel-Or E, Huflejt M, Packer L. (1985) The role of glutathione and ascorbate in hydroperoxide removal in cyanobacteria. Biochem Biophys Res Commun 132: 533–539 [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Avenson TJ, Thomas JB, Herbert SK. (1998) A cyanobacterium lacking iron superoxide dismutase is sensitized to oxidative stress induced with methyl viologen but is not sensitized to oxidative stress induced with norflurazon. Plant Physiol 116: 1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy M, Vermaas W. (1999) In vivo role of catalase-peroxidase in Synechocystis sp. strain PCC 6803. J Bacteriol 181: 1875–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inzé D, et al. (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AM, Asoh S, Ohsawa I, Ohta S. (2008) Imaging mitochondrial redox environment and oxidative stress using a redox-sensitive fluorescent protein. J Nippon Med Sch 75: 66–67 [DOI] [PubMed] [Google Scholar]
- Zechmann B, Tomašić A, Horvat L, Fulgosi H. (2010) Subcellular distribution of glutathione and cysteine in cyanobacteria. Protoplasma 246: 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.