Abstract
PEX5 and PEX7 are receptors required for the import of peroxisome-bound proteins containing one of two peroxisomal targeting signals (PTS1 or PTS2). To better understand the role of PEX5 in plant peroxisomal import, we characterized the Arabidopsis (Arabidopsis thaliana) pex5-10 mutant, which has a T-DNA insertion in exon 5 of the PEX5 gene. Sequencing results revealed that exon 5, along with the T-DNA, is removed in this mutant, resulting in a truncated pex5 protein. The pex5-10 mutant has germination defects and is completely dependent on exogenous Suc for early seedling establishment, based on poor utilization of seed-storage fatty acids. This mutant also has delayed development and reduced fertility, although adult pex5-10 plants appear normal. Peroxisomal metabolism of indole-3-butyric acid, propionate, and isobutyrate also is disrupted. The pex5-10 mutant has reduced import of both PTS1 and PTS2 proteins, and enzymatic processes that occur in peroxisomes are disrupted. To specifically study the import and importance of PTS1 proteins, we made a truncated PEX5 construct lacking the PTS1-binding region (PEX5454). Transformation of this construct into pex5-10 resulted in the rescue of PTS2 import, thereby creating a line with PTS1-specific import defects. The pex5-10 (PEX5454) plants still had developmental defects, although restoring PTS2 import resulted in a less severe mutant phenotype. Comparison of pex5-10 and pex5-10 (PEX5454) phenotypes can separate the import mechanisms for enzymes acting in different peroxisomal processes, including indole-3-butyric acid/2,4-dichlorophenoxybutyric acid oxidation, isobutyrate and propionate metabolism, and photorespiration.
Peroxisomes house a number of critical processes in plants (Kaur et al., 2009). They are the primary site for fatty acid β-oxidation and play an essential role during germination and early seedling establishment in oilseed plants. Prior to photosynthetic initiation, stored lipids are β-oxidized in peroxisomes to provide the necessary energy and metabolites for development. Peroxisomes also play a role in plant hormone β-oxidation. They are the site for the conversion of an auxin storage form, indole-3-butyric acid (IBA), to its active signaling counterpart, indole-3-acetic acid (IAA; Zolman et al., 2000; Strader et al., 2010). The latter reactions of the jasmonic acid (JA) synthesis pathway also take place in peroxisomes (Wasternack and Kombrink, 2010). Metabolism of intermediates such as propionate, isobutyrate, acetate, and benzoic acid occurs in peroxisomes (Zolman et al., 2001; Hooks et al., 2007; Lucas et al., 2007; Ibdah and Pichersky, 2009). As a result of these metabolic processes, peroxisomes affect important physiological processes, including senescence, starvation, photorespiration, and photomorphogenesis (Wanner et al., 1991; Graham et al., 1992; Hu et al., 2002; Foyer et al., 2009; Kunz et al., 2009).
Since peroxisomes lack genetic material, proteins acting in peroxisomal processes are synthesized on free ribosomes in the cytosol and imported posttranslationally. These proteins contain one of two well-characterized peroxisomal targeting signals (PTS1 and PTS2) that direct target proteins to the peroxisome matrix. PTS1 is a C-terminal tripeptide consisting of the consensus sequence (S/A/C)(K/R/H)(L/M); in plants, the preferred targeting signal is Ser-Lys-Leu (SKL; Reumann et al., 2007). This signal is not removed after import. PTS2 is at the N terminus of target proteins and consists of nine amino acids; the most common plant PTS2 is RL-X5-HL (Reumann et al., 2004). The N-terminal signal sequence is cleaved after import. Approximately 400 genes in the Arabidopsis (Arabidopsis thaliana) genome code for proteins with either PTS1 or PTS2 signals (Kamada et al., 2003; Reumann, 2004; Reumann et al., 2007; Kaur et al., 2009). A majority of these proteins contain a PTS1, with about one-quarter having the PTS2 sequence (Kamada et al., 2003; Reumann, 2004; Reumann et al., 2007).
Proteins that play a role in peroxisome biogenesis are referred to as PEROXINS (PEX). PEX5 and PEX7 function as receptors that recognize PTS1 and PTS2 proteins, respectively, for matrix protein import. PTS1 protein import occurs when the PTS1-containing protein is bound by PEX5 in the cytoplasm. The receptor-cargo complex interacts with the peroxisome membrane proteins PEX13 and PEX14 (Hayashi et al., 2000; Nito et al., 2002; Mano et al., 2006). The complex then translocates the matrix protein into the peroxisome in a process that requires the RING-finger proteins PEX2, PEX10, and PEX12 (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003; Fan et al., 2005). After translocation into the matrix, the cargo is released and the receptor is recycled to the cytosol for additional rounds of import (Dodt and Gould, 1996; Dammai and Subramani, 2001; Nair et al., 2004). Receptor recycling requires the PEX1 and PEX6 ATPases and the PEX4 ubiquitin-conjugating enzyme, which is anchored to the peroxisome membrane by PEX22 (Geisbrecht et al., 1998; Kiel et al., 1999; Zolman and Bartel, 2004; Zolman et al., 2005). In yeast, monoubiquitination of PEX5 by PEX12 targets the receptor for recycling back to the cytosol, allowing additional rounds of import (Platta et al., 2009). PTS2-containing proteins are imported into peroxisomes by the PEX7 receptor. PEX7-mediated import requires PEX5 for peroxisomal membrane protein interactions (Nito et al., 2002; Hayashi et al., 2005; Woodward and Bartel, 2005). Recent evidence also indicates that PEX5 requires PEX7 for optimal peroxisomal import in Arabidopsis (Ramón and Bartel, 2010).
Arabidopsis PEX5 contains three regions critical for its function: the tetratricopeptide repeat (TPR) region, the pentapeptide repeat (PPR) region, and a PEX7-binding domain (Fig. 1A). In its C terminus, PEX5 has six TPRs, which are 34-amino acid conserved repeats. The TPR interacts with the PTS1 sequence of proteins destined for import (Dodt et al., 1995; Terlecky et al., 1995; Gatto et al., 2000). In the N terminus, PEX5 contains 11 PPRs, which interact with peroxisomal membrane proteins during translocation (Nito et al., 2002). This region also affects the recycling of PEX5 back to the cytosol (Costa-Rodrigues et al., 2004). PEX5 has a central PEX7-binding domain (Nito et al., 2002), allowing it to bind the PTS2 receptor.
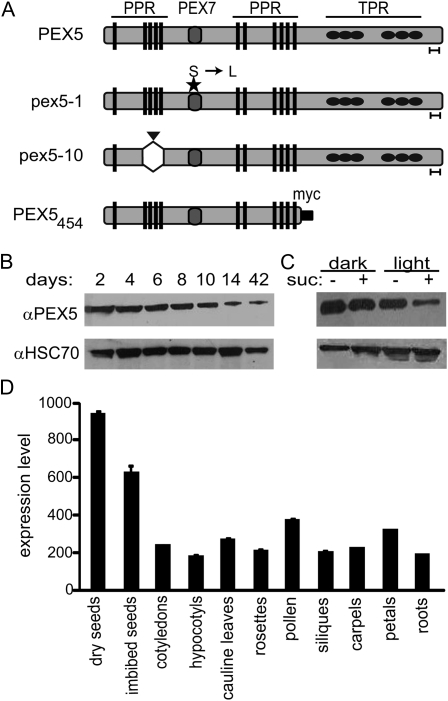
Figure 1.
PEX5 gene expression and protein concentration. A, Cartoon representation of the PEX5 protein, the pex5 mutant proteins, and the PEX5454 truncation. The pex5-1 Ser-to-Leu point mutation (Zolman et al., 2000) and the pex5-10 insertion site and resulting deletion are indicated by a star and a triangle, respectively. Gray circles indicate the PEX7-binding domain (PEX7), black ovals indicate the TPR domains, and rectangles represent the PPR domains. The goalpost under the protein indicates the recognition site of the PEX5 antibody (Zolman and Bartel, 2004). B, Total protein was extracted from wild-type Col-0 seedlings and adult plants and analyzed by western blot for PEX5 levels. C, Total protein from 5-d-old wild-type seedlings grown in the dark or light in the presence or absence of Suc was extracted and analyzed by western blot for PEX5 levels. For both B and C, equal amounts of protein were loaded, as confirmed by immunoblotting with an HSC70 antibody (bottom). D, Levels of PEX5 mRNA expression (absolute values) in dry and imbibed seeds, cotyledons, hypocotyls, cauline leaves, rosette leaves, mature pollen, seeds/siliques, carpels, petals, and roots. The graph was constructed using Arabidopsis eFP Browser data from http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi (Winter et al., 2007). Data were retrieved on March 6, 2010.
Genetic analysis has indicated that numerous PEX proteins are required for correct peroxisomal matrix protein import in Arabidopsis. Strong loss-of-function mutations resulted in embryo-lethal mutants, as seen in pex2 and pex10 (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003). However, partial loss-of-function mutants have revealed characteristic phenotypes associated with decreased peroxisome function. Peroxisome-defective mutants are unable to efficiently metabolize long-chain fatty acids. This deficiency can be directly measured by monitoring the metabolism of C20:1 (eicosenoic acid), which is high in seeds but quickly metabolized (Lemieux et al., 1990). Alternatively, because mutants break down seed-storage fatty acid substrates more slowly, providing less energy, the plants show delayed development and a “Suc-dependent” phenotype: mutants are unable to establish into adults, unless provided exogenous Suc. Other processes commonly are disrupted in peroxisome-defective mutants, including the conversion of IBA to IAA (Zolman et al., 2000). This deficiency can be seen in a root elongation assay; peroxisome-defective mutants are resistant to the inhibitory effects of exogenous hormone.
Because PEX5 plays a critical role in peroxisomal matrix protein import, we further investigated the activity of this protein in Arabidopsis. Previously, the pex5-1 mutant was identified as an IBA-resistant mutant with weak Suc dependence and a slight decrease in seed-storage fatty acid metabolism (Zolman et al., 2000). pex5-1 has a C-to-T point mutation, causing a Ser-to-Leu amino acid conversion in the region of PEX5 responsible for binding PEX7 (Zolman et al., 2000). The pex5-1 mutant has a PTS2 protein import defect, indicated by reduced PTS2-GFP peroxisomal localization and normal GFP-PTS1 import (Woodward and Bartel, 2005). In this work, we characterized phenotypic abnormalities in the pex5-10 insertion mutant. pex5-10 exhibits a more severe phenotype than pex5-1, including disruption of both PTS1 and PTS2 import. However, we show that the insertion mutant is not a null allele and likely survives because of an unusual T-DNA processing that allows the production of a partial transcript. Finally, we created a PTS1-specific pex5 line by complementing pex5-10 with a partial PEX5 cDNA, restoring PTS2 import. Comparison of pex5-10 with mutant transgenic lines expressing the truncated PEX5454 protein allowed us to separate the effects of defects in PTS1, PTS2, or PTS1 and PTS2 import.
RESULTS
PEX5 Levels Are Highest Early in Development
We began our study of PEX5 by investigating how the protein accumulates in plants. Western blots using an antibody that recognizes the C terminus of PEX5 (Zolman and Bartel, 2004) revealed that the protein accumulated at high levels in seedlings and at lower levels as plants mature (Fig. 1B). Because peroxisomal metabolism, and hence PEX5, are critical during early seedling development when fatty acids are being highly metabolized, we also checked the accumulation of the protein under different developmental conditions. PEX5 was present at a higher level in the dark than in the light (Fig. 1C). In addition, we saw higher accumulation in the absence of Suc than when Suc was included in the medium (Fig. 1C). These protein patterns are consistent with β-oxidation increases during dark development, when photosynthesis cannot occur, and with decreases when Suc is present, indicating that photosynthesis is high. These protein studies are consistent with publicly available microarray data, which show that PEX5 gene expression is highest in seeds but relatively stable throughout plant development (Fig. 1D).
pex5-10 Encodes an Altered pex5 Protein
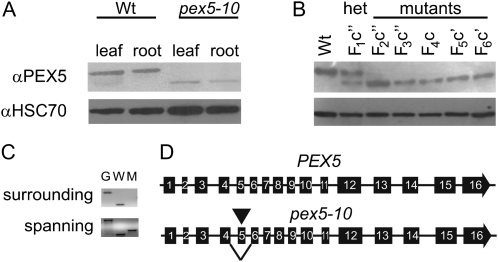
To study the role of PEX5 in plants and examine the effects of peroxisomal import during plant development, we wanted to examine a stronger mutant allele than the previously described pex5-1 point mutation (Zolman et al., 2000). The pex5-10 mutant has a T-DNA insertion in exon 5 (Fig. 1A), which we confirmed by PCR amplification using genomic and T-DNA-specific primers (data not shown). Sequence analysis of the mutant line revealed that the site of the insertion was at position 1,769 (with 1 being the A of the ATG initiator). Using western blots with a PEX5 antibody, we detected no full-length protein in the mutant (Fig. 2A). However, we saw a lower molecular weight product (Fig. 2A), which segregated in an F1 backcross plant (Fig. 2B). The smaller protein product typically was present at a lower abundance but was detectable consistently through several generations and following additional backcrosses to the wild type (Fig. 2B).
Figure 2.
T-DNA insertion effects in the pex5-10 mutant. A, Total protein extracted from leaves and roots of 12-d-old wild-type (Wt) and pex5-10 seedlings was analyzed by western blot with a PEX5 antibody. B, Total protein from 3-week-old wild-type and pex5-10 plants was analyzed by western blot for PEX5 levels. Seedlings were taken from the heterozygous progeny of a pex5-10 backcross to Col-0 (F1c′′) and pex5-10 homozygous mutant plants over four generations (F2 through F6) and three backcrosses (F4c for the first backcross; F5c′ and F6c′ for the second backcross; F2c′′ and F3c′′ for the third backcross). For all blots, equal amounts of protein were loaded, as confirmed by immunoblotting with an HSC70 antibody (bottom). C, To examine the effects of the insertion in exon 5, cDNAs from 3-week-old wild-type (W) and pex-10 (M) plants were amplified with PEX5 primers that directly surround (exons 5 + 6; expected cDNA size 340 bp/expected genomic size 774 bp) or span (exons 3 + 7; 780 bp/2,234 bp) the T-DNA. Genomic DNA (G) from wild-type plants was amplified to show the purity of cDNA synthesis. D, Cartoon representation of the PEX5 and pex5-10 genomic structures. Exon numbers are labeled. The pex5-10 insertion is shown by the triangle above the sequence, and the splice transition from the end of exon 4 to the beginning of exon 6 is indicated.
To determine the mutant protein sequence, we isolated cDNA from mutant plants and performed reverse transcription (RT)-PCR using PEX5-specific primers. We did not see a product using primers directly surrounding the insertion, consistent with our PCR data. However, we could see products amplified using either upstream or downstream primer sets (data not shown). Interestingly, we observed smaller than expected products when we used spanning primers around the insertion (Fig. 2C). We sequenced the spanning product and found a complete deletion of exon 5 along the normal exon/intron junctions in the mutant cDNA, with the insertion removed (Fig. 2D). This excision resulted in the production of a mutant protein lacking four of the PPRs (Fig. 1A). The abnormal protein accumulated in both leaves and roots of adult plants at similar levels, indicating that this alteration was not tissue specific (Fig. 2A).
pex5-10 Has Defects in Both PTS1 and PTS2 Import
One rationale for studying pex5 mutants was to determine the effects of losing PTS1 import into peroxisomes. However, the pex5-1 mutant has a PTS2-specific defect (Woodward and Bartel, 2005). To demonstrate the import defects in pex5-10, we crossed the mutant with lines containing GFP reporter genes targeted to peroxisomes using either the PTS1 (GFP-PTS1; Zolman and Bartel, 2004) or PTS2 (PTS2-GFP; Woodward and Bartel, 2005) sequence. Peroxisomal import in wild-type plants appears as a characteristic punctate patterning. Compared with the wild-type controls, pex5-10 showed a diffuse pattern indicative of cytosolic mislocalization for both PTS1- and PTS2-containing proteins (Fig. 3A). This result demonstrates that both peroxisomal import pathways are disrupted, consistent with previous studies showing disruptions in the import of PTS1 proteins (Lee et al., 2006; Lingard and Bartel, 2009; Lingard et al., 2009) and PTS2 proteins (Zolman et al., 2005; Lee et al., 2006; Lingard and Bartel, 2009; Ramón and Bartel, 2010).
Figure 3.
Peroxisomal import defects in pex5 mutant lines. A, Localization of peroxisomally targeted GFP reporters with either PTS1 (Zolman and Bartel, 2004) or PTS2 (Woodward and Bartel, 2005) sequence. Wild-type (Wt) lines (Col-0 background) containing these constructs were crossed into the pex5-10 or pex5-10 (PEX5454) background. Root hair cells for each line are shown, with all images taken at the same magnification. B, Total protein from wild-type and three independent transgenic Col-0 (PEX5454) plants were analyzed by western blot for PEX5454 accumulation using a myc antibody. C, Western blot of total protein from wild-type, pex5-10 (PEX5), pex5-1, pex5-1 (PEX5454), pex5-10, and pex5-10 (PEX5454) with PEX5, myc, and thiolase antibodies. Thiolase contains a PTS2 that is cleaved following import; protein size indicates if the protein is localized in the peroxisome (mature short form; two asterisks) or if it remains in the cytoplasm (unprocessed long form; asterisk). Four independent pex5-10 (PEX5454) lines (p126, p122, p109, and p111) are shown to indicate the range of responses. For all blots, equal amounts of protein were loaded, as confirmed by immunoblotting with an HSC70 antibody. D, Extracts from 4-d-old Col-0 (wild-type), pex5-1, pex5-1 (PEX5454), pex5-10, and pex5-10 (PEX5454) seedlings were tested for acyl-CoA oxidase activity on n-hexanoyl-CoA (C6:0) and lauroyl-CoA (C12:0). Data are presented as pmol H2O2 produced per min. Error bars represent se of the rates from three independent experiments. The control samples used water in place of plant extracts to account for background readings.
Overexpression of a Truncated PEX5 Rescues PTS2 Import Defects
To further investigate the role of PEX5, we wanted to create a pex5 mutant plant line with a PTS1-specific import defect. We designed six constructs to express truncated PEX5 proteins using the cauliflower mosaic virus 35S promoter. Each of these constructs was independently transformed into wild-type ecotype Columbia (Col-0) plants. However, five of the mutant proteins did not accumulate in wild-type plants selected to contain the transgene (data not shown). The PEX5454 construct encodes the first 454 amino acids of the protein, including the PPRs important for peroxisomal membrane protein binding and the PEX7 interaction domain; this construct does not have any of the C-terminal TPR domains known to bind the PTS1 of peroxisome-bound proteins (Fig. 1A). Because our PEX5 antibody recognizes the C terminus of the full-length protein, we added a myc epitope tag to PEX5454. We analyzed wild-type Arabidopsis transformants for mutant protein accumulation using αmyc (Fig. 3B). Col-0 (PEX5454) plants with a strong αmyc signal then were crossed to pex5-1 and pex5-10 mutants. We selected mutant lines containing the transgene by PCR, then confirmed the accumulation of the PEX5454 protein (Fig. 3C). Because we saw some variability in pex5-10 (PEX5454) responses, four independent lines (designated px) are shown for most assays to represent the range of phenotypes. As a control for phenotypic experiments, we used pex5-10 complemented with a full-length PEX5 cDNA.
Our hypothesis was that PEX5454 would restore PTS2 import in the pex5 mutants, given that the PEX5454 protein retains the regions necessary for PEX7 binding and peroxisomal membrane interactions. To determine the effects of the transgene on PTS2 import, we examined the import of the GFP reporter genes and compared the responses with the mutant parents. We saw restored PTS2-GFP import in pex5-10 (PEX5454) compared with the pex5-10 parent line (Fig. 3A). As hypothesized, PTS1 protein import was similarly impaired in the transgenic lines and the pex5-10 parent line (Fig. 3A). The pex5-1 (PEX5454) line showed a similar recovery of PTS2 import compared with the pex5-1 control (data not shown).
We further examined the import and processing of the PTS2 protein 3-ketoacyl-CoA thiolase, which acts in the final step of β-oxidation (Hayashi et al., 1998). PTS2 proteins are synthesized in the cytosol, and the N-terminal signal sequence is proteolytically cleaved by the DEG15 protease following import into the peroxisomal matrix, resulting in a smaller mature protein (Hijikata et al., 1987; Helm et al., 2007; Schuhmann et al., 2008). Analysis of thiolase processing from the larger cytosolic form to the smaller mature form by western blot can serve as one indicator of the degree of peroxisomal import. Both pex5-1 and pex5-10 showed defects in thiolase processing, with pex5-10 being more severe (Fig. 3C). However, inclusion of the PEX5454 transgene in pex5-1 mutants led to a complete rescue of thiolase processing. The pex5-10 (PEX5454) lines still showed defects in thiolase processing to varying degrees, but each showed improvement compared with the parent phenotype (Fig. 3C). The remaining processing defect in pex5-10 (PEX5454) may be due to decreased import of DEG15, which uses a PTS1, as we did not see residual defects in pex5-1 (PEX5454) lines or pex5-10 (PEX5) lines, in which PTS1 import was normal. These results further suggest that the PEX5454 protein can improve PTS2 import in both pex5-1 and pex5-10 mutant plants.
To characterize the consequences of the import defects on peroxisomal function, we measured the activity of two acyl-CoA oxidase (ACX) enzymes, ACX3 and ACX4. These enzymes act in the first step of β-oxidation (Graham, 2008). ACX3 has a PTS2 and ACX4 has a PTS1. Activity assays indicate that ACX3 primarily β-oxidizes medium-chain fatty acids, such as C12:0 (Eastmond et al., 2000; Froman et al., 2000; Adham et al., 2005) and that ACX4 β-oxidizes short-chain substrates of C4:0 to C6:0 (Hayashi et al., 1999; Rylott et al., 2003; Adham et al., 2005). Extracts prepared from pex5-1 had similar activity as the wild type on C6:0 substrates, but pex5-10 showed significantly reduced activity on this substrate (Fig. 3D). These data confirm that PTS1 protein activity is not disrupted in pex5-1 but is severely compromised in pex5-10. The pex5-1 and pex5-10 mutants were both impaired in β-oxidation of C12:0 fatty acid substrates, consistent with the defects that these mutants have in PTS2 import (Fig. 3D). Addition of PEX5454 to pex5-1 rescued C12:0 activity to wild-type levels and partially rescued the pex5-10 mutant (Fig. 3D). These results suggest that the pex5 PTS2 import defects are reduced by inclusion of the PEX5454 protein. The activity assays showed similar C6:0 fatty acid defects in pex5-10 and pex5-10 (PEX5454) (Fig. 3D). This result further suggests that the pex5-10 PTS1 protein import defect is not improved by the PEX5454 protein.
pex5-10 Mutants Have Severe Defects in β-Oxidation
The most striking phenotype for the pex5-10 mutant was a severe germination defect. Typical germination rates for pex5-10 lines ranged from 0% to 20%, which were seed line and harvest date dependent (data not shown). Any germination that occurred was delayed compared with the wild type; by day 3, 100% of wild-type and pex5-1 mutant seeds had germinated but none of the pex5-10 seeds had germinated (Table I). To quantify germination rates, we plated progeny seeds from an F2 segregating population of a pex5-10 × Col-0 backcross. After 4 d, approximately 75% of the seeds had germinated, but we never identified pex5-10 mutant seedlings (Table II). We saw the expected number of heterozygous plants (Table II), indicating that the mutation segregated in a recessive fashion. Interestingly, if we nicked the seed coats of seeds that did not germinate, we saw increased germination, as defined by radicle emergence. Genotyping results indicated that this population represented the homozygous mutant lines (Table II).
Table I. Unassisted germination rate of 100 seeds over time.
Data are shown as percentage germination.
| Seed Lines | Day 2 | Day 3 | Day 4 |
| Wild type | 7 | 100 | 100 |
| pex5-10 | 0 | 0 | 1 |
| pex5-10 (PEX5) | 9 | 100 | 100 |
| pex5-1 | 10 | 100 | 100 |
| pex5-1 (PEX5454) | 74 | 100 | 100 |
| pex5-10 (PEX5454) p111 | 0 | 38 | 100 |
| pex5-10 (PEX5454) p126 | 1 | 47 | 99 |
| pex5-10 (PEX5454) p109 | 8 | 50 | 100 |
| pex5-10 (PEX5454) p122 | 3 | 33 | 100 |
Table II. Germination responses of 100 segregating seeds from four progeny of a pex5-10 × Col-0 backcross.
Data are shown as number of individuals.
| Genotype | Parent 1 | Parent 2 | Parent 3 | Parent 4 |
| Wild type | 19 | 21 | 26 | 28 |
| Heterozygous | 52 | 54 | 52 | 49 |
| pex5-10 | 0 | 0 | 0 | 0 |
| After nicking | ||||
| Wild type | 0 | 0 | 0 | 0 |
| Heterozygous | 0 | 0 | 0 | 0 |
| pex5-10 | 24 | 25 | 20 | 23 |
Although pex5-10 seeds germinated following nicking, hypocotyl elongation still did not occur on medium lacking Suc (Fig. 4A). However, nicked pex5 seedling hypocotyls could elongate on Suc-supplemented medium in a concentration-dependent manner (Fig. 4A). This phenotype is consistent with a peroxisome-defective phenotype, as loss of fatty acid β-oxidation coincides with an inability to develop that can be rescued by the application of Suc (Hayashi et al., 1998). This phenotype is similar to, but more severe than, the pex5-1 phenotype (Fig. 4A).
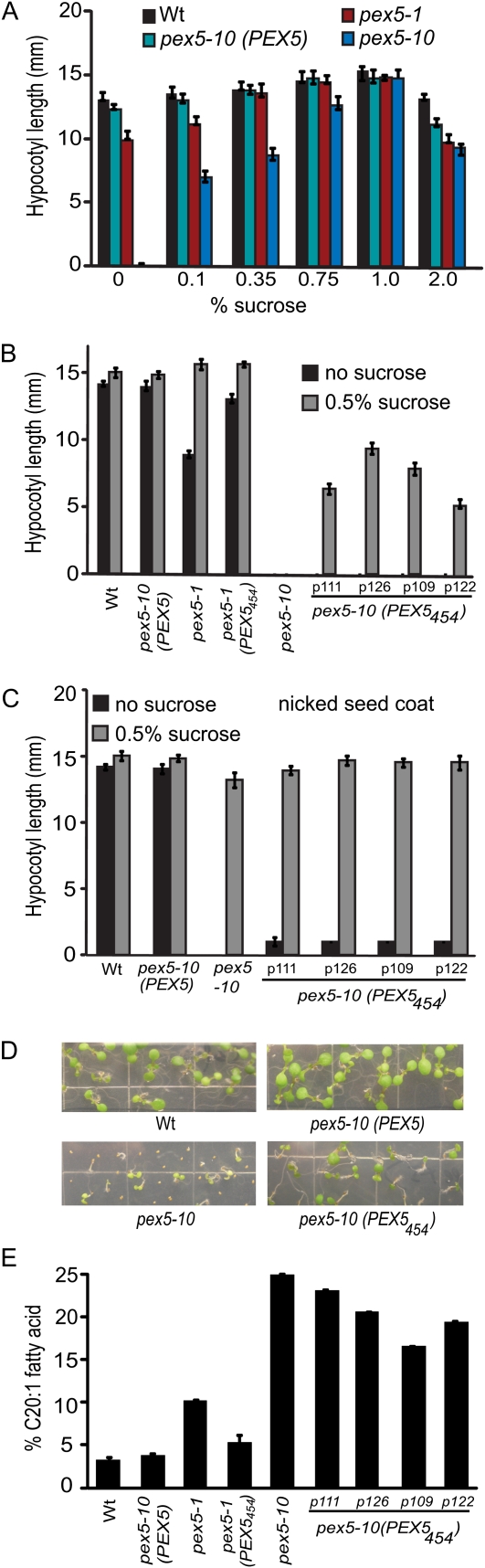
Figure 4.
Fatty acid metabolism in pex5 mutant seedlings. A, Hypocotyl length of seedlings grown for 24 h in light and 6 d in dark on medium supplemented with increasing concentrations of Suc. Error bars represent se (n ≥ 12). B, Hypocotyl length of seedlings grown for 24 h in light and 6 d in dark on medium without Suc or with 0.5% Suc. Error bars represent se (n ≥ 12). C, Hypocotyl length of seedlings grown for 24 h in light and 6 d in dark on medium without Suc or with 0.5% Suc. pex5-10 seed coats were nicked immediately after plating. Error bars represent se (n ≥ 12). D, Photographs depicting pex5-10 plant lines. Plants were grown on complete medium for 7 d. pex5-10 seed coats were nicked after plating. E, Percentage of C20:1 fatty acid (versus total fatty acids) in 6-d-old seedlings grown in light on 0.5% Suc medium. Error bars represent se of three biological replicates. Wt, Wild type.
To determine whether the PEX5454 protein rescued the germination defects in pex5-1 and pex5-10, we assayed the transgenic lines for reduced Suc dependence. The results showed that the PEX5454 protein restored pex5-1 growth without Suc (Fig. 4B). Although pex5-10 seeds did not germinate without nicking, seeds of pex5-10 (PEX5454) lines could germinate (Table I) and hypocotyls could elongate in medium supplemented with Suc (Fig. 4B). When seed coats were nicked, the pex5-10 (PEX5454) lines had enhanced hypocotyl elongation compared with pex5-10, although still reduced compared with the wild type (Fig. 4C). Additionally, the pex5-10 (PEX5454) seedlings established more robustly than the pex5-10 seedlings (Fig. 4D).
To directly examine fatty acid β-oxidation, we quantified levels of the seed-storage fatty acid eicosenoic acid in 6-d-old seedlings. Eicosenoic acid is high in seeds but quickly is degraded (Lemieux et al., 1990); wild-type seedlings show only approximately 4% eicosenoic acid after 6 d (Fig. 4E). However, eicosenoic levels remained at approximately 25% in pex5-10 seedlings and at 10% in pex5-1 (Fig. 4E). These results indicate that the pex5 mutants have reduced β-oxidation of seed-storage fatty acids, with phenotypic strength paralleling the genetic defects. In contrast to pex5-1, pex5-1 (PEX5454) levels were similar to wild-type levels (Fig. 4E). pex5-10 (PEX5454) lines showed improvements in metabolism from 25% eicosenoic acid to 23% to 17%, indicative of a somewhat increased β-oxidation rate in the transgenic lines compared with the mutant parent (Fig. 4E).
Taken together, these results indicate that the pex5 mutants have disruptions in peroxisomal function that can be visualized by developmental defects. The PEX5454 protein restored pex5-1 hypocotyl elongation and fatty acid β-oxidation back to wild-type levels. pex5-1 has a PTS2-specific defect, and the complete rescue of this phenotype in the transgenic lines is consistent with the PEX5454 protein increasing PTS2 import. The truncated protein only partially rescued the severe fatty acid β-oxidation defects in the pex5-10 mutant, which has both PTS1 and PTS2 import defects. Although this rescue is only partial, the PEX5454 addition does provide sufficient energy for the pex5-10 (PEX5454) seeds to germinate without physical assistance.
pex5-10 Mutants Have Reduced Responses in Peroxisomal Hormone Metabolism
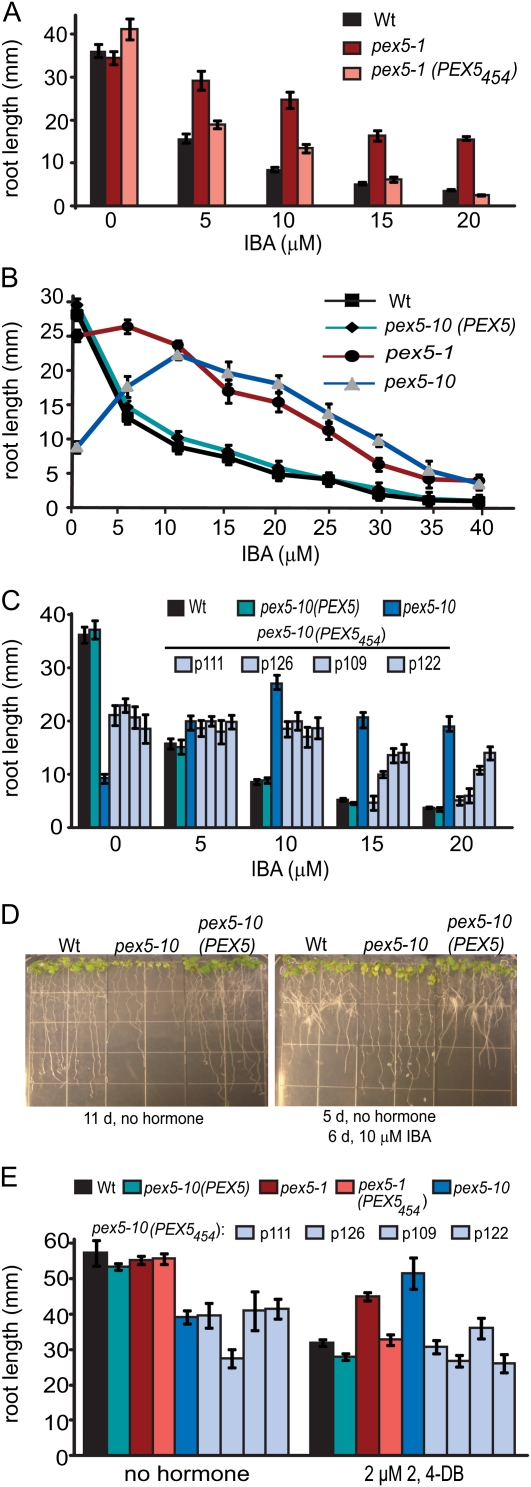
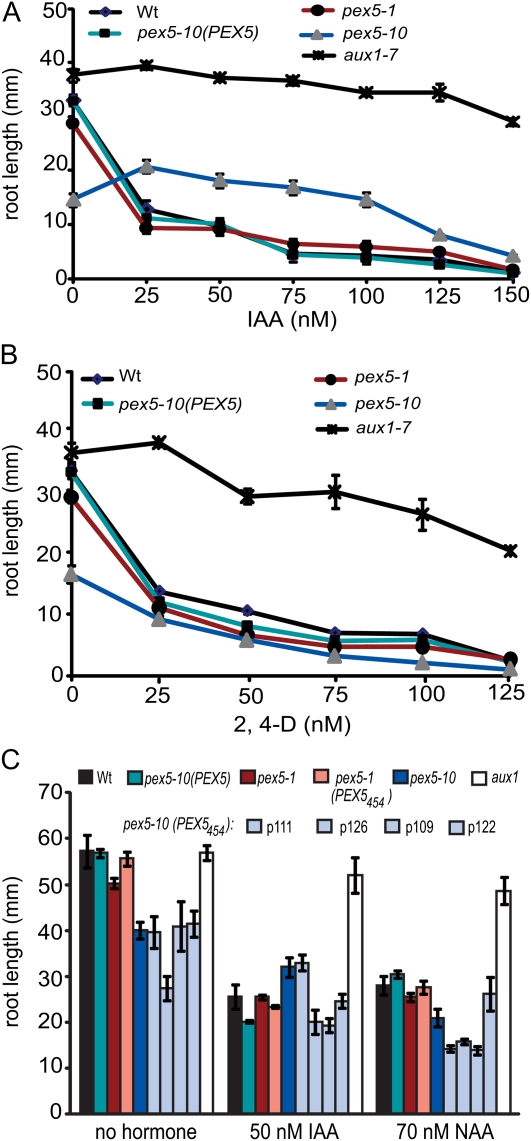
To investigate the extent of peroxisomal defects in the pex5 mutants, we next examined the responses of seedlings to IBA and 2,4-dichlorophenoxybutyric acid (2,4-DB), a synthetic IBA analog (Figs. 5 and 6). These compounds are metabolized in peroxisomes. Both pex5-1 and pex5-10 seedlings were insensitive to the inhibitory effects of IBA and 2,4-DB on root elongation (Fig. 5). pex5-10 mutants have short roots on unsupplemented medium, but root length actually increased on 5 to 10 μm IBA and remained longer than the wild type over a range of IBA concentrations (Fig. 5B). Expression of PEX5454 restored IBA sensitivity to pex5-1 mutants (Fig. 5A). However, this truncated PEX5 protein only rescued the IBA insensitivity of pex5-10 partially (Fig. 5C), and the degree of rescue varied among lines. In contrast, the pex5-1 (PEX5454) and pex5-10 (PEX5454) lines were all similarly sensitive to 2,4-DB (Fig. 5E).
Figure 5.
pex5 mutant responses to IBA or the IBA analog 2,4-DB. A to C, Root length of 10-d-old seedlings on medium supplemented with IBA. Error bars represent se (n ≥ 12). D, Photographs of 11-d-old wild-type (Wt), pex5-10, and pex5-10 (PEX5) seedlings grown without hormone or on 10 μm IBA. E, Root length of 10-d-old seedlings grown under continuous light on medium supplemented with 2,4-DB. Error bars represent se (n ≥ 12).
Figure 6.
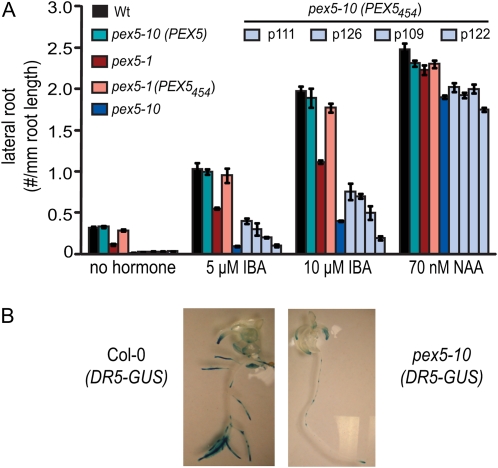
pex5 lateral root formation. A, Lateral root density of seedlings grown without hormone for 5 d and then transferred to medium with or without hormone for another 5 d. Data are shown as the number of lateral roots per mm root length. Error bars represent se (n ≥ 12). B, DR5-GUS expression in 8-d-old Col-0 and pex5-10 transgenic seedlings grown on complete medium with 0.5% Suc. Wt, Wild type.
The pex5 mutants and the PEX5454 lines responded normally to the synthetic auxins 2,4-dichlorophenoxyacetic acid (2,4-D) and naphthylacetic acid (NAA; Fig. 7). However, as seen with IBA and 2,4-DB, pex5-10 often showed increased growth on low concentrations of IAA (Fig. 7A), perhaps indicating that IAA levels are low in the mutant or that exogenous application increases the ability of the mutant to develop.
Figure 7.
pex5 mutant responses to IAA, 2,4-D, and NAA. Root length is shown for 10-d-old seedlings grown on medium with IAA (A and C), 2,4-D (B), or NAA (C). The auxin transport mutant aux1 (Pickett et al., 1990) was used as a control to demonstrate insensitivity to IAA. Error bars represent se (n ≥ 12). Wt, Wild type.
Because one role of IBA in plants is the initiation of lateral roots, we examined mutants for lateral root formation. Both pex5 mutants were compromised in lateral root formation on medium without hormone or with IBA (Fig. 6). On medium supplemented with IBA, the PEX5454 protein rescued pex5-1 seedling defects to wild-type levels, but lateral root formation was again only partially rescued in pex5-10 (PEX5454) seedlings (Fig. 6A). In contrast, pex5 mutants initiated lateral roots similar to the wild type on NAA (Fig. 6A), indicating that the defect is not in lateral root development but in lateral root stimulation. This defect can be visualized in pex5-10 (DR5-GUS) lines, which contain an auxin-inducible reporter (Guilfoyle, 1999) that highlights lateral root primordia. pex5-10 lines with this transgene show reduced GUS levels and fewer lateral roots than Col-0 (DR5-GUS) lines (Fig. 6B). Taken together, these data suggest that pex5 mutants are compromised in the peroxisomal conversion of IBA β-oxidation to IAA.
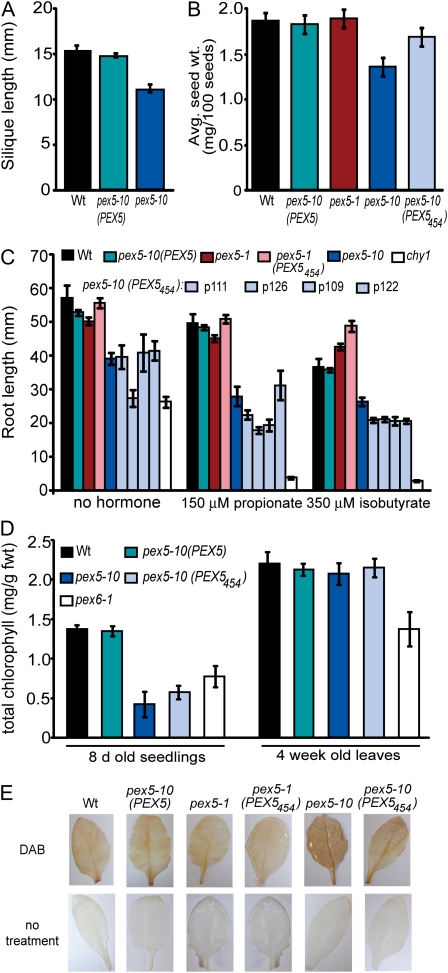
JA biosynthesis and perception are required for male fertility (Wasternack and Kombrink, 2010). Although JA biosynthesis is initiated in chloroplasts, the intermediate 12-oxo-phytodienoic acid is transported to the peroxisome for β-oxidation to JA. Measurement of siliques indicated that pex5-10 makes fewer seeds than the wild type (Fig. 8A). pex5-1 silique lengths were normal (data not shown). Furthermore, we determined the average seed weight of 100 seeds, and pex5-10 seeds weighed less than wild-type seeds (Fig. 8B) and frequently looked small and wrinkled (data not shown). However, seed weight of pex5-10 (PEX5454) was similar to that in the wild type (Fig. 8B). These data suggest that pex5-10 has minor fertility defects due to defects in PTS2 import. We also examined the mutants for JA insensitivity by assessing root elongation on medium supplemented with JA. None of the lines exhibited resistance to JA (data not shown).
Figure 8.
pex5-10 fertility, photorespiration, and H2O2 inactivation defects. A, Length of the fifth silique on the main stem. Error bars represent se (n ≥ 18). B, Average seed weight in mg per 100 seeds. Error bars represent se (n ≥ 4). C, Root length of 10-d-old seedlings grown on medium containing propionate or isobutyrate. Error bars represent se (n ≥ 12). D, Total chlorophyll content in mg per g fresh weight for 8-d-old seedlings and 4-week-old leaves. Error bars represent se (n ≥ 6). E, DAB staining showing the accumulation of H2O2 in 2-week-old leaves. Wt, Wild type.
pex5-10 Has Reduced Peroxisomal Metabolism
Propionate and isobutyrate catabolism happens in peroxisomes using the CHY1 β-hydroxybutyryl-CoA hydrolase enzyme. Mutants defective in CHY1 accumulate a toxic intermediate that blocks β-oxidation (Zolman et al., 2001). chy1 mutants exhibit Suc dependence and IBA/2,4-DB insensitivity (Zolman et al., 2001; Lange et al., 2004; Lucas et al., 2007), similar to other peroxisomal β-oxidation-defective mutants. In addition, the mutant lines have increased sensitivity to root elongation inhibition by propionate and isobutyrate compared with the wild type (Lucas et al., 2007). To examine if catabolism is compromised in pex5, we assayed wild-type and mutant seeds on propionate and isobutyrate. The chy1 mutant (Zolman et al., 2001) was used as a control and exhibited a high degree of root elongation inhibition from propionate and isobutyrate (Fig. 8C). In contrast, pex5-1 seedlings showed wild-type responses (Fig. 8C). The pex5-10 seedlings showed a stronger inhibition on these compounds than the wild type, although the defect was not as severe as in chy1 (Fig. 8C). Furthermore, the PEX5454 protein did not rescue this root elongation inhibition (Fig. 8C). These data suggest that these catabolism pathways are affected in the pex5-10 mutant, and this defect may be based on disruptions in PTS1 protein import.
pex5-10 seedlings had a pale green color (Fig. 5D). Leaf peroxisomes are involved in photorespiration because they metabolize a nonuseful photosynthetic side product, phosphoglycolate; Arabidopsis mutants with strong defects in photorespiration are a pale green color due to reduced chlorophyll accumulation (Reumann, 2002). We quantified total chlorophyll in both seedlings and adult leaves as an indicator of whether photorespiration was affected in the pex5 mutants. pex5-10 showed a reduction in total chlorophyll compared with the wild type (Fig. 8D). This chlorophyll defect was more severe than in the previously described pex6-1 mutant, which also has defects in PEX5 accumulation (Zolman and Bartel, 2004). The PEX5454 protein did not rescue the defect in pex5-10 seedlings (Fig. 8D), and total chlorophyll of pex5-1 seedlings was similar to that in the wild type (data not shown). These results demonstrate that enzymes in photorespiration require PTS1-mediated import, consistent with the fact that the major photorespiration enzymes each use a PTS1 (Foyer et al., 2009). Interestingly, total chlorophyll in both pex5-10 and pex5-10 (PEX5454) adult leaves were similar to the wild type, while the pex6-1 plants maintained this defect (Fig. 8D).
Peroxisomes house many metabolic processes that produce hydrogen peroxide (H2O2), which must be detoxified before accumulating to damaging levels. To examine the import of the detoxifying enzymes, we detected H2O2 accumulation in wild-type and mutant leaves using 3,3′-diaminobenzidine tetrahydrochloride (DAB; Thordal-Christensen et al., 1997). Both pex5-10 and pex5-10 (PEX5454) leaves accumulated a brown precipitate indicative of H2O2 accumulation (Fig. 8E). This result indicates that H2O2 is not efficiently inactivated in pex5-10. The PEX5454 protein only partially rescued the H2O2 inactivation defect in pex5-10 (Fig. 8E), suggesting both PTS1 and PTS2 import pathways are required for detoxification.
pex5-10 Has Developmental Defects But Normal Morphology
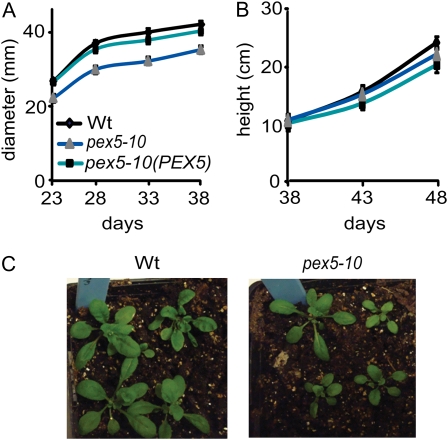
To determine the consequences of the pex5-10 insertion, we analyzed plant growth and development. We found initial issues with growth, as evidenced by low root growth and small aerial development (Fig. 5, B and D). pex5-10 plants showed reduced rosette diameter (Fig. 9, A and B). Once the plants had begun photosynthetic growth, they seemed to recover, and overall plant architecture remained fairly normal. pex5 and wild-type plants have the same numbers of stems produced, plant height, and date to bolting (Fig. 9C; data not shown).
Figure 9.
pex5-10 growth and development. A and B, Wild-type (Wt), pex5-10 (PEX5), and pex5-10 plants were grown under continuous white light at 22°C with normal watering two times per week. Plants were measured for rosette diameter (A) and plant height (B) on the indicated days. Error bars represent se (n ≥ 18). C, Photographs of 3-week-old soil-grown wild-type and pex5-10 plants.
DISCUSSION
pex5-10 is a loss-of-function mutant resulting from a T-DNA insertion in the PEX5 gene. The isolation of this mutant was unexpected; our original hypothesis was that a pex5 insertion mutant would be embryo lethal, similar to several other peroxisome-defective mutants (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003; Fan et al., 2005). When we analyzed the effect of the insertion at the genetic and protein levels, however, we found that the insertion effects were atypical. The insertion does lead to a loss of gene expression, as seen using primers in the exon in which the insertion is located (Fig. 2C). However, at some frequency, this exon and the insertion are removed in the Salk T-DNA line (Fig. 2D). This excision results in a protein with a deletion (Fig. 2, A and B); the pex5-10 mutant protein is missing four of the first five PPRs (Fig. 1A). In addition to the deletion, the mutant protein is at a reduced concentration compared with the wild-type protein (Fig. 2A). Although this excision does occur at the normal splicing boundaries, we found no evidence for this splice form in the wild type, either in the cDNA collection or in our RT-PCR trials (data not shown). In monocots, a PEX5 short form (PEX5pS) has been identified, which also is missing an exon compared with the full-length long form (PEX5pL); however, the pex5-10 removal of exon 5 does not correspond to the exon 7 deletion in rice (Oryza sativa), and the pex5-10 and PEX5pS alterations affect different domains in the protein (Lee et al., 2006).
This type of response by the plant to a T-DNA insertion is rare; a global analysis of the effects of insertions in exons revealed that only 2% of those studied resulted in truncated proteins being formed, compared with 88% of the samples with a loss of protein accumulation (Wang, 2008). However, isolation of this splicing event in the insertion mutant was fortuitous, as it allowed the study of PEX5 function when a true pex5 null likely would be embryo lethal. The mutant pex5-10 protein must retain just enough activity to maintain the mutant plant growth and development, particularly in adult plants. The defects in pex5-10 are stronger than pex5-1 but fall in a similar spectrum. This allelic spectrum allows us to draw conclusions about the function of PEX5 in plant growth and development. PEX5 is clearly required for the import of peroxisome matrix proteins, as pex5-10 showed mislocalization of both PTS1- and PTS2-containing GFP reporters (Fig. 3A), consistent with previous studies (Lee et al., 2006; Lingard et al., 2009). In addition, pex5-10 has altered localization of thiolase (Fig. 3C; Zolman et al., 2005; Lingard and Bartel, 2009; Ramón and Bartel, 2010), catalase (Lingard and Bartel, 2009; Lingard et al., 2009), isocitrate lyase (Lingard et al., 2009), malate synthase (Lingard and Bartel, 2009), and peroxisomal malate dehydrogenase (Lingard and Bartel, 2009; Ramón and Bartel, 2010). However, the pex5-10 mutant is able to correctly localize the peroxisomal membrane protein PEX14 (Lingard and Bartel, 2009), indicating that peroxisomal structures are intact and that membrane protein import occurs normally. This loss of matrix import can be seen in reduced peroxisomal matrix protein enzymatic activity (Fig. 3D). Therefore, this T-DNA mutant has enabled us to score the effects of loss of peroxisomal function, as the mutant strongly disrupts all peroxisome matrix processes by decreased import of the required proteins.
Despite the numerous functions of peroxisomes throughout the plant life cycle (Kaur et al., 2009), perhaps PEX5 is most critical in early seedlings, when peroxisomal fatty acid β-oxidation is required for development. The pex5-10 mutant has a low degree of germination (0%–20%) without assistance (Tables I and II) and a strict requirement for exogenous Suc during seedling development (Fig. 4; Lee et al., 2006, 2007; Lingard and Bartel, 2009; Ramón and Bartel, 2010). The requirement for seed coat disruption by nicking is similar to several other strong peroxisome-defective mutants (Footitt et al., 2002; Pracharoenwattana et al., 2005; Hooks et al., 2007; Kanai et al., 2010). Despite severe germination and seedling developmental phenotypes, some mutant seedlings survive and develop into adult plants. Seedlings also are unable to metabolize IBA/2,4-DB, resulting in alterations in secondary root development (Figs. 5 and 6; Lee et al., 2006, 2007; Lingard and Bartel, 2009; Ramón and Bartel, 2010). As plants develop and start photosynthesis, PEX5 decreases (Fig. 1B). This trend may explain the strong seedling defects in pex5-10 seedlings but less severe phenotypes of the adult plants (Fig. 9). Our enzymatic studies of peroxisomal processes (Fig. 3D) and examination of β-oxidation using eicosenoic acid as a marker (Fig. 4E) demonstrate the effects of decreased peroxisomal import in pex5-10 and explain the above defects in plant development. Hayashi et al. (2005) used RNA interference to decrease PEX5 levels, resulting in a dwarf phenotype and photorespiration defects. In addition, the silenced lines were Suc dependent and resistant to 2,4-DB, with defects in both PTS1 and PTS2 import (Hayashi et al., 2005). These phenotypes are similar to pex5-10. Each of these defective processes reveals the importance of PEX5 in peroxisomal function and the various roles of peroxisomes in plant growth and development.
Although pex5-1 and pex5-10 mutants allowed us to study defects in peroxisomal matrix protein import, neither was appropriate to specifically study PTS1 import. We wanted to generate a line defective specifically in PTS1 import so we could study the consequences of disruptions in PTS1 and PTS2 import separately. The pex5-10 (PEX5454) construct expresses the N-terminal region of PEX5, including the PPR domains required for binding to peroxisomal membrane proteins and the PEX7-binding domain. These domains should allow normal peroxisomal import for the PEX7-bound PTS2 proteins via PEX5. However, PEX5454 is missing the TPR domains required for binding the tripeptide PTS1 at the C terminus of the cargo protein. We crossed this construct into the pex5-1 and pex5-10 mutants. We hypothesized that PEX5454 would restore pex5-1 defects and partially complement pex5-10. Indeed, PTS2-GFP import was restored in both pex5 transgenic lines to a wild-type pattern (Fig. 3A; data not shown). However, GFP-PTS1 remained cytosolic in pex5-10 (PEX5454), similar to the mutant parent lines (Fig. 3A). Although pex5-1 and pex5-10 retain much of thiolase (a PTS2 model protein) in the unprocessed, cytosolic form, the transgenic lines exhibited increased peroxisomal import (Fig. 3C). In addition, PEX5454 restored the activity of the peroxisomally localized ACX enzymes. ACX4 has a PTS1 and is known to be the primary enzyme required for the oxidation of short-chain fatty acid substrates (Hayashi et al., 1999). The pex5 mutants cannot oxidize C6:0 substrates, a defect that is not affected by insertion of the PEX5454 construct (Fig. 3D). ACX3 is the major enzyme affecting C12:0 oxidation; this enzyme has a PTS2 (Eastmond et al., 2000; Froman et al., 2000). ACX3 activity is reduced in both mutants but is recovered partially in the transgenic lines (Fig. 3D), indicating that the PEX5454 protein is increasing PTS2 responses. Therefore, the defects in pex5-10 (PEX5454) result from a specific defect in PTS1 import.
Our examination of the pex5 and pex5 (PEX5454) lines allowed us to separate which import pathways are required for several peroxisomal processes (summarized in Supplemental Table S1). Comparison of the pex5-1 mutant and the pex5-1 (PEX5454) lines reveals a fairly complete phenotypic rescue under all conditions, consistent with the idea that the pex5-1 defect primarily disrupts PTS2 import and that PEX5454 restores PTS2 activity. In the pex5-1 background, the PEX5454-mediated recovery of PTS2 activity is evident with increased seed storage fatty acid utilization (Fig. 4E), increased development in the absence of Suc (Fig. 4B), and responses to IBA (Figs. 5A and 6A). Similarly, the pex5-10 (PEX5454) plants show improved growth and development, although the transgenic lines remain impaired. Expression of PEX5454 in pex5-10 promotes increased germination without physical stimuli compared with pex5-10, although substantial delays remain compared with the wild type (Table I). Improvements also include an increased ability to grow without Suc supplementation (Fig. 4, B–D), higher β-oxidation rates (Fig. 4E), greater responses to IBA stimuli (Figs. 5C and 6A), and an improved ability to make seeds (Fig. 8B). pex5-10 (PEX5454) plants show only a partial rescue of the pex5-10 phenotypes, consistent with the idea that PTS1 import still is disrupted in these lines. Although the import of PTS2-containing proteins restores some peroxisomal processes, most pathways require enzymes with both PTS1 and PTS2 signals.
Interestingly, two assays did not show any changes between the mutant parent and the PEX5454 transgenic line, allowing us to speculate about the types of import required. First, an inability of plants to metabolize either propionate and isobutyrate results in a hypersensitive phenotype, likely due to the accumulation of toxic intermediates in the pathway that inhibit growth (Lucas et al., 2007). pex5-10 was sensitive to both of these compounds, which was not affected by PEX5454 expression (Fig. 8C). This result suggests that the enzymes (or at least the key rate-limiting enzymes) involved in processing these metabolites have a PTS1. This mechanism is consistent with the suggested enzymatic pathways (Lucas et al., 2007). Also, photorespiration rates were essentially the same in pex5-10 and pex5-10 (PEX5454) lines (Fig. 8D). The four key peroxisomal enzymes involved in photorespiration each use PTS1 import pathways. The fact that these defects are not affected by PEX5454 expression and, therefore, may specifically require PTS1 import is further supported by the result that pex5-1 does not exhibit a defective phenotype in any of these assays. In contrast, responses to 2,4-DB (Fig. 5E) were completely rescued by PEX5454 expression, revealing that the disruption in pex5-10 may be caused by mislocalization of a key PTS2-containing protein. A recent study indicated that 2,4-DB β-oxidation to 2,4-D may occur using a distinct and/or overlapping set of enzymes as IBA β-oxidation to IAA, although AAE18 is the only protein with known differential activity and this enzyme has a PTS1 (Wiszniewski et al., 2009).
In this paper, we determined the effect of the pex5-10 mutation on PEX5 gene expression and protein accumulation. We then examined the importance and function of PEX5 in peroxisomal targeting by observing the phenotypes of pex5 mutants. The defects shown here confirm the critical role of PEX5 in peroxisomal metabolism. Additional comparisons of the three pex5 lines will be useful for enhanced studies of the protein complements of plant peroxisomes and for studies of the mechanism of plant peroxisomal matrix protein import.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) pex5-1 has been previously published with a Ser-to-Leu mutation at amino acid 318 in At5g56290 (Zolman et al., 2000). pex5-10 (SALK_124577) was obtained from the Salk collection of insertion mutants (Alonso et al., 2003) and was backcrossed at least twice prior to analyses. The mutant can be tracked experimentally using PCR amplification with a combination of genome-specific and insertion-specific primer combinations (Supplemental Table S2). chy1-3 (At5g65940; Zolman et al., 2001), pex6-1 (At1g03000; Zolman and Bartel, 2004), and aux1-7 (At2g38120; Pickett et al., 1990) were used as controls. All mutants are in the Col-0 background.
Growth Conditions and Phenotypic Assays
Seeds were surface sterilized with 30% bleach:0.1% Triton solution for 10 min and rinsed four times with sterile water. All seedling assays were done on plates wrapped with gas-permeable Micropore surgical tape, incubated at 22°C under continuous light for the indicated number of days. If plates contained auxins, yellow-filtered light was used (Stasinopoulos and Hangarter, 1990). Plants grown to maturity were placed in pots of Sun-Gro Metro Mix 360 soil and incubated at 22°C to 25°C under continuous white light.
For germination assays, 100 seeds were plated on solid plant nutrient medium (PN; Haughn and Somerville, 1986) supplemented with 0.5% Suc (PNS). F2 seeds were the second (segregating) generation of a backcross to the Col-0 parent. Seeds were incubated at 22°C under continuous white light. Plates were examined with a dissecting microscope for the number of seeds that germinated every 24 h for 4 d. Seeds that did not germinate by day 4 were transferred in a sterile manner to a fresh PNS plate, and the seed coats were nicked using a Leica Zoom 2000 dissecting microscope with sterile forceps. After 10 d, DNA was extracted from all seedlings. The genotype of each seedling was determined by PCR amplification with gene-specific PEX5 primers and T-DNA primers (Supplemental Table S1).
To assay the Suc dependence of pex5-10, seeds were plated on PN medium alone or on medium supplemented with different concentrations of Suc. After plating, seeds were nicked with sterile forceps and then incubated at 22°C under continuous white light for 24 h. Plates were transferred to the dark for 6 d and hypocotyl length was measured.
For root elongation assays, seeds were plated on PNS alone or with the indicated hormone concentrations. After plating, all pex5-10 seeds were nicked. After 10 d, seedlings were removed from plates and root length was measured.
To determine lateral root initiation, seeds were plated on PNS medium and pex5-10 seeds were nicked. After 5 d, seedlings were transferred to fresh PNS plates supplemented with or without hormone and incubated for an additional 5 d. The number of lateral roots was counted with a Leica Zoom 2000 dissecting microscope, and the root length measured. Data are represented as lateral root density or lateral root number per root length (mm).
For the adult phenotypic assays, seeds were surface sterilized and plated on solid PNS medium. pex5-10 seeds were nicked immediately after plating. Ten-day-old seedlings were transferred to soil. Plants were watered once weekly, and measurements were taken at regular intervals for rosette diameter, number of stems, and plant height. Each plant was observed daily for date of bolting. The fifth silique from the main stem was measured for each plant. The weight of 100 wild-type and mutant seeds was recorded, and photographs were taken.
RNA Isolation and RT-PCR Analysis
To examine the effects of the T-DNA insertion on PEX5 gene expression, we isolated RNA from wild-type and 3-week-old mutant plants. Total RNA was isolated with the RNeasy Plant Kit (Qiagen). RNA was treated with DNase (Fisher Scientific) to remove contaminating genomic DNA, and cDNA was synthesized with SuperScript III Reverse Transcriptase (Invitrogen). One microliter of cDNA was used for PCR amplification with gene-specific primer sets (Supplemental Table S2). For all PCR amplifications, Col-0 genomic DNA was used as a control. PCR products were resolved on a 2% agarose gel.
GFP Examination of Peroxisomal Import
To examine the import of proteins into peroxisomes, we used the GFP-PTS1 construct, which includes the SKL import signal at the C terminus of the GFP (Zolman and Bartel, 2004), and PTS2-GFP, which contains the targeting signal from the Arabidopsis PED1/KAT2 thiolase enzyme fused to the N terminus of GFP (Woodward and Bartel, 2005). The GFP parent lines in the Col-0 background were crossed to the mutant lines, and plants homozygous for the appropriate mutation and construct combinations were identified by PCR and immunoblotting. GFP analysis was done on root hair cells using a Zeiss LSM700 confocal microscope.
Protein Extraction and Immunoblot Analysis
For western-blot analysis, samples were ground on dry ice and mixed with 2× NuPAGE LDS Sample Buffer (Invitrogen). The mixture was centrifuged, and the supernatant containing the protein was collected. Protein was quantified with Bradford reagent (Sigma-Aldrich), and 40 μg of total protein was electrophoresed on NuPAGE 10% Bis-Tris gels with NuPAGE MOPS SDS Running Buffer (Invitrogen). Proteins were transferred to Hybond-ECL nitrocellulose membranes (Amersham Biosciences). Membranes were blocked for 2 h in 10% (w/v) nonfat dry milk in Tris-buffered saline plus Tween 20 buffer. Membranes were incubated with primary antibodies recognizing the C-terminal region of PEX5 (Zolman and Bartel, 2004; 1:1,000 dilution), the myc epitope (Sigma; 1:1,000), thiolase (Lingard et al., 2009; 1:5,000), or HSC70 (Stressgen Bioreagents; 1:5,000). Secondary detection involved horseradish peroxidase-linked goat anti-rabbit antibodies (PEX5 and PED1; Southern Biotech; 1:10,000) and rabbit anti-mouse antibodies (myc and HSC70; Sigma; 1:10,000). All antibodies were visualized using SuperSignal West Dura extended duration substrate (Thermo Scientific).
Fatty Acid Extraction and GC Analysis of C20:1 Content
Seeds were surface sterilized, plated on solid PNS medium covered with filter paper, and incubated for 6 d. Fatty acid extraction and esterification were done (Devaiah et al., 2007) using 50 seedlings. Prior to esterification, 50 μm heptadecanoic acid (C17:0) was added to each sample as an internal standard. Analysis was done using an automated Shimadzu gas chromatograph (Shimadzu GC-17A) equipped with a silica capillary column (DB-5MS, 30 m, 0.25 mm, 0.25 mm). The temperatures of the injector column and flame ionization detector were 220°C, 170°C, and 220°C.
DAB and DR5-GUS Staining
To visualize the accumulation of H2O2, we used DAB stain, which reacts with H2O2 to produce a brown precipitate (Thordal-Christensen et al., 1997). Briefly, leaves were collected from 2-week-old plants and vacuum infiltrated with 1 mg mL−1 DAB solubilized in Tris-acetate buffer, pH 3. Following infiltration, leaves were incubated with the stain for 24 h in the dark, then boiled in 80% ethanol to remove chlorophyll for visualization.
For DR5-GUS visualization, Col-0 and pex5-10 transgenic seedlings were stained at 37°C for 2 d in 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Bartel and Fink, 1994).
Chlorophyll Quantification
Chlorophyll extraction and quantification were performed using modifications of a previously published protocol (Wintermans and de Mots, 1965). Five milligrams of 8-d-old seedlings or 4-week-old leaves was boiled in 96% ethanol at 80°C until all the chlorophyll was removed. The chlorophyll content was measured with a spectrophotometer at 654, 665, and 649 nm, and total chlorophyll content was calculated.
PEX5454 Transgenic Plants
The 35S-PEX5 overexpression construct has been described previously (Zolman et al., 2000). pex5 mutant lines transformed with this construct can be selected by resistance to the herbicide Basta.
We created a truncated gene construct of the first 454 amino acids of the PEX5 protein. The truncated PEX5 cDNA was amplified with primers PEX5-Sal and PEX5 454 B/N (Supplemental Table S2) using Ex-Taq (TaKaRa), the PCR product was run on a 1% guanosine gel, and the DNA was purified with the Qiaex II Gel Extraction Kit (Qiagen). The purified product was ligated into a TOPO vector (Invitrogen), and the insert was verified by sequencing prior to subcloning into pBluescript KS+ vector (pKS) using SalI and NotI. For myc ligation to PEX5454, pT26 myc and pKS PEX5454 were digested with BamHI. The pT26 myc digestion product was electrophoresed, and the myc fragment was extracted. The BamHI-digested pKS PEX5454 was dephosphorylated using SAP (Promega). The myc fragment was ligated at the C terminus of the PEX5454 gene. For cloning of the PEX5454 insert (with the myc epitope) into the plant transformation vector 35S pBARN (LeClere and Bartel, 2001), pKS PEX5454 was digested with ApaI and blunt ends were created using T4 DNA polymerase (New England Biolabs) followed by digestion with NotI. 35S pBARN was digested with SmaI and NotI. The 35S-PEX5454 construct was transformed into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986) and transformed into wild-type plants (Clough and Bent, 1998). Transformants were selected by Basta resistance. The Col-0 (PEX5454) line was crossed to pex5-1 and pex5-10, which were selected first by genotyping the mutation and then by identifying lines containing the cDNA.
ACX Enzyme Assays
Col-0 and mutant seeds were surface sterilized and plated on solid PNS medium covered with filter paper. ACX assays were performed similar to previously described protocols (Hryb and Hogg, 1979; Gerhardt, 1987; Adham et al., 2005). A total of 0.3 g of 4-d-old seedlings was immersed in liquid nitrogen and ground to powder in a mortar. Ground seedlings were incubated for 15 min at 4°C in 800 μL of cold extraction buffer containing 50 mm KPO4, 50 μm FAD, and 0.01% Triton X-100 supplemented with 10 μL of protease inhibitor cocktail (Sigma). After incubation, extracts were centrifuged for 10 min at 13,200 rpm, and the supernatant was loaded on quick-spin protein columns (Roche) and processed according to the manufacturer’s instructions. For the enzymatic reaction, 200 μL of extract was added to 200 μL of reaction mix (50 mm KPO4, 0.025% Triton X-100, 50 mm p-hydroxybenzoic acid, 100 μg mg−1 bovine serum albumin, 50 μm FAD, 2 mm 4-aminoantipyrine, and 110 units of horseradish peroxidase) and 125 μm C6:0 and C12:0 substrates (Sigma). The reaction was measured spectrophotometrically at 500 nm for H2O2 production, and a standard H2O2 curve was created to calculate H2O2 concentration.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Summary of phenotypes in pex5 mutant lines.
Supplemental Table S2. Primers used in this work.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Xuemin (Sam) Wang and Amanda Tawfall in the performance of the gas chromatography analysis and Liang Guo for assistance making the PEX5 deletion constructs. We thank the Arabidopsis Biological Resource Center at Ohio State University for seeds, the Salk Institute Genomic Analysis Laboratory for generating the sequence-indexed Arabidopsis T-DNA insertion mutants, and Bonnie Bartel for the thiolase antibody. We are grateful to Robert Barlow, Shelly Boyer, Lisa Schechter, and Gretchen Spiess for critical comments on the manuscript.
References
- Adham AR, Zolman BK, Millius A, Bartel B. (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β-oxidation. Plant J 41: 859–874 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR. (1994) Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA 91: 6649–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Costa-Rodrigues J, Carvalho AF, Gouveia AM, Fransen M, Sá-Miranda C, Azevedo JE. (2004) The N terminus of the peroxisomal cycling receptor, Pex5p, is required for redirecting the peroxisome-associated peroxin back to the cytosol. J Biol Chem 279: 46573–46579 [DOI] [PubMed] [Google Scholar]
- Dammai V, Subramani S. (2001) The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105: 187–196 [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Pan X, Hong Y, Roth M, Welti R, Wang X. (2007) Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. Plant J 50: 950–957 [DOI] [PubMed] [Google Scholar]
- Dodt G, Braverman N, Wong C, Moser AB, Moser HW, Watkins PA, Valle D, Gould SJ. (1995) Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet 9: 115–125 [DOI] [PubMed] [Google Scholar]
- Dodt G, Gould SJ. (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol 135: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Hooks MA, Williams D, Lange P, Bechtold N, Sarrobert C, Nussaume L, Graham IA. (2000) Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. J Biol Chem 275: 34375–34381 [DOI] [PubMed] [Google Scholar]
- Fan J, Quan S, Orth T, Awai C, Chory J, Hu J. (2005) The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol 139: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M. (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21: 2912–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G. (2009) Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Froman BE, Edwards PC, Bursch AG, Dehesh K. (2000) ACX3, a novel medium-chain acyl-coenzyme A oxidase from Arabidopsis. Plant Physiol 123: 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto GJJ, Jr, Geisbrecht BV, Gould SJ, Berg JM. (2000) A proposed model for the PEX5-peroxisomal targeting signal-1 recognition complex. Proteins 38: 241–246 [DOI] [PubMed] [Google Scholar]
- Geisbrecht BV, Collins CS, Reuber BE, Gould SJ. (1998) Disruption of a PEX1-PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc Natl Acad Sci USA 95: 8630–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt B. (1987) Peroxisomes and fatty acid degradation. Methods Enzymol 148: 516–525 [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Graham IA, Leaver CJ, Smith SM. (1992) The induction of malate synthase gene expression in senescent and detached organs of cucumber. Plant Cell 4: 349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ. (1999) Auxin-regulated genes and promoters. Hooykaas PJJ, Hall MA, Libbenga KR, eds, Biochemistry and Molecular Biology of Plant Hormones Elsevier, Amsterdam, pp 423–459 [Google Scholar]
- Haughn GW, Somerville CR. (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Hayashi H, De Bellis L, Ciurli A, Kondo M, Hayashi M, Nishimura M. (1999) A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J Biol Chem 274: 12715–12721 [DOI] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Yamaguchi K, Kato A, Hayashi M, Nishimura M. (1998) Molecular characterization of a glyoxysomal long chain acyl-CoA oxidase that is synthesized as a precursor of higher molecular mass in pumpkin. J Biol Chem 273: 8301–8307 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Toriyama-Kato K, Kondo M, Yamaya T, Nishimura M. (2000) AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J 19: 5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M. (2005) Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J Biol Chem 280: 14829–14835 [DOI] [PubMed] [Google Scholar]
- Helm M, Lück C, Prestele J, Hierl G, Huesgen PF, Fröhlich T, Arnold GJ, Adamska I, Görg A, Lottspeich F, et al. (2007) Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA 104: 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M, Ishii N, Kagamiyama H, Osumi T, Hashimoto T. (1987) Structural analysis of cDNA for rat peroxisomal 3-ketoacyl-CoA thiolase. J Biol Chem 262: 8151–8158 [PubMed] [Google Scholar]
- Hooks MA, Turner JE, Murphy EC, Johnston KA, Burr S, Jarosławski S. (2007) The Arabidopsis ALDP protein homologue COMATOSE is instrumental in peroxisomal acetate metabolism. Biochem J 406: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hryb DJ, Hogg JF. (1979) Chain length specificities of peroxisomal and mitochondrial beta-oxidation in rat liver. Biochem Biophys Res Commun 87: 1200–1206 [DOI] [PubMed] [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J. (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405–409 [DOI] [PubMed] [Google Scholar]
- Ibdah M, Pichersky E. (2009) Arabidopsis Chy1 null mutants are deficient in benzoic acid-containing glucosinolates in the seeds. Plant Biol (Stuttg) 11: 574–581 [DOI] [PubMed] [Google Scholar]
- Kamada T, Nito K, Hayashi H, Mano S, Hayashi M, Nishimura M. (2003) Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol 44: 1275–1289 [DOI] [PubMed] [Google Scholar]
- Kanai M, Nishimura M, Hayashi M. (2010) A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J 62: 936–947 [DOI] [PubMed] [Google Scholar]
- Kaur N, Reumann S, Hu J. (2009) Peroxisome biogenesis and function. Somerville CR, Meyerowitz EM, eds, The Arabidopsis Book American Society of Plant Biologists, Rockville, MD, pp 1–41 [Google Scholar]
- Kiel JAKW, Hilbrands RE, van der Klei IJ, Rasmussen SW, Salomons FA, van der Heide M, Faber KN, Cregg JM, Veenhuis M. (1999) Hansenula polymorpha Pex1p and Pex6p are peroxisome-associated AAA proteins that functionally and physically interact. Yeast 15: 1059–1078 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kunz HH, Scharnewski M, Feussner K, Feussner I, Flügge UI, Fulda M, Gierth M. (2009) The ABC transporter PXA1 and peroxisomal β-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21: 2733–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange PR, Eastmond PJ, Madagan K, Graham IA. (2004) An Arabidopsis mutant disrupted in valine catabolism is also compromised in peroxisomal fatty acid beta-oxidation. FEBS Lett 571: 147–153 [DOI] [PubMed] [Google Scholar]
- LeClere S, Bartel B. (2001) A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol 46: 695–703 [DOI] [PubMed] [Google Scholar]
- Lee JR, Jang HH, Park JH, Jung JH, Lee SS, Park SK, Chi YH, Moon JC, Lee YM, Kim SY, et al. (2006) Cloning of two splice variants of the rice PTS1 receptor, OsPex5pL and OsPex5pS, and their functional characterization using pex5-deficient yeast and Arabidopsis. Plant J 47: 457–466 [DOI] [PubMed] [Google Scholar]
- Lee JR, Park SC, Kim MH, Jung JH, Shin MR, Lee DH, Cheon MG, Park Y, Hahm KS, Lee SY. (2007) Antifungal activity of rice Pex5p, a receptor for peroxisomal matrix proteins. Biochem Biophys Res Commun 359: 941–946 [DOI] [PubMed] [Google Scholar]
- Lemieux B, Miquel M, Somerville C, Browse J. (1990) Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor Appl Genet 80: 234–240 [DOI] [PubMed] [Google Scholar]
- Lingard MJ, Bartel B. (2009) Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol 151: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Monroe-Augustus M, Bartel B. (2009) Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA 106: 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas KA, Filley JR, Erb JM, Graybill ER, Hawes JW. (2007) Peroxisomal metabolism of propionic acid and isobutyric acid in plants. J Biol Chem 282: 24980–24989 [DOI] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Nito K, Kondo M, Nishimura M. (2006) The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J 47: 604–618 [DOI] [PubMed] [Google Scholar]
- Nair DM, Purdue PE, Lazarow PB. (2004) Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J Cell Biol 167: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K, Hayashi M, Nishimura M. (2002) Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol 43: 355–366 [DOI] [PubMed] [Google Scholar]
- Pickett FB, Wilson AK, Estelle M. (1990) The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol 94: 1462–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, El Magraoui F, Bäumer BE, Schlee D, Girzalsky W, Erdmann R. (2009) Pex2 and Pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol 29: 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17: 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón NM, Bartel B. (2010) Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol Biol Cell 21: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S. (2002) The photorespiratory pathway of leaf peroxisomes. Baker A, Graham IA, eds, Plant Peroxisomes Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 141–190 [Google Scholar]
- Reumann S. (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 135: 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O. (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Ma C, Lemke S, Babujee L. (2004) AraPerox: a database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol 136: 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA. (2003) Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development. J Biol Chem 278: 21370–21377 [DOI] [PubMed] [Google Scholar]
- Schuhmann H, Huesgen PF, Gietl C, Adamska I. (2008) The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol 148: 1847–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C. (2003) AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA 100: 9626–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A. (2003) An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol 133: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP. (1990) Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Culler AH, Cohen JD, Bartel B. (2010) Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol 153: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlecky SR, Nuttley WM, McCollum D, Sock E, Subramani S. (1995) The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO J 14: 3627–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Wang YH. (2008) How effective is T-DNA insertional mutagenesis in Arabidopsis? Biochem Tech 1: 11–20 [Google Scholar]
- Wanner L, Keller F, Matile PH. (1991) Metabolism of radiolabelled galactolipids in senescent barley leaves. Plant Sci 78: 199–206 [Google Scholar]
- Wasternack C, Kombrink E. (2010) Jasmonates: structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem Biol 5: 63–77 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermans JF, de Mots A. (1965) Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim Biophys Acta 109: 448–453 [DOI] [PubMed] [Google Scholar]
- Wiszniewski AA, Zhou W, Smith SM, Bussell JD. (2009) Identification of two Arabidopsis genes encoding a peroxisomal oxidoreductase-like protein and an acyl-CoA synthetase-like protein that are required for responses to pro-auxins. Plant Mol Biol 69: 503–515 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B. (2004) An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. (2005) Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SPT, Bartel B. (2001) chy1, an Arabidopsis mutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl-CoA hydrolase. J Biol Chem 276: 31037–31046 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.