Abstract
Understanding the genetic basis of nitrogen and carbon metabolism will accelerate the development of plant varieties with high yield and improved nitrogen use efficiency. A robotized platform was used to measure the activities of 10 enzymes from carbon and nitrogen metabolism in the maize (Zea mays) intermated B73 × Mo17 mapping population, which provides almost a 4-fold increase in genetic map distance compared with conventional mapping populations. Seedling/juvenile biomass was included to identify its genetic factors and relationships with enzyme activities. All 10 enzymes showed heritable variation in activity. There were strong positive correlations between activities of different enzymes, indicating that they are coregulated. Negative correlations were detected between biomass and the activity of six enzymes. In total, 73 significant quantitative trait loci (QTL) were found that influence the activity of these 10 enzymes and eight QTL that influence biomass. While some QTL were shared by different enzymes or biomass, we critically evaluated the probability that this may be fortuitous. All enzyme activity QTL were in trans to the known genomic locations of structural genes, except for single cis-QTL for nitrate reductase, Glu dehydrogenase, and shikimate dehydrogenase; the low frequency and low additive magnitude compared with trans-QTL indicate that cis-regulation is relatively unimportant versus trans-regulation. Two-gene epistatic interactions were identified for eight enzymes and for biomass, with three epistatic QTL being shared by two other traits; however, epistasis explained on average only 2.8% of the genetic variance. Overall, this study identifies more QTL at a higher resolution than previous studies of genetic variation in metabolism.
Plant growth and development are largely dependent on nitrogen and carbon metabolism. Carbon metabolism produces storage carbohydrates, provides accessible energy, and creates primary building blocks for other metabolism, including phosphorylated intermediates and organic acids that provide the carbon skeletons for the assimilation of nitrate and ammonium and amino acid synthesis. Certain enzymes have been shown to play key roles in those reactions (Nunes-Nesi et al., 2010; Stitt et al., 2010), among which are ADP-Glc pyrophosphorylase (AGPase; EC 2.7.7.27) for starch synthesis (Preiss, 1982; Neuhaus and Stitt, 1990), Fru-bisP aldolase (ALD; EC 4.1.2.13; Haake et al., 1999) and phosphoglucomutase (PGM; EC 5.4.2.2; Gibon et al., 2004b) in glycolysis, and citrate synthase (CS; EC 2.3.3.1; Sienkiewicz-Porzucek et al., 2008) in the tricarboxylic acid cycle.
Nitrogen is an important constituent of many biomolecules in plants and is a limiting factor affecting yield in a variety of agricultural systems. Following its uptake from soil via specific transporters, nitrate is reduced to ammonium by the concerted action of nitrate reductase (NR; EC 1.7.1.1) and nitrite reductase (EC 1.7.7.1). Ammonium then enters the Gln synthetase (GS; EC 6.3.1.2) and Glu synthase (EC 1.4.7.1) cycles, where it is converted to Gln and Glu (Andrews et al., 2004). The amino group of Glu can be transferred to amino acids by a number of different aminotransferases (Lam et al., 1996), such as Asp aminotransferase (AspAT; EC 2.6.1.1) and Ala aminotransferase (AlaAT; EC 2.6.1.2). AspAT and AlaAT catalyze the reversible transfer of the amino group from the second position of Glu to oxaloacetate and pyruvate, respectively, to yield 2-oxoglutarate and Asp and Ala, respectively. AspAT has been proposed to play several metabolic roles, including providing precursors for the biosynthesis of the Asp family of amino acids (Sentoku et al., 2000). Recently, it was reported that transgenic Brassica napus and rice (Oryza sativa) plants overexpressing a barley (Hordeum vulgare) AlaAT cDNA showed increased biomass and seed yield (Good et al., 2007; Shrawat et al., 2008). NAD-Glu dehydrogenase (GDH; EC 1.4.1.2) also catalyzes a reversible reaction, but it is thought to oxidize Glu into 2-oxoglutarate in vivo (Masclaux-Daubresse et al., 2006; Forde and Lea, 2007). Although its role is not fully understood, GDH has been shown to play an important regulatory function in carbon and nitrogen metabolism (Robinson et al., 1991) and to be of major importance in the control of plant growth and productivity (Dubois et al., 2003). Shikimate dehydrogenase (SDH; EC 1.1.1.25) catalyzes the reduction of 3-dehydroshikimate by NADPH to shikimate. The shikimate pathway leads to the three aromatic amino acids Phe, Tyr, and Trp. They are important precursors for auxin-type plant hormones and various secondary compounds, including phenylpropanoids (Strack, 1997).
Despite the importance of carbon and nitrogen metabolism, little is known about the genetic variation of activities of enzymes involved in the pathways. Quantitative trait loci (QTL) mapping can provide information on the chromosomal locations of unknown genes that influence the quantitative variation of complex traits (Mitchell-Olds and Pedersen, 1998). To date, some studies have been published on mapping QTL for enzyme activities in maize (Zea mays) with recombinant inbred lines (RILs) from a cross between the northern Flint line F2 and Iodent line Io (Causse et al., 1995; Hirel et al., 2001; Limami et al., 2002; Thévenot et al., 2005). In one of these, GS and NR QTL were found on the same chromosomal positions and GS and NR activities correlated with grain yield (Hirel et al., 2001). Another study detected a coincidence of QTL for germination efficiency and its components with genes encoding cytosolic GS and the corresponding enzyme activity (Limami et al., 2002). Colocations between activity QTL and a structural locus were observed for Suc phosphate synthase, soluble acid invertase, and AGPase (Causse et al., 1995; Prioul et al., 1999; Thévenot et al., 2005). In a study of 10 glycolytic enzymes in Arabidopsis (Arabidopsis thaliana), Mitchell-Olds and Pedersen (1998) found significant activity QTL for most of the enzymes, and three QTL mapped very close to known enzyme-encoding loci (e.g. hexokinase, phosphoglucose isomerase, and PGM). Moreover, they found a QTL that may be a joint regulator of Glc-6-P dehydrogenase, phosphoglucose isomerase, and Glc-6-phosphatase. Recently, Keurentjes et al. (2008) investigated transcript QTL, enzyme activity QTL, and metabolite QTL in an Arabidopsis Landsberg erecta × Cape Verde Islands RIL population and identified examples where the structural gene colocated with transcript QTL and enzyme activity QTL, and other cases where the enzyme activity QTL was unrelated to the position of the structural gene and the genetic variation in the encoding transcript.
However, most QTL mapping resources are derived from F2 populations, in which gametes have undergone only a single cycle of recombination. Due to limited opportunities for recombination, the relatively small size of many mapping populations, and insufficient marker density, these QTL mapping efforts have been rather imprecise. Because the QTL delimit large genomic regions, considerable effort will be needed to identify the causal genes and polymorphisms. Furthermore, colocation of QTL and candidate genes (e.g. structural genes for a specific enzyme) and colocation of QTL for different metabolic and structural traits are likely to be fortuitous, which decreases the usefulness of such data for the identification of coregulated traits or the analysis of epistasis.
Studies in the outcrossing species maize (Beavis et al., 1992) and the self-fertilizing species Arabidopsis (Liu et al., 1996) demonstrate that the genetic resolution can be improved significantly by providing additional opportunities for recombination prior to the development of the mapping progeny. In maize, the intermated B73 × Mo17 (IBM) population has been developed by randomly intermating plants for four generations following the F2 generation, prior to the derivation of mapping progeny (Lee et al., 2002). The increased opportunity for recombination in IBM has resulted in an almost 4-fold increase in the genetic map distance compared with conventional nonintermated RIL populations, allowing more precise definition of QTL (Balint-Kurti et al., 2007). Recently, the Maize Genome Sequencing Consortium published the B73 Reference Genome Sequence (Schnable et al., 2009) and generated a golden path (AGP) of chromosome-based pseudomolecules, referred to as the B73 Reference Genome Sequence version 1 (Wei et al., 2009). Consequently, we are able to use the QTL-flanking marker information to identify the genetic and physical locations of QTL derived from B73-involved mapping populations, such as the IBM population. Moreover, maize is the most diverse model crop species, with the genetic diversity between any two maize lines being similar to the divergence between humans and chimpanzees (Buckler et al., 2006).
Thus, maize provides an ideal system to study the genetics of carbon and nitrogen metabolism. In this project, we use the maize IBM population to study genetic variation for activities of four enzymes from carbon metabolism and six enzymes from nitrogen metabolism as well as seedling/juvenile biomass. Enzyme activity traits may be genetically simpler than other complex quantitative traits (Mitchell-Olds and Pedersen, 1998). The genes controlling enzyme activities will belong to either structural genes, such as enzyme-encoding loci, or regulatory genes, such as loci that influence the expression of structural genes (Holton and Cornish, 1995; Mitchell-Olds and Pedersen, 1998). Maize IBM mapping, therefore, could provide a novel opportunity to gain new insights into the genetic variance of carbon and nitrogen metabolism and its relationship with plant growth.
RESULTS
Quantitative Variation of Enzyme Activities and Seedling Biomass
Enzyme activities were measured in a robotized platform, using optimized assays with saturating substrate concentrations. In a given genotype, Piques et al. (2009) showed that the measured enzyme activities highly correlate with the protein abundance measured by mass spectrometry for Arabidopsis. In this study, we did not measure individual protein abundances, so we used leaf mass to calculate enzyme activities, whose unit is expressed as nanomoles of substrate converted per gram of fresh maize leaf tissue per minute. Across a set of genotypes, the measured activities will be indicative of changes in the enzyme concentration and/or enzyme properties resulting from nonsynonymous polymorphisms in members of the gene family that encodes a particular enzymatic activity. All 10 enzyme activities and biomass showed quantitative genetic variation in the IBM intermated recombinant inbred line (IRIL) population (Table I). There were a wide range of values for all measured traits among the IRILs. Significant differences among the IRILs for all the traits were detected at the 5% level using ANOVA, although GS (P = 0.038) did not pass the sequential Bonferroni test (Holm, 1979). Among the 10 enzymes measured, the IRIL population displayed a 1- to 2-fold difference between the high and low lines for four enzymes, namely NR, AlaAT, ALD, and PGM, while it was only 13% for GS. Broad-sense heritability (H2) was 0.65, on average. For enzyme activities, GDH and PGM were the highest, H2 = 0.82, while GS was the lowest, H2 = 0.20. For biomass, H2 was determined to be 0.87.
Table I. Means for enzyme activities and biomass.
| Trait | B73 | Mo17 | IRI Lines |
Genetic Effect P | H2 | ||
| Mean | Minimum | Maximum | |||||
| Biomass | 376a | 282 | 363 | 204 | 451 | 2.2E-18b | 0.87 |
| NR | 66 | 98 | 74 | 43 | 126 | 4.6E-14 | 0.74 |
| SDH | 307 | 343 | 353 | 289 | 444 | 9.9E-07 | 0.77 |
| AlaAT | 1,940 | 2,379 | 2,139 | 1,162 | 3,264 | 1.8E-06 | 0.76 |
| GDH | 785 | 736 | 762 | 604 | 963 | 1.3E-05 | 0.82 |
| PGM | 7,289 | 7,079 | 7,009 | 4,556 | 10,934 | 0.0017 | 0.82 |
| ALD | 12,222 | 9,971 | 10,292 | 6,433 | 18,226 | 0.0020 | 0.68 |
| CS | 242 | 217 | 216 | 178 | 259 | 0.0028 | 0.62 |
| AspAT | 11,815 | 14,761 | 13,721 | 11,013 | 18,994 | 0.0036 | 0.47 |
| AGPase | 3,710 | 4,073 | 3,964 | 3,495 | 4,466 | 0.0051 | 0.42 |
| GS | 133 | 143 | 132 | 125 | 141 | 0.0380 | 0.20 |
The enzyme activity unit is expressed as nanomoles of substrate converted per gram of fresh maize leaf tissue per minute; the biomass unit is grams.
Those tests with P ≤ 0.025 passed the sequential Bonferroni test at α = 0.05.
Correlations among Enzyme Activities and Seedling Biomass
We examined patterns of correlations among activity levels of pairs of enzymes and between biomass and enzymes (Table II). For the 10 enzymes measured, 58% (26 out of 45) of the pairwise correlations were significantly different from zero; moreover, 29% (13 out of 45) passed the sequential Bonferroni test at α = 0.05. The number of significant correlations to other enzymes was between eight (ALD and PGM) and three (GDH, AspAT, and CS). All significant correlations among pairs of enzymes were positive. The positive correlations found between most enzyme activities appear to suggest some sort of coregulation acting on these enzymes.
Table II. Pearson correlations among enzyme activity levels and biomass.
Asterisks indicate significance as follows: * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
| Trait | GDH | NR | GS | AlaAT | AspAT | SDH | CS | AGPase | ALD | PGM |
| NR | −0.05 | |||||||||
| GS | 0.03 | 0.13 | ||||||||
| AlaAT | 0.05 | 0.55****a | 0.28** | |||||||
| AspAT | 0.16 | 0.12 | 0.13 | 0.12 | ||||||
| SDH | 0.11 | 0.19 | 0.12 | 0.21* | 0.17 | |||||
| CS | 0.26* | 0.11 | 0.23* | 0.28** | 0.12 | −0.02 | ||||
| AGPase | 0.20 | 0.43**** | 0.28** | 0.45**** | 0.30** | 0.38*** | 0.17 | |||
| ALD | 0.36*** | 0.26* | 0.32** | 0.28** | 0.36*** | 0.29** | 0.15 | 0.48**** | ||
| PGM | 0.34*** | 0.26* | 0.26* | 0.50**** | 0.42**** | 0.22* | 0.17 | 0.51**** | 0.70**** | |
| Biomass | −0.05 | −0.21* | −0.31** | −0.27* | −0.11 | −0.32** | −0.09 | −0.36*** | −0.24* | −0.19 |
Those tests with |r|> 0.31 passed the sequential Bonferroni test at α = 0.05 and are shown in boldface.
Juvenile/seedling biomass was correlated to six out of the 10 enzymes, NR, GS, AlaAT, SDH, AGPase, and ALD; moreover, the correlation to GS, SDH, and AGPase also passed the sequential Bonferroni test at α = 0.05 (Table II). All of these biomass-enzyme correlations were negative. The six enzymes would account for 22.1% of the biomass variation, and three of them on their own (GS, SDH, and AGPase) could explain 21.5% of the biomass variation.
To identify if the positive correlations among the 10 enzymes resulted from the correlations between the biomass and the enzymes, we performed a partial correlation analysis among the 10 enzymes that accounted for the biomass effect. Plotting the original Pearson correlation coefficients against the corresponding partial correlation coefficients revealed a very tight correlation (r = 0.99). This result shows that the positive correlation did not result from variation in sample preparation, which is consistent with Mitchell-Olds and Pedersen (1998).
We performed principal component analysis to determine a possible common factor that explains the observed correlations. The first principal component (PC1) explained 32% of the variation for the activity levels of 10 enzymes and biomass. The remaining variance spread over the remaining different component vectors, which individually explained only a small part of the total variation (Supplemental Fig. S1A). Hence, PC1 captured the joint activity signal information and was used as a new trait to conduct QTL mapping. In PC1, enzyme activities showed positive values, while biomass showed a negative value (Supplemental Fig. S1B). This is in line with the observed correlations between these traits (Table II). Among the 10 enzymes, CS and PGM have the lowest and highest weighting in PC1, respectively.
QTL for Enzyme Activities and Seedling Biomass
Composite interval mapping (CIM; Jansen and Stam, 1994; Zeng, 1994) detected 73 QTL for activities of the 10 enzymes and eight QTL for biomass (Table III). The number of QTL per enzyme ranged from six to eight, with an average of 7.3 QTL per enzyme. The r2 for the individual QTL ranged from 0.034 for QTL42 to 0.242 for QTL17. The total summed r2 for a given enzyme was over 0.5 for eight of the 10 enzymes and ranged from 0.395 for AGPase to 0.622 for AlaAT (Table III), roughly 83% of the heritability. For the eight biomass QTL, the total r2 was 0.548, with individual QTL contributions (r2) ranging from 0.042 for QTL74 to 0.169 for QTL80 of the total variance. The size of the genetic map interval for enzyme activity QTL ranged from 0.8 centimorgan (cM) for QTL47 to 20.3 cM for QTL55, and that for biomass QTL ranged from 2.4 cM for QTL80 to 14.8 cM for QTL77 (Table III). Median resolution for the QTL interval was 5.6 cM, or about 1 cM on a regular F2 map; in terms of physical position, median resolution was 1.8 Mb or about 26 genes (note that there is a large variance of the number of genes per megabase sequence throughout the maize genome). The best parental allele changed from one QTL to the other for all the traits (Table III). QTL for activities of enzymes and biomass spread all over the 10 chromosomes.
Table III. Enzyme activity, biomass, and PC1 QTL.
| QTL | Chromosome | Flanking Markersa | Intervalb | AGP Coordinate Intervalc | LOD | r2 | Allele Effect |
| % | |||||||
| GDH | |||||||
| 1 | 1 | bnl5.59a-php20682 | 522.2–524.4 | 183,652,506–184,708,080 | 4.55 | 6.5 | −18.91 |
| 2d | 1 | umc1553-bon110 | 1,008.1–1,014.2 | 285,915,476–287,632,595 | 4.37 | 6.4 | −17.85 |
| 3 | 4 | bnlg490-agrr301 | 239.3–245.5 | 31,198,257–32,104,770 | 8.84 | 19.2 | 30.78 |
| 4 | 4 | chb102-mbd116 | 612.8–613.8 | 234,931,379–237,336,780 | 2.99 | 4.1 | 15.41 |
| 5 | 5 | ago108-umc2293 | 187.6–193.9 | 13,528,340–14,981,920 | 3.26 | 4.4 | −14.72 |
| 6 | 6 | uaz121a-rz444d | 366.4–374.3 | 151,057,971–152,276,209 | 8.05 | 12.8 | −24.55 |
| 7 | 7 | bnlg1070-npi394 | 322.7–329.8 | 127,401,766–131,675,168 | 4.60 | 9.8 | 21.58 |
| 8 | 9 | psr160d-chr120 | 217.7–221.4 | 25,731,588–31,827,160 | 7.73 | 11.9 | −25.42 |
| Totale | 57.6 | ||||||
| NR | |||||||
| 9 | 1 | lim78-mmp165 | 933.6–935.7 | 274,753,840–277,362,236 | 5.46 | 8.9 | −11.50 |
| 10 | 1 | mmp195g-npi238 | 1k001.3–1k003.8 | 285,765,172–28,5915,476 | 3.70 | 5.5 | 4.70 |
| 11 | 3 | umc1030-umc2000 | 161.5–166.2 | 14,687,115–17,051,214 | 5.29 | 8.5 | 7.16 |
| 12 | 4 | mmp115-bnl5.24b | 421.2–430.1 | 174,462,939–177,509,810 | 3.03 | 5.1 | −4.24 |
| 13 | 5 | nfe101-mmp47 | 395.9–401.1 | 172,689,557–174,848,983 | 6.08 | 9.3 | −5.71 |
| 14 | 6 | AW036917-bnlg1732 | 354.3–370.9 | 149,647,086–151,962,415 | 5.69 | 9.2 | −5.56 |
| 15 | 7 | bnlg434-mbd108 | 326.1–336.6 | 128,404,335–134,999,219 | 3.10 | 5.3 | −4.38 |
| Total | 57.3 | ||||||
| GS | |||||||
| 16 | 1 | csu374b-AY111834 | 643.6–656.7 | 202,618,140–206,940,320 | 6.14 | 11.9 | 1.12 |
| 17 | 1 | umc1147-bnlg1564 | 715.1–718.3 | 224,358,016–226,272,023 | 10.74 | 24.2 | −1.69 |
| 18 | 1 | lim78-mmp165 | 932.1–936.6 | 274,753,840–277,362,236 | 7.42 | 16.2 | −1.46 |
| 19 | 2 | AY109592-bnlg469b | 626.2–643.8 | 222,216,549–229,414,867 | 3.56 | 7.0 | 0.88 |
| 20 | 4 | umc169-cat3 | 736–744.1 | 243,667,899–245,611,438 | 2.76 | 5.1 | 0.71 |
| 21 | 5 | AY111142-umc1705 | 235.6–240.4 | 28,707,869–30,962,415 | 5.21 | 11.0 | 1.04 |
| 22 | 5 | umc1822-ufg18 | 396.9–404 | 174,799,585–174,962,356 | 3.14 | 6.0 | 0.81 |
| 23 | 7 | AY110439-csu8 | 473-475.4 | 157,530,949–157,928,364 | 3.17 | 5.7 | −0.77 |
| AlaAT | |||||||
| 24 | 1 | AY109096-umc1744 | 1,038.5–1,048.4 | 291,034,442–291,738,717 | 5.52 | 12.1 | 158.01 |
| 25 | 2 | bnlg1940-gpm16 | 577.6–581.7 | 216,054,373–216,832,936 | 2.69 | 4.0 | 93.60 |
| 26 | 3 | umc1030-umc2000 | 161.5–165.4 | 14,687,115–17,051,214 | 7.29 | 12.3 | 170.51 |
| 27 | 4 | AY110310-rz567b(klc) | 363–370.3 | 156,689,269–158,606,432 | 6.29 | 10.4 | −149.54 |
| 28 | 5 | umc2301-BE639933 | 316.1–319.8 | 92,564,391–136,661,511 | 4.47 | 7.0 | 131.33 |
| 29 | 9 | chr113-AY103622 | 103.8–112.1 | 13,331,063–14,210,373 | 2.66 | 4.0 | 91.21 |
| 30 | 9 | umc1714-brd102 | 567.3–573.4 | 146,862,896–147,007,976 | 4.69 | 10.0 | 141.99 |
| 31 | 9 | dmt103a-bnlg1129 | 626.3–633.4 | 149,505,376–149,617,582 | 5.46 | 9.7 | 151.04 |
| Total | 62.2 | ||||||
| AspAT | |||||||
| 32 | 2 | ufg55-umc1696 | 711.7–715.3 | 231,339,404–234,076,935 | 2.60 | 5.0 | 366.99 |
| 33 | 3 | bnlg1144-AY109549 | 77.3–91.4 | 5,150,373–7,409,294 | 4.97 | 16.0 | −657.95 |
| 34 | 3 | mmp79-mmp186 | 139.3–144.8 | 11,850,113–12,284,825 | 3.87 | 10.1 | 549.45 |
| 35 | 3 | lim424-AY111125 | 503–505.9 | 188,168,358–188,830,505 | 3.34 | 6.4 | −420.03 |
| 36 | 4 | asg33-umc1667 | 439.6–447.2 | 177,509,810–180,074,862 | 5.83 | 12.0 | 566.62 |
| 37 | 8 | umc2154-AY110056 | 302–310.3 | 106,931,390–111,334,026 | 5.18 | 11.6 | 587.04 |
| 38 | 9 | umc1921-AW257883 | 250.8–253.7 | 92,862,141–96,413,788 | 5.50 | 12.1 | −579.38 |
| Total | 54.7 | ||||||
| SDH | |||||||
| 39 | 1 | mmp93-umc2224 | 104.4–110.4 | 10,195,439–12,125,507 | 7.49 | 10.7 | −12.22 |
| 40 | 2 | AY110485-bnlg1018 | 293.2–294.2 | 41,764,243–42,358,008 | 8.11 | 12.4 | 14.23 |
| 41 | 6 | umc1653-chr118 | 536–537.9 | 166,377,513–167,373,672 | 8.12 | 12.0 | −14.22 |
| 42 | 8 | umc89a-umc1889 | 369.7–374.9 | 132,646,661–133,582,418 | 2.60 | 3.4 | 7.51 |
| 43 | 8 | phi233376-umc1916 | 614.8–625.3 | 172,506,802–174,104,145 | 3.80 | 4.9 | −9.40 |
| 44 | 10 | php06005-bnlg210 | 178.6–182.7 | 23,420,566–26,790,198 | 9.21 | 20.0 | 17.16 |
| 45 | 10 | jpsb527c-AY110248 | 198.5–200.4 | 62,094,704–66,490,944 | 13.42 | 22.5 | 17.95 |
| 46 | 10 | mmp63-bnlg1079 | 205.8–211.1 | 70,076,248–77,021,285 | 9.86 | 18.0 | 16.03 |
| CS | |||||||
| 47 | 1 | umc1590-AY110566 | 517.2–518 | 182,674,328–183,625,873 | 9.27 | 21.5 | 8.94 |
| 48 | 1 | asg62-umc2239 | 612.9–624.7 | 198,751,409–201,314,250 | 4.16 | 10.6 | −6.02 |
| 49 | 2 | b1-psr901 | 201.9–206.8 | 19,108,477–20,722,480 | 4.89 | 10.2 | −6.35 |
| 50 | 5 | umc1597-psr544 | 217.8–227.2 | 19,559,027–23,223,419 | 3.06 | 7.3 | 4.98 |
| 51 | 6 | nfa102-mlg3 | 447.9–452.4 | 161,776,932–162,048,016 | 3.53 | 7.0 | −4.84 |
| 52 | 7 | umc1450- bnlg434 | 315.9–323.3 | 126,449,934–127,869,820 | 3.54 | 7.0 | 4.97 |
| 53 | 9 | jpsb596-mmp171a | 571.7–576.2 | 146,862,896–147,007,976 | 5.50 | 10.9 | 6.41 |
| Total | 52.9 | ||||||
| AGPase | |||||||
| 54 | 2 | umc1165-umc1542 | 48.5–57.6 | 4,135,834–4,719,432 | 2.64 | 4.9 | 53.78 |
| 55 | 3 | AY109934-umc2174 | 646.4–666.7 | 212,041,263–214,728,696 | 2.64 | 5.1 | 54.14 |
| 56 | 5 | ufg25-rz474a(dnaj) | 192.6–203.7 | 14,550,787–16,208,692 | 4.27 | 8.9 | 73.72 |
| 57 | 5 | bnlg1902-umc2298 | 301.4–307.7 | 79,724,746–84,076,624 | 5.88 | 14.0 | 106.77 |
| 58 | 6 | AY104289-bnlg1740 | 501.9–510.3 | 163,933,189–164,778,873 | 5.08 | 9.4 | −74.41 |
| 59 | 8 | umc1889-hda103 | 375.9–378.7 | 133,582,418–137,623,043 | 9.71 | 20.3 | 252.45 |
| 60 | 8 | mmc0181-npi268a | 453.9–459 | 162,848,464–164,377,373 | 7.04 | 13.7 | −92.27 |
| Total | 39.5 | ||||||
| ALD | |||||||
| 61 | 5 | mmp47-umc2303 | 402.6–406.5 | 174,848,983–178,760,993 | 9.12 | 20.9 | 1,286.53 |
| 62 | 5 | npi288a-AY110182 | 635.3–643.1 | 211,860,408–212,486,849 | 2.88 | 5.6 | 643.26 |
| 63 | 6 | psr160a-uck1 | 88.7–95.2 | 54,450,298–57,531,067 | 7.49 | 17.8 | 1,286.53 |
| 64 | 6 | umc1350-umc62 | 510.2–513.8 | 164,373,190–164,899,991 | 4.92 | 10.2 | −964.90 |
| 65 | 7 | umc1324-isu150 | 388.5–393.1 | 147,131,709–149,709,978 | 5.58 | 11.9 | 1,179.32 |
| 66 | 9 | umc1809-umc1588 | 69.9–81.2 | 9,596,965–11,409,891 | 3.24 | 7.4 | −750.47 |
| 67 | 9 | ufg66-AY110782 | 300–306.4 | 112,637,702–113,561,806 | 2.59 | 5.1 | 643.26 |
| Total | 52.0 | ||||||
| PGM | |||||||
| 68 | 1 | an1-umc1383 | 791.1–803.1 | 237,739,763–246,969,159 | 3.75 | 8.6 | 350.43 |
| 69 | 1 | AY111936-BE639426 | 905.1–907.1 | 272,696,181–273,640,938 | 6.55 | 13.4 | −560.69 |
| 70 | 4 | jpsb527b-agrr301 | 231.1–242.9 | 27,604,019–32,104,770 | 6.06 | 13.0 | 420.52 |
| 71 | 4 | umc1132-rz596b | 535.9–538.9 | 201,145,140–206,105,272 | 7.34 | 15.1 | −490.61 |
| 72 | 6 | php20599-chr118 | 533.4–538.3 | 166,175,304–167,373,672 | 7.65 | 16.7 | −560.69 |
| 73 | 7 | npi380-mmp17 | 543.4–547.2 | 164,659,875–164,849,264 | 5.02 | 9.5 | −420.52 |
| Total | 43.9 | ||||||
| Biomass | |||||||
| 74 | 1 | csu374b-bcd98a | 642.3–649.5 | 202,618,140–205,117,912 | 2.54 | 4.2 | −8.74 |
| 75 | 1 | uaz130a(tlk)-mmp165 | 934.9–939.6 | 275,339,855–277,362,236 | 3.90 | 6.0 | 11.13 |
| 76 | 1 | phi064-umc1819 | 1108.6–1118.7 | 295,590,053–297,468,124 | 7.30 | 11.0 | 14.00 |
| 77 | 3 | csu303-jpsb107c | 699.5–714.3 | 214,770,502–216,749,570 | 3.09 | 4.3 | 9.09 |
| 78 | 5 | bnlg1902-umc2298 | 298.8–306.5 | 79,724,746–84,076,624 | 3.10 | 4.4 | −8.92 |
| 79 | 7 | AY109968-umc2142 | 239.5–245.3 | 89,791,900–103,092,643 | 3.58 | 4.7 | 8.76 |
| 80 | 7 | umc2331-umc1251 | 409.3–411.7 | 150,413,852–151,629,292 | 10.61 | 16.9 | 17.58 |
| 81 | 9 | asg44-mmp131 | 444.2–453.5 | 138,514,686–140,366,458 | 3.45 | 5.8 | −10.13 |
| Total | 54.8 | ||||||
| PC1 | |||||||
| 82 | 1 | umc1076-AY110396 | 440.4–441.3 | 142,567,141–147,063,178 | 6.71 | 12.3 | 1.48 |
| 83 | 1 | umc1676-umc2231 | 451.8–453.5 | 148,630,198–156,340,584 | 8.43 | 15.8 | −1.58 |
| 84 | 2 | npi254a-umc1265 | 57.4–77.1 | 4,298,278–5,516,306 | 4.01 | 9.0 | 0.58 |
| 85 | 2 | umc1454-psr666 | 339.3–344.2 | 69,623,391–82,665,401 | 5.16 | 8.6 | 0.60 |
| 86 | 3 | umc1730-bnl10.24a | 399.1–402 | 169,757,743–170,791,380 | 5.39 | 9.1 | 0.61 |
| 87 | 6 | php20599-agp2 | 533.4–536.4 | 166,175,304–166,702,812 | 4.89 | 8.1 | −0.59 |
| 88 | 9 | dmt103a-AW216329 | 621.5–638.6 | 149,505,376–150,584,039 | 3.15 | 5.1 | 0.48 |
| Total | 60.0 |
Markers that flank the 1 − LOD confidence interval.
The position that defines the 1 − LOD confidence interval around the position of peak likelihood for the QTL.
The genome location of each genetic marker was obtained through the integration of the IBM linkage map (http://www.maizegdb.org/qtl-data.php) and the maize B73 genome sequence (Maize Genome AGP version 1, release 4a53; www.maizesequence.org).
The three cis-QTL are shown in boldface.
Total r2 was obtained by fitting all the significant QTL in the model simultaneously.
Seven QTL were detected from PC1, with total r2 equaling 0.60 and individual r2 ranging from 0.051 for QTL88 to 0.158 for QTL83. The seven QTL located on five chromosomes and their positive alleles were again either from B73 or Mo17 (Table III).
Shared QTL among Different Enzymes and between Enzymes and Seedling Biomass
We found 14 chromosome regions comprising in total 44 intervals (each interval is flanked by a pair of consecutive markers) where enzyme and/or biomass QTL were colocalized (Table IV). For example, three regions on chromosomes 1, 5, and 6 and one region on chromosomes 3, 4, 7, 8, and 9 were shared by QTL from different traits. The most frequent colocation was for GDH and NR, which shared five QTL with other traits. The least was for AlaAT, SDH, ALD, and PGM, which shared only two QTL with other traits, and AspAT, which did not share any QTL with other traits. Three cases were found where biomass QTL colocated with enzyme activity QTL: with GS activity on chromosome 1, with GS and NR activity on chromosome 1, and with AGPase activity on chromosome 5.
Table IV. Genomic positions of QTL shared by different traits.
Triangle symbol represents a QTL in that region.
| Chromosome No. | AGP Coordinate Intervala | GDH | NR | GS | AlaAT | SDH | CS | AGPase | ALD | PGM | BM | LRb | PC1 |
| 1 | 182,674,328–-184,708,080 | ▲ | ▲ | 2.1 | |||||||||
| 202,618,140–-206,940,320 | ▲ | ▲ | 2.4 | ||||||||||
| 274,753,840–-277,362,236 | ▲ | ▲ | ▲ | 3.2 | |||||||||
| 2 | 4,135,834–-5,516,306 | ▲ | ▲ | ||||||||||
| 3 | 14,687,115-–17,051,214 | ▲ | ▲ | 2.6 | |||||||||
| 4 | 27,604,019–-32,104,770 | ▲ | ▲ | 1.8 | |||||||||
| 5 | 13,528,340–-16,208,692 | ▲ | ▲ | 1.1 | |||||||||
| 79,724,746–-84,076,624 | ▲ | ▲ | 2.0 | ||||||||||
| 172,689,557-–178,760,993 | ▲ | ▲ | ▲ | 1.8 | |||||||||
| 6 | 149,647,086–-152,276,209 | ▲ | ▲ | 2.3 | |||||||||
| 163,933,189–-164,899,991 | ▲ | ▲ | 2.1 | ||||||||||
| 166,175,304–-167,373,672 | ▲ | ▲ | 3.0 | ▲ | |||||||||
| 7 | 126,449,934–-134,999,219 | ▲ | ▲ | ▲ | 3.1 | ||||||||
| 8 | 132,646,661–-137,623,043 | ▲ | ▲ | 2.0 | |||||||||
| 9 | 146,862,896–-147,007,976 | ▲ | ▲ | 1.3 | |||||||||
| 149,505,376–-150,584,039 | ▲ | ▲ |
The interval location in the maize genome was obtained through the integration of the IBM linkage map (http://www.maizegdb.org/qtl-data.php) and the maize B73 genome sequence (Maize Genome AGP version 1, release 4a53; www.maizesequence.org).
LR represents the log likelihood ratio value for colocalization without PC1, and the significance threshold for LR is 3.0.
Given the high density of markers and the large number of traits analyzed, a high portion of overlaps of QTL is expected by chance. To determine whether there was functional relevance to the overlap, we developed a novel permutation test based on the recombination and gene density across the genome. Our permutation test indicated that, on average, nearly 21 out of the 44 observed overlaps could possibly be due to chance alone (Supplemental Fig. S2). This suggests that most colocation is still due to a lack of genetic resolution, even with this very high-resolution map.
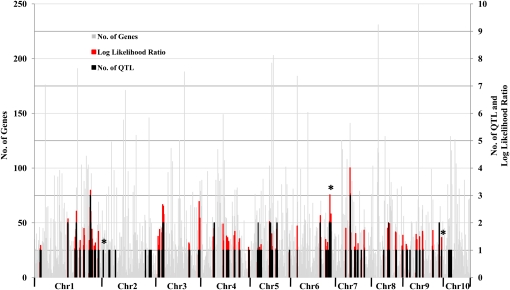
Information about the gene content within each interval (Schnable et al., 2009) allows a more detailed analysis of QTL colocalization in a particular map interval. Briefly, the significance of QTL colocalization in a given map interval will depend on the number of genes in the corresponding genome region, which is known from the whole genome sequence. A Bayesian approach for testing QTL colocalization (see “Materials and Methods”) provided a coherent support to specify genomic positions where multiple QTL colocate (Table IV; Fig. 1). Overall, there are three regions that most likely control the activity of several enzymes: the region on chromosome 1 (AGP: 274,753,840–277,362,236) was shared by QTL from NR, GS, and BM; the region on chromosome 6 (AGP: 166,175,304–167,373,672) was shared by QTL from SDH and PGM; and the region on chromosome 7 (AGP: 126,449,934–134,999,219) was shared by QTL from GDH, NR, and CS (Table IV).
Figure 1.
Distribution of QTL, gene content, and likelihood ratio for QTL colocalization. The gray bars in the background are numbers of genes estimated per map interval, the black bars are numbers of QTL identified in the region, the red bars are the likelihood ratio for QTL colocalization, and the asterisks above the columns are indicative of a colocalization with a PC1 QTL.
The above analysis investigated colocalization of QTL for individual traits. We also looked at overlap between PC1 QTL and QTL for individual traits. Of the seven PC1 QTL, three overlapped with at least one other trait (Table IV). For example, a PC1 QTL shared a region on chromosome 6 (AGP: 166,175,304–167,373,672) with two enzymes, SDH and PGM. The activities of the two enzymes were correlated with each other, and the region is one of the three regions that most likely control the activity of multiple enzymes (see above). Therefore, this result confirmed our QTL colocalization testing results. The other four PC1 QTL did not colocate with QTL for individual enzymes or biomass (Table IV). However, these PC1 QTL did overlap with weak enzyme activity and/or biomass QTL, which fell below threshold in the regions (data not shown). The results were consistent with the principal component analysis finding that PC1 captured the common factor (Supplemental Fig. S1).
Epistasis
Metabolic networks consist of multiple interconnected metabolic pathways. For this reason, prevalent epistatic interactions are expected to occur among QTL that influence various enzymatic steps in single or multiple pathways (Lisec et al., 2008). Therefore, we conducted a two-way epistatic interaction analysis. Multiple interval mapping was applied to analyze the activity levels of the 10 enzymes, seedling biomass, and PC1. To do this, we fitted the initiation model with the QTL detected with CIM and then refined the model based on Bayesian information criterion and also searched for all possible two-way epistatic interactions among the significant additive QTL for each trait. The detected additive QTL (data not shown) were very similar to those detected using CIM. The detected epistatic interactions are shown in Table V.
Table V. Digenic epistatic interactions.
| Trait | QTL aa | Shared QTL ab | QTL ba | Shared QTL bb | Effect | r2 |
| % | ||||||
| GDH | 3 | No | 8 | No | −9.41 | 3.4 |
| NR | 11 | No | 15 | Yes | 4.29 | 1.5 |
| GS | 19 | No | 23 | No | −0.54 | 2.9 |
| AlaAT | 25 | No | 28 | No | 57.61 | 1.2 |
| AspAT | 32 | No | 36 | No | 246.64 | 3.0 |
| SDH | 44 | No | 46 | No | 15.90 | 1.1 |
| CS | 49 | No | 50 | No | −3.75 | 2.7 |
| 47 | No | 52 | Yes | 4.45 | 3.6 | |
| 51 | No | 52 | Yes | −4.24 | 3.3 | |
| AGPase | 54 | No | 55 | No | −43.85 | 2.8 |
| 56 | No | 57 | No | 43.56 | 3.5 | |
| 55 | No | 59 | No | −80.38 | 6.9 | |
| 58 | No | 59 | No | −54.83 | 3.1 | |
| Biomass | 75 | Yes | 76 | No | −4.0743 | 3.2 |
| 75 | Yes | 77 | No | 4.8573 | 1.0 | |
| PC1 | 84 | Yes | 86 | No | 0.38 | 2.1 |
| 87 | Yes | 88 | Yes | 0.35 | 2.6 |
QTL are numbered the same as in Table III.
QTL shared by at least two traits.
In total, 17 epistatic interactions were detected. They affected all traits except for ALD and PGM, with one to four interactions for each trait. In total, 26 out of 88 detected additive QTL were involved in the epistatic interactions. The average r2 for the specific members of the pairs that interact epistatically was 9.7%, and the average r2 for all of the QTL from the traits was 10.1%. Compared with these additive QTL, epistatic effects were rather small, on average explaining only 2.8% of the phenotypic variation and improving 5.4% of the basic model fitting. The strongest epistatic interaction (r2 = 6.9%) was determined between two AGPase QTL (QTL55 and QTL59). Among the 26 significant epistatic interaction QTL, three were shared by two other traits (Table V).
cis-QTL and Structural Genes for Enzymes
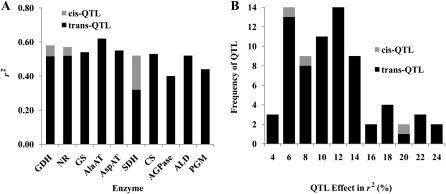
Because the enzyme activities measured here are the products of the corresponding structural genes, it is likely that we might detect candidate genes within QTL. With the information of the annotated maize genome DNA sequence (Schnable et al., 2009), we were able to search for all the genes encoding the 10 enzymes we studied here. We identified a gene encoding GDH that was located within the GDH activity QTL2 region (AGP: 285,915,476–287,632,595) on chromosome 1 (Table III), a gene encoding NR that was located within the NR activity QTL12 region (AGP: 174,462,939–177,509,810) on chromosome 4, and a gene encoding SDH that was located within the SDH activity QTL44 region (AGP: 23,420,566–26,790,198) on chromosome 10 (Table III). The r2 values for these cis-QTL lay in the range of the r2 values for the trans-QTL and represented only a small fraction of the total summed QTL r2 values for these three enzymes (Fig. 2). No cis-QTL were identified for the remaining seven enzymes.
Figure 2.
Effect and frequency comparison between cis- and trans-QTL for the activities of the 10 enzymes. A, r2 of cis- and trans-QTL. B, Frequency of cis- and trans-QTL for the effect in r2.
DISCUSSION
In this study, we examined quantitative genetic variation in the maximum activities of four carbon and six nitrogen metabolism enzymes and in seedling/juvenile biomass in the maize IBM population. This is more enzymes than in any previous study, except for two reports on Arabidopsis (Mitchell-Olds and Pedersen, 1998; Keurentjes et al., 2008). There is evidence that use of the intermated mapping population improves the resolution at least four times compared with conventional mapping populations (Lee et al., 2002; Balint-Kurti et al., 2007). Moreover, with the sequencing of the maize genome (Schnable et al., 2009), our ability to resolve QTL down to genes is much enhanced. Our median QTL covers about 26 genes (of the 32,000 genes in the genome), which is a vast improvement, although still not at the single gene level. Mapping in the IBM population improves the resolution of enzyme activity QTL by 23-fold, compared with prior mapping in maize completed about a decade ago (Hirel et al., 2001), and by 14-fold compared with a recent Arabidopsis study (Keurentjes et al., 2008) in terms of the number of genes per QTL interval, although comparing one population in one species with a different structured population in another species is very complicated.
Overall, we found more QTL per enzyme than previous studies. This is likely to result from the tremendous genetic variation within maize, which is represented in this particular cross, and, moreover, the increased opportunity for recombination in this intermated mapping population. The QTL did not colocalize with previous reports for QTL mapping in maize (Hirel et al., 2001; Gallais and Hirel, 2004). This resembles other high-diversity traits in maize, where there may be 50 to 100 QTL for given traits but only 20% are likely to be segregating in any one cross (Buckler et al., 2009). It suggests that a great deal can be learned about the regulation of this pathway by evaluating many crosses, such as in the maize nested association mapping (NAM) population (Buckler et al., 2009; McMullen et al., 2009).
The improvement in resolution of QTL mapping and the novel statistical approaches applied in this study allowed us to rigorously evaluate the importance of coordinate regulation. Because so many QTL were mapped for numerous traits, there was overlap between them. However, the vast majority of the colocalization was shown to be merely coincidental. Of the 81 QTL regions identified, perhaps three are likely to be true regions of coordinate regulation. None of the structural genes for the enzymes themselves or known regulators is in these regions. These results indicate that some previous reports of colocalization of QTL with candidate genes (Hirel et al., 2001; Gallais and Hirel, 2004) need to be reevaluated with current knowledge of recombination, maize genes, and the maize genome. Another way to test QTL colocalization is by taking advantage of the maize NAM population, in which pleiotropy could be investigated by correlating the allelic effects on multiple traits of each QTL across a robust sample of founders (Buckler et al., 2009).
One of the key issues being wrestled with in genetics today is the relative importance of cis- versus trans-regulation of networks. Since enzyme activities were measured in a robotized platform in this study, using optimized assays with saturating substrate concentrations, the likely causes for trans-regulation are the effects of transcription factors or differential enzyme degradation through proteasome and ubiquitination pathways. Since reaction conditions are with saturating concentrations in vitro, trans-effects are less likely to be caused by allosteric regulation of the enzyme activity. It is also unlikely that changes in enzyme phosphorylation are retained, because the extraction and assay were not performed in the presence of protein phosphatase inhibitors. Thus, cis-effects detected using this enzyme activity analysis assay are likely to be caused by expression differences, nonsynonymous polymorphisms that affect activity, and mRNA polymorphisms that affect translation and degradation rates. Although we have proven the effect of cis-regulation in the activity of maize NAD-dependent isocitrate dehydrogenase (EC 1.1.1.41) using the association mapping approach (Zhang et al., 2010), this study permitted an opportunity to evaluate the importance of cis- versus trans-regulation for 10 enzymes on the genome-wide scale. The number of structural genes in the B73 genome for the enzymes in this study varies from one to seven, with a total of 36 locations for all 10 enzymes. We only mapped three cis-QTL, and the effects of these cis-QTL lay in the range of the trans-QTL. Thus, cis-regulation appears less important than trans-regulation.
Although the regulation is trans-acting, it does not appear to be statistically epistatic. Statistical epistasis is simply a nonlinear response between two loci. In other words, a lack of statistical epistasis does not mean that there is no biochemical or genetic interaction. In this study, we found that the contribution of the epistatic interactions was quite small. This lack of statistical epistasis may be due to the difference between outcrossing versus inbreeding species (Holland, 2007; Buckler et al., 2009; McMullen et al., 2009). Although many other studies of enzyme activities and metabolites detected epistatic interactions (Keurentjes et al., 2008; Rowe et al., 2008; Kliebenstein, 2009), this study, and probably many other studies of enzyme activities (Causse et al., 1995; Mitchell-Olds and Pedersen, 1998; Hirel et al., 2001; Limami et al., 2002) and metabolites (Meyer et al., 2007; Lisec et al., 2008), lack the statistical power to rigorously detect all of the epistatic interactions that may be present.
Several of the enzyme activities correlated negatively with seedling biomass in the IBM population, which is consistent with previous studies in the maize association panel for isocitrate dehydrogenase (Zhang et al., 2010) and in tomato (Solanum lycopersicum) for mitochondrial malate dehydrogenase (Nunes-Nesi et al., 2005) and aconitase (Carrari et al., 2003). The negative correlation between enzyme activity and plant growth may be due to increases in photosynthesis (Zhang et al., 2010), a compensation for the reduction in energy production by respiration (Nunes-Nesi et al., 2005), or “overcompensation” by other isoforms of that enzyme that would not be detected by the current assay (Zhang et al., 2010). We found that one biomass QTL on chromosome 1 colocated with QTL for GS and NR activities in this study. The colocated QTL resulted in more biomass but less enzyme activities. While this might indicate an influence of these enzyme activities on biomass, as already discussed, further analysis is needed to assess whether the colocations are fortuitous or causal. In addition, it needs to be pointed out that seedling biomass is not a proxy for grain yield. In the field at this stage, plants are competing with weeds and developing canopies. Although bigger seedling plants (greater seedling biomass) are more likely to win in the competition, plant grain yield (sink) is shaped by many other factors in addition to the plant biomass (source). Therefore, the relationship between enzyme activity and biomass is likely to change through ontogeny.
In summary, we found heritable variation in enzyme activity for all 10 enzymes under study and for seedling biomass. Strong positive correlations were found among activity levels of different enzymes, indicating that carbon and nitrogen enzymes are coregulated. Compared with other studies, we found more QTL for each enzyme, and for each QTL we have higher resolution due to the use of the intermated mapping population. While colocation of QTL for different enzymes might be taken as evidence for coordinate regulation, our statistical analysis indicates that many of these colocations are likely to be fortuitous even in our intermated population, which gives a much higher genetic resolution than conventional mapping populations. While three QTL influencing NR, GDH, and SDH activities were mapped close to their encoding loci, overall, we found that cis-regulation was relatively unimportant versus the trans-regulation of networks. In agreement, we mapped some QTL regions shared by different enzymes and biomass and further regions that colocated with principal component QTL and that may represent a joint regulator of these enzymes. Nevertheless, these conclusions are limited by current knowledge about which polymorphisms are present in the genes and cis-regulatory sequences residing on these chromosome intervals and by our incomplete understanding of gene regulation. Using the maize NAM population specifically designed for dissecting complex quantitative traits with high resolution and statistical power (McMullen et al., 2009), along with the Maize HapMap designed for finding millions of single nucleotide polymorphisms among the maize NAM founder lines (Gore et al., 2009), we may be able to identify the actual loci responsible for quantitative trait variation. Candidate gene association mapping (Harjes et al., 2008; Zhang et al., 2010) and genome-wide association studies in the future may provide a route to verify the results obtained from this study, especially for the three cis-QTL, and to predict polymorphisms that could underlie the QTL.
MATERIALS AND METHODS
Plant Materials and Greenhouse Experiment
We used 94 IRILs of maize (Zea mays) from the B73 × Mo17 cross (IBM-94; www.maizegdb.org) for QTL mapping. The IRILs were derived from four times intermating after the F2 stage (Lee et al., 2002).
Plants were grown in five replications in cell packs in the greenhouse in a completely randomized design. Three seeds from each line were sown in each cell and thinned 5 d after germination to one plant per cell to ensure uniform germination across the experiment. Tissue was collected from the youngest expanded leaf at 35 d after germination and immediately frozen in liquid nitrogen. Immediately before the tissue collection, above-soil whole plant weight was taken as the biomass trait. Tissue was stored at −80°C until analysis.
Enzyme Measurements
Enzyme extracts were prepared as described previously (Gibon et al., 2004a), except that Triton X-100 was used at a concentration of 1% (v/w) and glycerol at 20% (v/w) and maximal activities were determined in batches of 40 extracts on flat-bottom microplates (Sartstedt) using EP3 and Janus 96 tip-head robots (Perkin-Elmer) and ELX-800, ELX-808, or Synergy readers (Bio-Tek). AGPase, AlaAT, ALD, AspAT, GDH, GS, NR, and SDH were determined as described by Gibon et al. (2004a), CS was determined as described by Nunes-Nesi et al. (2007), and PGM was determined as described by Manjunath et al. (1998). Except for AspAT, ALD, and PGM, with three replications, the other seven enzymes were measured with five biological replications.
Statistical Analyses
Mixed linear model analyses for enzyme activities and biomass were conducted using the PROC MIXED procedure of SAS version 9.1. For enzyme activities, mixed linear models were fit, including genotypic effects of lines, batch effects of enzyme activity assay, and effects of blocks and flats of the greenhouse designs. For biomass, the same mixed linear model as the enzymes was fit, except that batch effect of enzyme activity assay was not included in the model. We treated the line, block, and flat as random effects and the batch as a fixed effect. A correction for multiple tests was applied using the sequential Bonferroni test (Holm, 1979) at α = 0.05 for the genetic effects of the 11 traits. Best linear unbiased predictors (BLUPs) for each trait of each line were predicted from the analysis of the above models. H2 was calculated for each trait as described previously by Holland et al. (2003). All enzyme activity and biomass correlation analyses were based on BLUPs and conducted using the PROC CORR procedure of SAS. To make a correction for multiple statistical tests, the sequential Bonferroni test at α = 0.05 was also conducted for the correlations of the 11 traits. Principal component analysis was performed using the PROC PRINCOMP procedure of SAS. To identify how different BLUPs and LSMEANS are in this study, we treated the line as a fixed effect and calculated LSMEANS for each IRIL in each of the 11 traits and plotted it against the corresponding BLUPs. The results revealed very tight correlations (r ranges from 0.993–0.999 in the 11 traits and average r equal to 0.995). Therefore, the conclusions using LSMEANS will not change compared with BLUPs.
QTL Mapping
The Windows QTL cartographer version 2.51 was used to conduct QTL analysis (Wang et al., 2010). We performed CIM to identify QTL for enzyme activities, biomass, and PC1. BLUP values were calculated for each line over the five replications and used for mapping. For each trait analysis, 10 control markers were chosen as the background, and a window size of 5 cM and a walk speed of 0.5 cM were used to identify QTL. Permutation testing of 1,000 times was used to determine the likelihood ratio for a 5% significance level of identified QTL. It was considered as colocation when the 1 − LOD (for log of the odds) confidence interval overlapped between two QTL. Moreover, we performed multiple interval mapping to detect epistatic interactions among main-effect QTL. For the analysis, initial multiple interval mapping models were constructed with QTL identified that were significant in CIM. The models then were refined based on Bayesian information criterion (Piepho and Gauch, 2001). Each pair of QTL in the refined model was tested for epistatic interactions. Epistatic interactions were chosen if they decreased the Bayesian information criterion. We recalculated the r2 of each epistatic interaction the same way as in CIM, so the new r2 is the proportion of the variance explained by the epistatic interaction term conditioned on all additive QTL and other epistatic interactions.
The genotypic data for the IBM population were available publicly on the MaizeGDB Web site (http://www.maizegdb.org/qtl-data.php; Lawrence et al., 2005). We used the 2,200 markers spaced over the maize genome of 10 chromosomes for the QTL mapping. Map distances were based on the IBM2 map (http://www.maizegdb.org/map.php). The sequences of genetic markers in the IBM map were obtained from MaizeGDB (http://www.maizegdb.org). There were 1,623 unique genetic positions after blasting against the maize B73 reference genome sequence (Maize Genome AGP version 1, release 4a53; www.maizesequence.org), and 1,242 loci were left after removing loci for which the genetic and physical maps were in disagreement. The map positions of genetic markers and their locations in the genome were obtained through the integration of the IBM linkage map (http://www.maizegdb.org/qtl-data.php) and the maize genome sequence (Maize Genome AGP version 1, release 4a53; www.maizesequence.org). The AGP coordinates of these 1,242 markers were used to calculate the numbers of genes under intervals (Schnable et al., 2009).
QTL Colocalization
To test the significance of QTL colocalization in different traits, we adopted the randomization test of Lisec et al. (2008). The QTLs of each trait except for PC1 were first randomly distributed over 2,200 genetic marker positions. We then counted the number of markers that are responsible for more than one enzyme activity trait. This procedure was repeated 10,000 times to obtain the expected number of QTL colocalizations, giving the total number of genetic markers on the map, traits, and markers that underlie QTL regions. The mean and the 95% quantile of the distribution are 21.5 and 28.0, respectively (Supplemental Fig. S2).
Summed from the number of gene identifiers in the current version of maize gene annotation, there are in total 32,540 genes in the maize AGP genome. Sj denotes the number of genes contained in chromosome 1 to chromosome 10 (j = 1, 2, … 10). Given the number of genes (n) residing within the ith map interval, the probability of finding QTL putatively underlying at least one of the 11 traits in the given region is:
Here, we assume that genes in a map interval have an equal chance to code for any given enzyme activity variation and that the variations of enzyme activity are independent. For each chromosome (j), the probability of defining enzyme activity QTL in the ith interval is given as:
if there are QTL identified within the map interval, and
if no QTL are identified.
The prior probabilities of QTL colocalization [Pr(colocalized QTL)] are calculated by the proportion of the chromosome intervals for which there are more than one QTL identified in the region. Posterior probability of QTL colocalization is then estimated as:
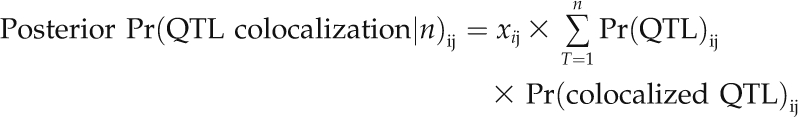 |
where T indicates the number of traits analyzed in the study. This provides a simple likelihood ratio test to examine the significance of QTL colocalization for each chromosome interval, with the comparison of the probability of QTL colocalization by permutations.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A, Proportion of variation explained by each individual principal component; B, contribution of different traits to the first principal component.
Supplemental Figure S2. Permutations of the number of QTL colocalizations that occurred by chance.
Supplementary Material
Acknowledgments
We thank Linda Rigamer Lirette and Sara Miller for their editorial assistance and Peter Bradbury for his comments and useful suggestions on the revision of the manuscript.
References
- Andrews M, Lea PJ, Raven JA, Lindsey K. (2004) Can genetic manipulation of plant nitrogen assimilation enzymes result in increased crop yield and greater N-use efficiency? An assessment. Ann Appl Biol 145: 25–40 [Google Scholar]
- Balint-Kurti PJ, Zwonitzer JC, Wisser RJ, Carson ML, Oropeza-Rosas MA, Holland JB, Szalma SJ. (2007) Precise mapping of quantitative trait loci for resistance to southern leaf blight, caused by Cochliobolus heterostrophus race O, and flowering time using advanced intercross maize lines. Genetics 176: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis WD, Lee M, Grant D, Hallauer AR, Owens T, Katt M, Blair D. (1992) The influence of random mating on recombination among RFLP loci. Maize Genet Coop News Lett 66: 52–53 [Google Scholar]
- Buckler ES, Gaut BS, McMullen MD. (2006) Molecular and functional diversity of maize. Curr Opin Plant Biol 9: 172–176 [DOI] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, Fernie AR. (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causse M, Rocher JP, Henry AM, Charcosset A, Prioul JL, Devienne D. (1995) Genetic dissection of the relationship between carbon metabolism and early growth in maize, with emphasis on key-enzyme loci. Mol Breed 1: 259–272 [Google Scholar]
- Dubois F, Terce-Laforgue T, Gonzalez-Moro MB, Estavillo JM, Sangwan R, Gallais A, Hirel B. (2003) Glutamate dehydrogenase in plants: is there a new story for an old enzyme? Plant Physiol Biochem 41: 565–576 [Google Scholar]
- Forde BG, Lea PJ. (2007) Glutamate in plants: metabolism, regulation, and signalling. J Exp Bot 58: 2339–2358 [DOI] [PubMed] [Google Scholar]
- Gallais A, Hirel B. (2004) An approach to the genetics of nitrogen use efficiency in maize. J Exp Bot 55: 295–306 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. (2004a) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios-Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Höhne M, Günther M, Stitt M. (2004b) Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Good AG, Johnson SJ, De Pauw M, Carroll RT, Savidov N. (2007) Engineering nitrogen use efficiency with alanine aminotransferase. Can J Bot 85: 252–262 [Google Scholar]
- Gore MA, Chia J-M, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. (2009) A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Haake V, Geiger M, Walch-Liu P, Engels C, Zrenner R, Stitt M. (1999) Changes in aldolase activity in wild-type potato plants are important for acclimation to growth irradiance and carbon dioxide concentration, because plastid aldolase exerts control over the ambient rate of photosynthesis across a range of growth conditions. Plant J 17: 479–489 [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET, et al. (2008) Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Bertin P, Quilleré I, Bourdoncle W, Attagnant C, Dellay C, Gouy A, Cadiou S, Retailliau C, Falque M, et al. (2001) Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol 125: 1258–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JB. (2007) Genetic architecture of complex traits in plants. Curr Opin Plant Biol 10: 156–161 [DOI] [PubMed] [Google Scholar]
- Holland JB, Nyquist WE, Cervantes-Martínez CT. (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22: 9–111 [Google Scholar]
- Holm S. (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6: 65–70 [Google Scholar]
- Holton TA, Cornish EC. (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Stam P. (1994) High resolution of quantitative traits into multiple loci via interval mapping. Genetics 136: 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJB, Sulpice R, Gibon Y, Steinhauser MC, Fu JY, Koornneef M, Stitt M, Vreugdenhil D. (2008) Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biol 9: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ. (2009) A quantitative genetics and ecological model system: understanding the aliphatic glucosinolate biosynthetic network via QTLs. Phytochem Rev 8: 243–254 [Google Scholar]
- Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM. (1996) The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 569–593 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Seigfried TE, Brendel V. (2005) The maize genetics and genomics database: the community resource for access to diverse maize data. Plant Physiol 138: 55–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Sharopova N, Beavis WD, Grant D, Katt M, Blair D, Hallauer A. (2002) Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol Biol 48: 453–461 [DOI] [PubMed] [Google Scholar]
- Limami AM, Rouillon C, Glevarec G, Gallais A, Hirel B. (2002) Genetic and physiological analysis of germination efficiency in maize in relation to nitrogen metabolism reveals the importance of cytosolic glutamine synthetase. Plant Physiol 130: 1860–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SC, Kowalski SP, Lan TH, Feldmann KA, Paterson AH. (1996) Genome-wide high-resolution mapping by recurrent intermating using Arabidopsis thaliana as a model. Genetics 142: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath S, Lee CHK, VanWinkle P, Bailey-Serres J. (1998) Molecular and biochemical characterization of cytosolic phosphoglucomutase in maize: expression during development and in response to oxygen deprivation. Plant Physiol 117: 997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M, Jouglet T, Suzuki A. (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. (2009) Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, Becher M, Witucka-Wall H, Törjék O, Fiehn O, Eckardt A, Willmitzer L, Selbig J, et al. (2007) The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 4759–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Pedersen D. (1998) The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis. Genetics 149: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus HE, Stitt M. (1990) Control analysis of photosynthate partitioning: impact of reduced activity of ADP-glucose pyrophosphorylase or plastid phosphoglucomutase on the fluxes to starch and sucrose in Arabidopsis thaliana (L) Heynh. Planta 182: 445–454 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AMO, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant (in press) [DOI] [PubMed] [Google Scholar]
- Piepho HP, Gauch HG., Jr (2001) Marker pair selection for mapping quantitative trait loci. Genetics 157: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Hohne M, Usadel B, Gibon Y, Rohwer J, Stitt M. (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J. (1982) Regulation of the biosynthesis and degradation of starch. Annu Rev Plant Physiol Plant Mol Biol 33: 431–454 [Google Scholar]
- Prioul JL, Pelleschi S, Sene M, Thevenot C, Causse M, de Vienne D, Leonardi A. (1999) From QTLs for enzyme activity to candidate genes in maize. J Exp Bot 50: 1281–1288 [Google Scholar]
- Robinson SA, Slade AP, Fox GG, Phillips R, Ratcliffe RG, Stewart GR. (1991) The role of glutamate dehydrogenase in plant nitrogen metabolism. Plant Physiol 95: 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. (2008) Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sentoku N, Taniguchi M, Sugiyama T, Ishimaru K, Ohsugi R, Takaiwa F, Toki S. (2000) Analysis of the transgenic tobacco plants expressing Panicum miliaceum aspartate aminotransferase genes. Plant Cell Rep 19: 598–603 [DOI] [PubMed] [Google Scholar]
- Shrawat AK, Carroll RT, DePauw M, Taylor GJ, Good AG. (2008) Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol J 6: 722–732 [DOI] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A, Nunes-Nesi A, Sulpice R, Lisec J, Centeno DC, Carillo P, Leisse A, Urbanczyk-Wochniak E, Fernie AR. (2008) Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol 147: 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Lunn J, Usadel B. (2010) Arabidopsis and primary photosynthetic metabolism: more than the icing on the cake. Plant J 61: 1067–1091 [DOI] [PubMed] [Google Scholar]
- Strack D. (1997) Phenolic metabolism. Dey PM, Harborne JB, eds, Plant Biochemistry Academic Press, San Diego, pp 387–416 [Google Scholar]
- Thévenot C, Simond-Côte E, Reyss A, Manicacci D, Trouverie J, Le Guilloux M, Ginhoux V, Sidicina F, Prioul JL. (2005) QTLs for enzyme activities and soluble carbohydrates involved in starch accumulation during grain filling in maize. J Exp Bot 56: 945–958 [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng Z-B. (2010) Windows QTL Cartographer 2.51. Department of Statistics, North Carolina State University, Raleigh. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
- Wei F, Zhang J, Zhou S, He R, Schaeffer M, Collura K, Kudrna D, Faga BP, Wissotski M, Golser W, et al. (2009) The physical and genetic framework of the maize B73 genome. PLoS Genetics 5: e1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang NY, Gur A, Gibon Y, Sulpice R, Flint-Garcia S, McMullen MD, Stitt M, Buckler ES. (2010) Genetic analysis of central carbon metabolism unveils an amino acid substitution that alters maize NAD-dependent isocitrate dehydrogenase activity. PLoS ONE 5: e9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.