Abstract
In vascular plants, organelle RNAs are edited by C-to-U base modification. Hundreds of mitochondrial C residues are targeted for editing in flowering plants. In this study, we exploited naturally occurring variation in editing extent to identify Required for Efficiency of Mitochondrial Editing1 (REME1), an Arabidopsis (Arabidopsis thaliana) pentatricopeptide repeat protein-encoding gene belonging to the DYW subclass that promotes editing of at least two C residues on different mitochondrial transcripts. Positional cloning identified REME1 unambiguously as the gene controlling editing of nad2-558. Virus-induced gene silencing of REME1 confirmed its role in editing of nad2-558 and allowed us to identify orfX-552 as a second C whose editing is positively controlled by REME1. An unexpected outcome of REME1 silencing was the finding of a number of mitochondrial C targets whose editing extent exhibits a significant and reproducible increase in silenced tissues. That increase was shown to be partly due to the virus inoculation and partly to REME1-specific silencing. Analysis of an insertional T-DNA mutant within the REME1 coding sequence confirmed the findings of the virus-induced gene silencing experiments: decrease in editing extent of nad2-558 and orfX-552 and increase in editing extent of two sites, matR-1771 and rpl5-92. Transgenic complementation of the low-edited accession (Landsberg erecta) restored the editing of nad2-558 and orfX-552 to high-edited accession (Columbia)-type levels or to even higher levels than Columbia. There was no effect of the transgene on editing extent of matR-1771 and rpl5-92. The strategy and tools used in this report can be applied to identify additional genes that affect editing extent in plant mitochondria.
RNA editing is a process that alters the genetic information at specific sites on RNA molecules. Editing has been described in a wide range of organisms from viruses to animals and plants. Several systems involving unrelated mechanisms seem to have arisen separately during evolution (for review, see Gott and Emeson, 2000). In vascular plants, organelle transcripts are modified posttranscriptionally by C-to-U editing. While 30 to 40 C-to-U editing events are typically found in vascular plant chloroplast transcriptomes (Maier et al., 1995; Tsudzuki et al., 2001; Tillich et al., 2005), 1 order of magnitude more cytosine residues are subject to editing in the mitochondria of flowering plants: 427, more than 500, 491, and 357 sites have been reported, respectively, in rapeseed (Brassica napus), Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and sugar beet (Beta vulgaris; Notsu et al., 2002; Handa, 2003; Mower and Palmer, 2006; Bentolila et al., 2008). Conversion of a C to a U in a mitochondrial RNA usually results in a codon encoding an amino acid similar to that found in other plants or microorganisms at the homologous amino acid position, although silent editing events also occur (Covello and Gray, 1990; Gray and Covello, 1993). The main purpose of RNA editing has been thought to be the correction of deleterious mutations that would otherwise hamper the proper function of the encoded product. Indeed, an unedited ATP9 protein variant was shown to be not functional in tobacco (Nicotiana tabacum) because its presence disturbed mitochondrial function, resulting in a male sterility phenotype (Hernould et al., 1993).
The first trans-acting factor involved in plant organelle editing was found by screening Arabidopsis mutants defective in NADH dehydrogenase activity (Kotera et al., 2005). CHLORORESPIRATORY REDUCTION4 (CRR4) is required for editing of the start codon of the Arabidopsis chloroplast ndhD mRNA, which codes for a subunit of the chloroplast NAD dehydrogenase. CRR4 is a member of the pentatricopeptide repeat (PPR) protein family in plants (Small and Peeters, 2000) that comprises 450 members in Arabidopsis and 477 in rice (O’Toole et al., 2008), most of which are predicted to be targeted to either mitochondria or plastids (Lurin et al., 2004). Plant PPR proteins can be separated into two major subfamilies based on the nature of their PPR motifs, the P and PLS subfamilies. The P subfamily contains the canonical motif of 35 amino acids, while the PLS subfamily is characterized by the presence of longer (L) or shorter (S) variant PPR motifs within the tandem arrays. The PLS subfamily, which is specific to the plant kingdom, can be further separated into two groups, a subclass containing the DYW domain and a subclass lacking this domain (Lurin et al., 2004). All the chloroplast PPR motif-containing trans-factors identified so far belong to the PLS-E subgroup (Kotera et al., 2005; Okuda et al., 2007; Chateigner-Boutin et al., 2008; Hammani et al., 2009) or the PLS-E-DYW subgroup (Zhou et al., 2008; Cai et al., 2009; Hammani et al., 2009; Okuda et al., 2009, 2010; Robbins et al., 2009).
Similarly, the vast majority of the mitochondrial PPR motif-containing trans-factors belong to the PLS subfamily. MEF1, the first mitochondrial editing factor identified, is involved in editing of three specific C residues in different mitochondrial genes of Arabidopsis (Zehrmann et al., 2009). MEF1 belongs to the DYW subclass of PPR proteins, as does rice OGR1 (Kim et al., 2009). Mutants in the OGR1 gene fail to modify seven C residues edited in the wild type and exhibit various morphological phenotypes, including delayed seed germination, retarded growth, dwarfism, and sterility (Kim et al., 2009). Recently, a moss PPR protein with a DYW domain was shown to be responsible for editing of mitochondrial ccmFc transcript (Tasaki et al., 2010). Like chloroplast editing trans-factors, members of the PLS-E subgroup that lack the DYW motif have also been shown to be necessary for mitochondrial editing (Sung et al., 2010; Takenaka, 2010; Takenaka et al., 2010). PPR596 is the only example so far of a member of the P subfamily that is involved in mitochondrial editing (Doniwa et al., 2010).
Rather than analysis of mutants, our work on mitochondrial RNA editing relies on the use of natural variation in Arabidopsis coupled with a genetic approach to identify components of the RNA/protein complex responsible for the C-to-U modification (Bentolila et al., 2005, 2008). This strategy can potentially allow the identification of factors whose disruption would result in embryo lethality. We report here the identification of the gene Required for Efficiency of Mitochondrial Editing1 (REME1), which belongs to the DYW subclass of PPR-containing genes. By assaying all 523 mitochondrial C targets of editing in REME1-silenced leaf tissue in comparison with the wild type, we unexpectedly found that REME1 promotes RNA editing at some C targets while inhibiting the editing at other specific sites. This result was confirmed by the analysis of a T-DNA insertional mutant within the REME1 coding sequence.
RESULTS
Differential Editing of nad2-558 in Columbia and Landsberg erecta Is Controlled by a Single Gene
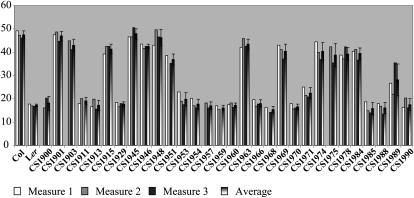
Study of mitochondrial editing polymorphisms between Columbia (Col) and Landsberg erecta (Ler) mitochondrial transcripts led to the first report on the genetic architecture of mitochondrial editing in Arabidopsis (Bentolila et al., 2008). We mapped 12 major quantitative trait loci (QTLs) for 11 of the 13 editing traits analyzed, demonstrating that efficiency of editing of individual mitochondrial C targets is generally governed by a major factor. The largest effect attached to a QTL was detected for editing extent of nad2-558. The QTL controlling this trait colocalizes with the marker m246 on chromosome 2; its effect could explain 95% of the phenotypic variation. The segregation of editing extent of nad2-558 observed in recombinant inbred lines (RILs; Fig. 1) indicates that the differential editing between Col and Ler is controlled by a single gene.
Figure 1.
Distribution of nad2-558 editing extent in 30 RILs and their progenitors, Col and Ler. The average of editing values was obtained for each inbred line by assaying three different plants, except for CS1900, CS1903, CS1911, CS1951, CS1957, CS1959, CS1968, and CS1975, where only two measures are available.
Positional Cloning of REME1, Which Promotes the Editing of nad2-558
We chose the QTL controlling the editing of nad2-558 for map-based cloning because of its high heritability and the subsequent simplicity of following its segregation by evaluation of editing extent (Fig. 1). Coarse mapping with 30 RILs delineated a log of the odds 1 interval for this QTL of 9.4 centimorgans between 7.75 and 17.15 centimorgans on chromosome 2 (Bentolila et al., 2008). By definition, the likelihood of finding the QTL in this interval is 10 times higher than finding the QTL outside the interval. The physical coordinates corresponding to this genetic interval were estimated to be 559 and 2,478 kb, thus determining a region of 1,919 kb comprising 503 genes. In order to reduce the size of this interval, we found recombinants in the pool of 70 RILs that had not been used in the original QTL mapping. The additional RILs considerably shortened the interval to 107 kb. We further restricted this interval by selecting 11 recombinants among a population of 691 F2 plants through the use of cleaved-amplified polymorphism markers (Fig. 2).
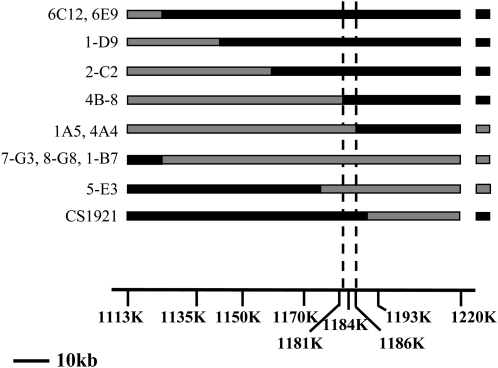
Figure 2.
Positional cloning of REME1, which controls nad2-558 editing. The graphical genotypes of the recombinants between 1113K and 1220K are represented by squares with either a black background (Col) or a gray background (Ler). The names of the recombinants are given on the left of each graphical genotype, with the nad2-558 editing value symbolized by a square at the right of the genotype (black for Col, gray for Ler). Below the graphical genotypes is drawn a map of the area with markers named after their physical coordinates. The location of the gene At2g03880 (REME1) that controls nad2-558 editing was bracketed by the recombination points occurring in 4B8 and 1A5 and 4A4.
As a result of this fine mapping, the interval containing the QTL controlling nad2-558 editing could be precisely bracketed between the crossover that took place in the individuals 4B8 on the left side and 1A5 and 4A4 on the right side (Fig. 2). Sequencing the PCR products containing the crossovers for these recombinants demonstrated that the interval containing the editing factor could be delineated by the physical coordinates 1182057 and 1185817. At this position lies only one annotated gene, At2g03880, which we have named REME1.
One of the fine-mapping recombinants we obtained, 4B8, is recombined inside the REME1 gene; the first 5′ 498 nucleotides were inherited from Ler, while the remaining nucleotides from position 666 to the end of the gene were inherited from Col, with crossing over taking place in between positions 498 and 666. This recombinant, which exhibits the Col editing phenotype, reveals which part of the gene is involved in the control of nad2-558 editing. Of the 15 nonsynonymous single nucleotide polymorphisms (SNPs) found between the Col and the Ler allele for REME1, one or more of the seven SNPs located downstream of the crossover found in the recombinant are implicated in differential editing of the parental alleles. Any one of the individual SNPs or any combination of them could be responsible for the differential editing of the parental alleles.
Confirmation of the Role of REME1 in nad2-558 Editing by Virus-Induced Gene Silencing
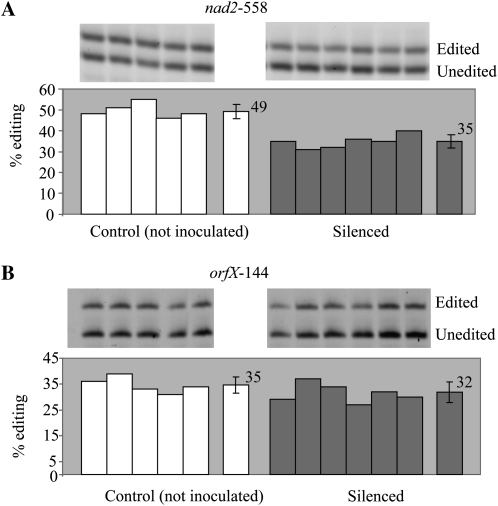
The positional cloning identified REME1 unambiguously as the nad2-558 editing QTL. As expected, virus-induced gene silencing (VIGS) of REME1 resulted in a significant decrease of nad2-558 editing (P < 5 × 10−5; Fig. 3A). The silencing vector (Robbins et al., 2009) contains a sequence of the gene to be silenced and a GFP sequence, so that we can visually screen for silenced plants after inoculating Arabidopsis plants carrying a GFP transgene. Under UV light, the silenced plants show a characteristic red phenotype resulting from chlorophyll autofluorescence, while unsilenced plants fluoresce green. Poisoned primer extension (PPE) assay on transcripts from uninoculated versus silenced plants shows that silencing of REME1 results on average in a 29% decrease in nad2-558 editing [(49 – 35)/49 = 0.29; Fig 3A]. Because the QTL controlling the editing of nad2-558 roughly colocalizes with the one controlling the editing of orfX-144 (Bentolila et al., 2008), we tested whether REME1 was also involved in the editing of orfX-144. No significant reduction of orfX-144 editing was observed in REME1-silenced plants (Fig. 3B; P = 0.16), thus indicating that orfX-144 editing is under the control of another gene.
Figure 3.
VIGS of REME1 results in decreased editing of nad2-558 (A) but does not affect the editing extent of orfX-144 (B). The gel of the PPE products is shown on top of each panel with the quantification of editing below each corresponding lane. On the right of each group is the average with sd. In A, the intensity of the top band, representing editing products, is lower in silenced plants than in the control plants.
REME1 Controls the Editing of Other Mitochondrial Sites
Because editing of nad2-558 is silent (editing does not change the encoded amino acid sequence), this site is likely to have no physiological importance. Why should silent editing occur? One possibility is that the trans-factor encoded by REME1 operates on a nonsilent site and also happens to interact with nad2-558 because of a fortuitous similarity in recognition cis-sequences between the two sites. This hypothesis was tested by comparing the electrophoretograms obtained from the bulk sequencing of reverse transcription (RT)-PCR products of a silenced plant and a control plant for the entire set of mitochondrial transcripts. In order to have enough material to produce the RT-PCR products for the whole set of mitochondrial genes, the RNA that was assayed for editing extent came from either a mixture of two control plants or a mixture of two silenced plants. Before conducting the bulk sequencing screen, we verified that a mixture of two silenced plants could be distinguished from a mixture of two control plants when sequencing electrophoretograms around nad2-558 were compared (Supplemental Fig. S1). As shown in Supplemental Figure S1, there is a noticeable difference between the T peaks of control versus silenced plants. Thus, we can conclude that the bulk sequencing assay can detect a 30% differential in editing extent between RNA samples.
Using RT-PCR and bulk sequencing, we screened the entire 439 C residues known to be edited in suspension cell transcripts of 34 mitochondrial genes for possible differences between the control and the silenced plants (Table I). We also analyzed C targets reported to lie in untranslated regions (UTRs) and in introns (Table I). As we reported previously (Bentolila et al., 2008), tissue specificity of editing results in the absence of editing in rosette leaves for 43 sites previously reported to be edited in suspension cells (Giegé and Brennicke, 1999). Conversely, 56 new sites were found to be edited in rosette leaves that were not reported in suspension cells, leading to the survey of a total of 452 mitochondrial sites in coding sequences (Table I). If a putative difference in editing extent between control and silenced pools was detected in the screen, editing efficiency at that site was then assayed by PPE, which is more time consuming but more reliable and quantitative than bulk sequencing. Because of the labor associated with the PPE assay, only a subset of the differentially edited sites was further checked by PPE. The lower accuracy of bulk sequencing for measuring editing extent is illustrated in Supplemental Figure S1, where the heights of the T peaks for nad2-558 for the control and silenced pools show a difference that does not reflect the real contrast in editing extent by PPE (53% versus 31%, respectively). Our PPE measurements were performed on several independent silenced and control plants in order to obtain sufficient data for testing the statistical significance of the differential editing extent (Table I).
Table I. Number and position of sites showing a differential editing extent in REME1-silenced Arabidopsis rosette leaves.
Sites in boldface are nonsilent sites whose editing results in an amino acid change. NS, Not significant.
| Gene | Sites Edited | Control > Silenced |
Silenced > Control |
Checked by PPE |
|||
| No. | Positiona | No. | Positiona | Position | P | ||
| Coding sequences | |||||||
| atp1 | 4 | 0 | 0 | ||||
| atp6-1 | 1 | 0 | 0 | ||||
| atp9 | 4 | 0 | 0 | ||||
| ccb203 | 11 | 2 | 208, 320 | 208 | NS | ||
| ccb206 | 37 | 1 | 406 | 1 | 80 | 80 | NS |
| ccb256 | 29 | 1 | 624 | 4 | 184, 252, 421, 673 | 184 | 2 × 10−2 |
| 421 | 4 × 10−3 | ||||||
| ccb382 | 14 | 0 | 1 | 709 | 709 | NS | |
| ccb452 | 19 | 0 | 2 | 1,246, 1,280 | 1,246 | NS | |
| 1,280 | 5 × 10−2 | ||||||
| cob | 10 | 0 | 0 | ||||
| cox2 | 13 | 0 | 0 | ||||
| cox3 | 7 | 0 | 0 | ||||
| matR | 10 | 0 | 7 | 374, 461, 1,593, 1,731, 1,771, 1,807 | 374 | 5 × 10−5 | |
| 461 | 5 × 10−4 | ||||||
| 1,771 | 8 × 10−5 | ||||||
| 1,807 | 3 × 10−5 | ||||||
| nad1 | 22 | 0 | 0 | ||||
| nad2 | 28 | 1 | 558b | 0 | 558 | 4 × 10−6 | |
| nad3 | 11 | 1 | 149 | ||||
| nad4 | 38 | 1 | 1401 | 0 | |||
| nad4L | 10 | 0 | 0 | ||||
| nad5 | 30 | 0 | 0 | ||||
| nad6 | 11 | 0 | 0 | ||||
| nad7 | 31 | 1 | 1137 | 0 | |||
| nad9 | 7 | 0 | 0 | ||||
| orf114 | 2 | 0 | 1 | 327 | 327 | 5 × 10−2 | |
| orf240A | 1 | 1 | 199 | 0 | 199 | NS | |
| orf25 | 9 | 0 | 0 | ||||
| orfB | 0 | 0 | 0 | ||||
| orfX | 34 | 2 | 552, 586 | 1 | 97 | 552 | 3 × 10−6 |
| rpl16 | 6 | 0 | 0 | ||||
| rpl2 | 3 | 1 | 212 | 212 | 5 × 10−2 | ||
| rpl5 | 10 | 6 | 58, 64, 92, 169, 317, 329 | 92 | 5 × 10−5 | ||
| 329 | 3 × 10−4 | ||||||
| rps12 | 8 | 0 | 0 | ||||
| rps14 | 2 | 1 | 99, 194 | 194 | 2 × 10−4 | ||
| rps3 | 12 | 1 | 603 | 4 | 64, 887, 1,344, 1,534 | ||
| rps4 | 17 | 0 | 10 | 77, 88, 175, 235, 967, 992, 1,042, 1,043, 1,052, 1,057 | 967 | 2 × 10−3 | |
| rps7 | 1 | 0 | 0 | ||||
| Total | 452 | 9 | 42 | ||||
| UTRs and introns | |||||||
| cox3 trailer | 4 | 0 | 0 | ||||
| nad5 intron | 2 | 1 | 803 | 0 | 803 | NS | |
| nad6 leader | 1 | 0 | 0 | ||||
| nad7 leader | 1 | 1 | −39 | 0 | −39 | NS | |
| nad7 second intron | 1 | 0 | 0 | ||||
| rpl5 trailer | 1 | 0 | 1 | +195 | +195 | 2 × 10−2 | |
| rpl16 trailer | 2 | 0 | 0 | ||||
| rps7 leader | 1 | 0 | 0 | ||||
| Total | 13 | 2 | 1 | ||||
Position of the edited C in the coding sequence is given relative to the start codon. In leader and trailer, the position of the edited C is relative to the start codon and the stop codon, respectively.
nad2-558 is the site used to map REME1.
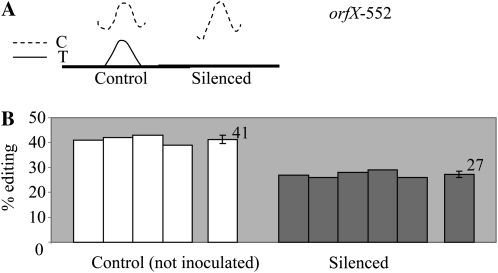
Among all the edited sites surveyed, one site showed a noticeable reduction in editing extent between control and silenced pools when comparing their respective electrophoretograms (Fig. 4A; Supplemental Fig. S2). When measured on the individual plants used for nad2-558, the C target of editing at position 552 on orfX, the tatC homolog, shows a very significant reduction in editing extent in REME1-silenced plants (P < 3 × 10−6; Table I; Fig. 4B). The average decrease in editing extent of orfX-552 found in REME1-silenced tissues, approximately 35%, is similar in range to the decrease observed for nad2-558. Like nad2-558, editing of orfX-552 results in a silent codon change. No nonsilent site was found to exhibit a reduction in editing extent in silenced leaf tissue. While editing of orf240A-199, a nonsilent site, appeared to exhibit lower editing on bulk-sequencing electrophoretograms, the reduction in editing extent was not statistically significant when assayed by PPE (Table I).
Figure 4.
VIGS of REME1 results in decreased editing of orfX-552. A, Comparison of the sequencing electrophoretograms of orfX RT-PCR products obtained from control and silenced pools shows a difference in the height of the T peak at position 552. B, PPE assay shows a significant (P < 3 × 10−6) difference in the editing extent of orfX-552 between several independent control and silenced plants. On the right of each group is the average value with sd.
A surprising result from the bulk-sequencing screen is the finding of 42 sites for which editing appeared to be higher in silenced tissues than in control tissues. Overall, 23 sites that appeared differentially edited in silenced versus uninoculated plants by bulk sequencing were further assayed by PPE; the difference in editing was observed to be not significant for seven of these sites, allowing an estimation of 30% as the rate of false positives in the bulk-sequencing screen (Table I). Using this correction factor, we conclude that REME1 silencing results in decreased editing in 1.4% of the sites and increased editing in 6.5% of the C targets known to exist in the mitochondrial transcripts. Strikingly, 81% (34 of 42) of the sites showing an increase in editing extent in REME1-silenced tissue result in codon changes that alter the encoded amino acid. This proportion closely fits the overall distribution of nonsilent sites in the population of edited sites in the coding sequences of mitochondrial genes (85% = 384 of 452). In contrast, no nonsilent sites were found to exhibit lower editing extent following VIGS of REME1.
The magnitude of the increase in editing of some C targets in VIGS tissue is rather pronounced. For example, the edited C at position 1,771 in matR exhibits an increase of 22% (13 of 60) in silenced tissues (P < 8 × 10−5; Table I; Fig. 5A). Not all the sites increased in editing are in coding regions; a site located in the 3′ UTR of rpl5 exhibited a significant increase in editing extent in VIGS plants (Table I).
Figure 5.
VIGS of REME1 results in increased editing of sites found in matR as detected by PPE assay. A, Editing extent of C at position 1,771 is significantly increased in silenced plants (P < 8 × 10−5). B, The increase in editing is less pronounced for the edited C at position 461 (P < 5 × 10−4). On the right of each group is the average editing extent with sd.
Increased editing extent in silenced tissues tends to affect several sites within transcripts of the same mitochondrial gene, although not all C targets in the same transcript exhibit increases to the same extent. For example, PPE demonstrated that matR possesses three sites with over a 10% increase in silenced tissues; nevertheless, another C target in this transcript, matR-461, exhibited only a 6% increase in silenced tissues (Fig. 5B). Similarly, the rpl5 site at position 92 exhibits a 15% increase in silenced tissues, while the increase is only 2% at position 329 (P < 5 × 10−5 versus P < 3 × 10−4, respectively; Table I; Supplemental Fig. S3).
Increased Editing Extent in REME1-Silenced Tissues Comes from the Virus Inoculation and from the Specific Silencing of REME1
We were concerned that the increase in editing extent observed in REME1-silenced tissues for certain mitochondrial sites might be an artifact caused by the VIGS technique; it is conceivable that the virus inoculation could stress the plant and as a result impact the editing machinery. The controls in the first set of silencing experiments were not inoculated plants and thus could not control for the possibility of an effect of virus inoculation on the editing extent. Therefore, we embarked on a new set of REME1-silencing experiments where controls for virus inoculation were performed by inoculating the plants with a vector harboring the GFP but no part of any plant gene (referred to empty vector in Fig. 6 and Supplemental Fig. S4; for more detail on the construction of the empty vector, see “Materials and Methods”). Because the PPE technique is rather time consuming, we focused our analysis on matR-1771, the site that showed the most pronounced increase in editing extent in the previous REME1-silencing experiments.
Figure 6.
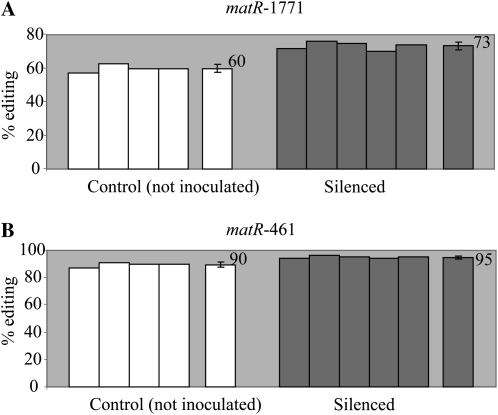
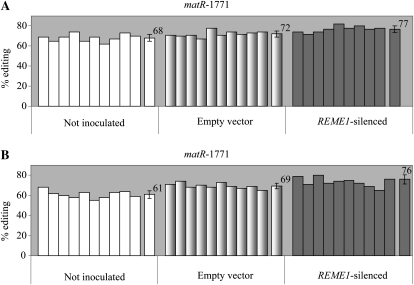
Increase in editing extent of matR-1771 in REME1-silenced plants comes from the virus inoculation and the specific silencing of REME1. Editing extents were assayed by PPE. The effect of virus inoculation can be estimated by comparing the noninoculated plants with the empty vector controls; the specific effect of REME1 silencing can be estimated by comparing the empty vector control with the REME1-silenced plants. A and B show data from two independent experiments. In both experiments, about half the increase of editing extent in REME1-silenced plants can be attributed to the specific silencing of REME1, the other half being the result of the virus inoculation. On the right of each group is the average editing extent with sd.
As shown in Figure 6A, the 13% [(77 – 68)/68] increase in editing extent for matR-1771 observed in REME1-silenced plants relative to noninoculated plants can be partitioned into two components: 6% is attributable to the virus inoculation (empty vector versus noninoculated; Fig. 6A; P < 2 × 10−2), while 7% comes specifically from REME1 silencing (empty vector versus REME1 silenced; Fig. 6A; P < 7 × 10−3). The main result from this experiment is the significance of the specific effect of REME1 silencing on the increase of matR-1771 editing extent; another outcome is the significant effect of the virus inoculation on the editing extent. The same observations were made for nad2-558, whose editing extent is decreased by REME1-VIGS: a part of the decrease can be specifically attributed to REME1 silencing, while another part comes from the virus inoculation itself (Supplemental Fig. S4A). The difference between nad2-558 and matR-1771 is the different role played by REME1-specific silencing in the overall difference between silenced plants and noninoculated plants (two-thirds for nad2-558 versus one-half for matR-1771; Fig. 6A; Supplemental Fig. S4A). All these observations were also found to be valid for orfX-552 editing extent (data not shown).
The efficiency of silencing in this new experiment seemed to be reduced when compared with the previous experiment, resulting in a variation in editing extent less pronounced when comparing the noninoculated control plants with the REME1-silenced plants. For instance, the increase in editing extent for matR-1771 is 13%, whereas it was observed to be 22% previously (compare Figs. 6A and 5A). Similarly, the decrease in editing extent for nad2-558 is less marked, 18% versus 29% (compare Supplemental Fig. S4A and Fig. 3A). The reason for this drop in silencing efficiency is unclear, but it led us to repeat this experiment in order to see whether results could be affected by the silencing efficiency.
In an independent experiment, the efficiency of silencing was found to be similar to the original REME1 silencing: the increase in editing of matR-1771 averaged 25% [(76 – 61)/61] in REME1-silenced plants versus noninoculated plants (Fig. 6B; P < 2 × 10−5), whereas the decrease for nad2-558 averaged 28% (Supplemental Fig. S4B; P < 3 × 10−7). We can consider the silencing efficiency of this latter experiment to be comparable to the original one because the variation in editing extents for both matR-1771 and nad2-558 are very close (25% versus 22% and 28% versus 29%, respectively). Again, the overall increase in editing extent of matR-1771 in REME1-silenced plants can be split into roughly equal parts between the effect of virus inoculation (13%; P < 2 × 10−5; Fig. 6B) and the specific silencing of REME1 (12%; P < 3 × 10−2; Fig. 6B). The same material was used to assess nad2-558 editing extent. Unlike matR-1771, there was no effect of virus inoculation on nad2-558 editing extent (compare not inoculated with empty vector controls in Supplemental Fig. S4B). In the previous experiment, the effect of virus inoculation on nad2-558 editing extent, although half the specific effect of REME1 silencing, was still significant (P < 2 × 10−2; Supplemental Fig. S4A). The reason for the absence of a virus inoculation effect on nad2-558 editing extent in the latter experiment is unclear but was also observed for orfX-552 (data not shown).
The main conclusion to be drawn from this new set of REME1-silencing experiments where an empty vector control was included is the reproducible and significant effect of REME1-specific silencing on the increase of matR-1771 editing extent. In addition, a decrease in editing extent of nad2-558 and orfX-552 was also confirmed in REME1-silenced plants.
REME1 Encodes a PPR-DYW Protein
REME1 encodes a PPR-DYW protein that is predicted to be targeted to mitochondria by both Predotar (Small et al., 2004) and TargetP (Emanuelsson et al., 2000) and thus is another example of a member of this subclass of PPR protein implicated in mitochondrial editing (Kim et al., 2009; Zehrmann et al., 2009; Verbitskiy et al., 2010). Although prediction softwares agree on REME1 targeting to mitochondria, and its effect on mitochondrial editing substantiates this prediction, REME1 (At2g03880) was reported to be targeted to chloroplasts (Lurin et al., 2004). Therefore, we examined REME1 subcellular localization in a transient assay where onion (Allium cepa) epidermal cells were bombarded with a vector expressing the first 100 amino acids from the REME1 N terminus attached to GFP. A mitochondrial marker was supplied by bombarding the epidermal cells with a vector expressing the cherry protein attached to a mitochondrial transit peptide. Our colocalization results clearly indicate that REME1 is targeted to mitochondria (Supplemental Fig. S5).
Like 80% of the Arabidopsis PPR-containing genes, the gene model for REME1 contains only one exon, a model that is supported by the isolation of R20567, a full-length cDNA in the RIKEN collection (Seki et al., 2002). The transit peptide length is predicted to be 76 amino acids by TargetP. The modular structure of REME1 (59-P-S-P-32-S-P-33-S-P-L2-S-4-E-E+-DYW) was defined by interrogating the PROSITE database (Fig. 7A) and is in very good agreement with the predicted structure by searching The Institute for Genomic Research (TIGR) protein families. The coordinates of the PPR motifs are shifted by two amino acids in the TIGRFAM output, because the PPR consensus used by TIGRFAM is shifted by two amino acids relative to the PROSITE consensus (Haft et al., 2001; de Castro et al., 2006).
Figure 7.
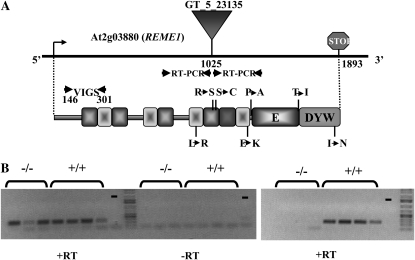
REME1 encodes a PPR-DYW protein. A, Localization of the T-DNA insertion in the At2g03880 locus and structure of the predicted REME1 protein. The 156-nucleotide region of At2g03880 RNA targeted by VIGS is delimited by facing arrows. Below the gene model is represented the modular structure of the predicted REME1 protein. Each PPR is symbolized by a square with different shading for each form of the PPR motif: P (gray), S (light gray), and L2 (dark gray). The coordinates of each PPR were defined by scanning the PROSITE and the TIGRFAM databases. Amino acid polymorphisms between the products encoded by Col and Ler alleles are indicated above and below the protein model. Only the amino acid polymorphisms implicated in the differential editing ability of the parental products are shown. The primers used to assess REME1 expression by RT-PCR in the GT_5_23135 mutant are represented by facing arrows upstream and downstream of the site of insertion. B, T-DNA insertion in GT_5_23135 prevents transcription from occurring downstream of the insertion, as shown by the absence of RT-PCR products in the homozygous mutants (right panel), whereas transcription is not affected upstream of the insertion (left panel).
Identification of a Ler Mutant with a T-DNA Insertion in REME1
We looked for an insertional mutation in At2g03880 (REME1) in collections of T-DNA mutants. Among several T-DNA insertional mutants present in the SALK T-DNA express browser (http://signal.salk.edu/cgi-bin/tdnaexpress), only GT_5_23135 presented an insertion within the REME1 coding sequence. Sequencing of the PCR amplicon obtained from the GT_5_23135 mutant allele confirmed that the insertion was located at nucleotide position 1,025, corresponding to the middle of the fourth P repeat (Fig. 7A). Monitoring REME1 expression by RT-PCR in GT_5_23135 homozygous mutants demonstrated that the T-DNA insertion prevents the accumulation of transcripts carrying REME1 sequence downstream of the insertion (Fig. 7B). If translated, the encoded product by the mutant allele will be shortened to a 384-amino acid protein versus 630 amino acids for the wild-type REME1 allele. The mutant plants show no detectable phenotypic defects and are absolutely indistinguishable from their wild-type siblings when grown in growth room conditions (data not shown).
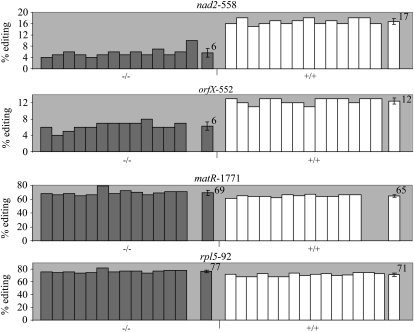
Assaying editing extent in the GT_5_23135 population showed a very significant reduction in editing extent for both nad2-558 and orfX-552 in homozygous mutants relative to the wild type (P < 9 × 10−19 and P < 2 × 10−15, respectively; Fig. 8). Two sites that showed the most pronounced increase in editing extent upon REME1 silencing, matR-1771 and rpl5-92 (Table I; Figs. 5 and 6), showed moderate but nevertheless significant increases in editing extent in homozygous mutants versus the wild type (P < 6 × 10−4 and P < 4 × 10−6, respectively; Fig. 8).
Figure 8.
Effect of T-DNA insertion in REME1 on editing extent in nad2-558, orfX-552, matR-1771, and rpl5-92. Editing extents were assayed by PPE. T-DNA insertion in REME1 coding sequence results in decreased editing of nad2-558 and orfX-552 in mutant GT_5_23135 (−/−; P < 9 × 10−19 and P < 2 × 10−15, respectively); an opposite effect of the insertion is observed on the editing extent of matR-1771 and rpl5-92, which are increased in GT_5_23135 (−/−; P < 6 × 10−4 and P < 4 × 10−6, respectively). On the right of each group is the average editing extent with sd.
The accession used to generate GT_5_23135 is Ler, which is the parent that exhibits low editing of nad2-558, Col being the more highly edited parent (Fig. 1). Knocking out the expression of the REME1 Ler allele, which has a weaker effect than the Col allele, has a lesser impact on editing extent than knocking down the expression of the Col allele by VIGS. Thus, the difference in editing extent between the mutant and the wild-type plants is smaller than the difference between the silenced plants and the control plants (11% versus 14% and 6% versus 14% for nad2-558 and orfX-552, respectively; Figs. 3, 4, and 8). Because the only available mutant is in the weaker Ler allele, we chose the silenced tissue over the mutant tissue in our screen for other mitochondrial sites controlled by REME1. The weaker REME1 Ler allele also made the mutant tissue less useful than silenced tissue for testing the inhibitory effect of REME1 on editing. Although the difference in editing extent observed between Col silenced tissues and control plants (empty vector) could reach 7% for matR-1771 (Fig. 6B), in GT_5_23135 homozygous mutants, the difference from wild-type plants is only 4% (Fig. 8). This rather moderate but significant increase in GT_5_23135 homozygous mutants can be rationalized based on the comparison of decrease in editing extent for nad2-558 and orfX-552 between silenced and mutant tissues (14% versus 11% and 14% versus 6%, respectively). The expected difference between the wild type and homozygous mutants for matR-1771 editing extent should range between 5.5% (7 × 11/14) and 3% (7 × 6/14), which is in rather good agreement with the observed 4%.
The REME1 Col Allele Boosts nad2-558 and orfX-552 Editing Extent in the Ler Background
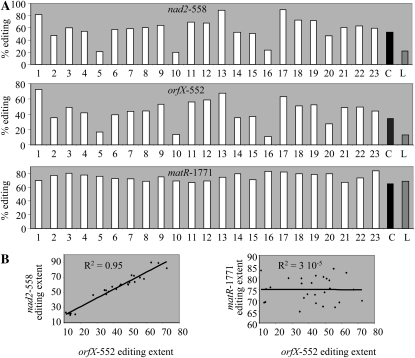
To further prove the function of REME1 in mitochondrial editing, we used a binary vector harboring the REME1 Col allele (high editor) under the control of a 35S promoter to transform the Ler accession (low editor). After floral dip transformation, the seeds were harvested and germinated on selective medium. Twenty-three putative transformants were transferred into soil and further assayed for editing extent in nad2-558, orfX-552, matR-1771, and rpl5-92. Among the 23 putative T0 Ler plants, three plants (individuals 5, 10, and 16) did not show a difference in editing extent for either nad2-558 or orfX-552 with the Ler parental accession (Fig. 9A). These plants represent either nontransformed material escaping from the selective medium or transgene-silencing events. The remaining 20 T0 plants show an editing extent for both nad2-558 and orfX-552 that exceeds the one exhibited by Ler, thus demonstrating that these plants are truly transgenic material and that both sites are controlled by REME1 (Fig. 9A). The very high and positive correlation (r2 = 0.95) between nad2-558 and orfX-552 editing extent in the T0 population supports the model of control of these two sites by the same mitochondrial editing factor (Fig. 9B).
Figure 9.
Overexpressing the REME1 Col allele in the Ler background has an effect on editing extent of nad2-558 and orfX-552 but not matR-1771. A, Editing extent was assayed by PPE on 23 T0 transgenic plants and the two parental accessions Col (C) and Ler (L). B, A very strong correlation between editing extent of nad2-558 and orfX-552 in T0 plants supports the control of both sites by REME1; the absence of correlation between editing extent of orfX-552 and matR-1771 does not support the inhibitory effect of REME1 on matR-1771 editing.
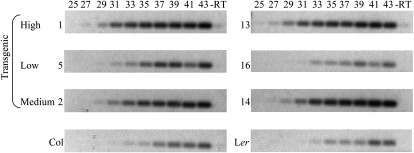
An interesting outcome of this experiment is the transgressive phenotype exhibited by the majority of the transgenic plants: 15 and 17 of the T0 plants show an editing extent that exceeds the one from Col for nad2-558 and orfX-552, respectively (Fig. 9A). Among those transgressive plants, individuals 1, 13, and 17 present the most extreme phenotype, with an orfX-552 editing extent twice the value of Col (Fig. 9A). The outperforming of the Col accession by the majority of the transgenic plants in relation to nad2-558 and orfX-552 editing extent could result from the 35S promoter used to drive the expression of the Col transgene. The 35S promoter sometimes drives high expression of the transgene, depending on the site of the transgene insertion in the genome. The variation in nad2-558 and orfX-552 editing extent observed in the T0 plants might thus result from variation in the level of expression of the transgene. We tested this hypothesis by assaying the level of REME1 expression by semiquantitative RT-PCR in some of the transgenic plants showing differential nad2-558 editing extent (Fig. 10). There is a very clear relationship between REME1 expression level and nad2-558 editing extent. The transgenic material exhibiting high nad2-558 editing extent exhibits a high level of REME1 expression; conversely, transgenic plants with a low nad2-558 editing extent also exhibit a low level of REME1 expression. Transgenic plants with an intermediate level of nad2-558 editing extent exhibit an intermediate level of REME1 expression (Fig. 10). A control experiment in which a reference gene expression was assayed in a similar way on the same material (Supplemental Fig. S6) validates the variation in REME1 expression level in transgenic material.
Figure 10.
Editing of nad2-558 is positively correlated with the level of REME1 expression in transgenic plants but not in the parental accessions. RT-PCR products of REME1 transcript were amplified and electrophoresed on 1% agarose gels after completion of the number of cycles indicated above each lane. –RT indicates a negative control in which the reverse transcriptase was omitted and was performed at 43 cycles. On the left of each gel is indicated the origin of the RNA used for the RT-PCR: 1, 13, 5, 16, 2, and 14 are T0 Ler plants expressing the REME1 Col allele and showing high, low, and medium levels of nad2-558 editing extent, as indicated. The same amount of template RNA (50 ng) was used for each RT reaction. In transgenic material, the correlation between the abundance of the REME1 RT-PCR product and the level of nad2-558 editing is readily observable at 29 and 31 cycles as follows: 1, 13 > 2, 14 > 5, 16. In contrast, no obvious difference in the abundance of REME1 RT-PCR is apparent between Col and Ler (similar intensity of the bands at 31 and 33 cycles).
We also assayed matR-1771 and rpl5-92 editing extent in the T0 population to test whether a decrease could be induced by the expression of the Col allele in the less-edited Ler background. A decrease in matR-1771 and rpl5-92 editing extent would be expected if REME1 has an inhibitory effect on these editing sites, as was suggested by our previous VIGS and mutant experiments. As apparent in Figure 9A, none of the T0 plants shows a matR-1771 editing extent less than what is observed in Ler. The same observation was made for rpl5-92 editing extent (data not shown). The absence of correlation between orfX-552 and matR-1771 editing extent casts doubt on a direct inhibitory function of REME1 on mitochondrial editing at that site (Fig. 9B). A negative correlation between orfX-552 and matR-1771 editing extent would be expected if the expression of REME1 results in inhibition of matR-1771.
REME1 Is Not Involved in nad2 trans-Splicing
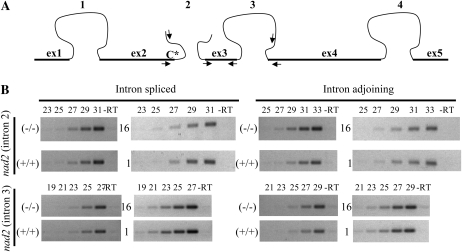
REME1 controls the editing of a C target in nad2, which encodes subunit 2 of the mitochondrial NADH ubiquinone oxidoreductase complex. Maturation of the nad2 transcript requires four RNA splicing events, one of which involves trans-splicing of independently transcribed RNAs and concerns the second intron (Fig. 11A). Given that the edited C at position 558 is only 20 nucleotides upstream of the trans-splicing site, we investigated whether editing of this C might affect trans-splicing efficiency. Two sets of plants with marked differential levels of nad2-558 editing extent were assayed for a possible difference in the amount of transcripts with or without the second intron spliced. Semiquantitative RT-PCR with appropriate primers was performed using RNAs from +/+ and −/− siblings from the GT_5_23135 population and two transgenic T0 plants, individuals 1 and 16, that exhibit a very different nad2-558 editing extent (Fig. 9A). The amount of cDNA used as a template for the quantitative PCR was adjusted to be similar between +/+ and −/− GT_5_23135 siblings (2.75 ng) and between T0 individuals 1 and 16 (3.5 ng). If a link existed between nad2-558 editing extent and trans-splicing of the second intron, then the amount of second intron-spliced cDNAs should be higher for +/+ versus −/− and for individuals 1 versus 16. Conversely, the amount of second intron-adjoining cDNAs should be lower for +/+ versus −/− and for individuals 1 versus 16. A more abundant template cDNA would in turn result in detection of the amplified product earlier during the cycles of amplification. No significant difference in the amount of intron-spliced and intron-adjoining cDNAs for the second intron was found between the low-edited and high-edited plants tested, indicating that REME1 does not indirectly control trans-splicing through its action on nad2-558 editing (Fig. 11B).
Figure 11.
Editing of nad2-558 and trans-splicing of the second intron are not coupled. A, Schematic representation of nad2 pre-mRNA and primers used in the semiquantitative RT-PCR. Exons are represented by thick bars and are drawn to scale. The maturation of nad2 involves three cis-splicing events (1, 3, and 4) and one trans-splicing event (2). Primers used in the semiquantitative RT-PCR are represented by facing arrows. Targeted C for editing at position 558 is represented by C*. B, Semiquantitative RT-PCR of different portions of nad2 transcript. Intron-spliced and intron-adjoining RT-PCR products for the second and third introns were amplified and electrophoresed on 2% agarose gels after a completed number of cycles indicated above each lane. –RT indicates a negative control in which the reverse transcriptase was omitted and was performed for each combination of primers at the maximum number of cycles (e.g. 31 for intron 2 spliced). On the left of each gel is indicated the origin of the RNA used for the RT-PCR: (−/−) and (+/+) are siblings from the GT_5_ 23135 population with or without the T-DNA insertion in REME1, and 16 and 1 are T0 Ler plants expressing the REME1 Col allele and showing low and high nad2-558 editing extent, respectively.
The similarity in the amount of starting template for the RT-PCR was validated by comparing the intron-spliced and intron-adjoining amplified products for the third intron of nad2. No detectable difference in the abundance of cDNAs with or without the third intron was observed between +/+ and −/− GT_5_23135 or between transgenic individuals 1 and 16 (Fig. 11B). Nevertheless, the slightly more abundant cDNA material for individuals 1 and 16 relative to +/+ and −/− (3.5 versus 2.75 ng, respectively) was enough to result in more abundant third intron-amplified products (e.g. intron-spliced product is absent at 19 cycles for −/− and +/+ but readily detectable for individuals 1 and 16; Fig. 11B). However, we cannot rule out an effect of REME1 on trans-splicing that is too small to be detected by semiquantitative RT-PCR. In mock experiments, we were able to detect, with good reproducibility, a 3- to 4-fold reduction of the trans-spliced product, a range comparable to the difference in nad2-558 editing extent between −/− and +/+ GT_5_23135 siblings and high-edited or low-edited transgenic individuals (Supplemental Fig. S7).
DISCUSSION
Mapping of Editing QTLs Can Lead to the Identification of Mitochondrial Site-Specific Editing Factors
Our identification of a mitochondrial RNA editing QTL in 2005 demonstrated the promise of natural variation for the identification of editing factors (Bentolila et al., 2005). Subsequently, our group (Bentolila et al., 2008) and another group (Zehrmann et al., 2008) identified a number of variations in the extent of RNA editing in plant mitochondria between ecotypes of Arabidopsis. Our identification of an editing QTL by precise mapping to a single gene, REME1, illustrates the power of this approach.
Because a number of PPR proteins have now been found to be chloroplast or mitochondrial editing factors (Kotera et al., 2005; Zehrmann et al., 2009), an alternative approach to identifying such factors is analysis of mutants in genes predicted to encode organelle-targeted PPR motif-containing proteins. Analyses of homozygous T-DNA PPR gene insertional mutants have identified a number of chloroplast editing factors (Hammani et al., 2009; Okuda et al., 2009; Robbins et al., 2009), as only 34 C targets need to be analyzed in putative mutants to determine if editing extent is affected. Reverse genetics is a far more cumbersome approach for the identification of editing factors in plant mitochondria, where there are over 500 C targets. PPR proteins are involved in other aspects of organelle gene regulation (Schmitz-Linneweber and Small, 2008; Stern et al., 2010); thus, laborious measurements of editing at hundreds of sites in a mutant could result in the unsatisfying conclusion that a particular PPR protein is not involved in editing. Recently, a reverse genetics approach identified five E-class PPR proteins involved in Arabidopsis mitochondrial RNA editing (Takenaka et al., 2010). However, the assay used in that study covers only 269 annotated editing sites, roughly just over half the total of the annotated mitochondrial sites. This incomplete coverage could explain why the mitochondrial editing factors identified were reported to control only one site. Furthermore, homozygous mutations in some PPR editing factors are likely to be embryo lethal. In contrast, by starting with a QTL that affects editing efficiency between Arabidopsis accessions, whatever gene is identified as encoding the QTL will definitely be involved in RNA editing, and even factors that are essential for embryo viability can be identified.
This study showed VIGS to be an excellent substitute for T-DNA insertional mutants whenever the background used to generate such mutants has a weak allele. VIGS should also become the method of choice to assay a number of candidate genes for mitochondrial editing, because it is much faster and less labor intensive than growing mutant populations and characterizing them. Furthermore, some putative T-DNA insertional mutants either lack the insertion or have a misannotated location.
REME1 Is a Site-Specific PPR-DYW Editing Factor
With the identification of REME1 as a PPR-DYW protein, the number of PPR proteins reported to be involved in organelle editing has now increased to 28. Among the 15 PPRs known to affect chloroplast editing, four belong to the E subclass (Kotera et al., 2005; Okuda et al., 2007; Chateigner-Boutin et al., 2008; Hammani et al., 2009) and 11 belong to the DYW subclass (Zhou et al., 2008; Cai et al., 2009; Hammani et al., 2009; Okuda et al., 2009; Robbins et al., 2009; Yu et al., 2009). All the mitochondrial editing factors identified so far but one also belong to the PLS subfamily: six mitochondrial editing factors are members of the DYW-PPR subclass (Kim et al., 2009; Zehrmann et al., 2009; Takenaka, et al., 2010; Tasaki et al., 2010; Verbitskiy et al., 2010; this study), while six members of the E subclass were shown to be necessary for mitochondrial editing (Takenaka, 2010; Takenaka et al., 2010). A member of the P subfamily denoted PPR596 was reported to control mitochondrial editing efficiency at partially edited sites (Doniwa et al., 2010).
Extensive fine mapping allowed us to identify a plant exhibiting a recombination event within the REME1 gene. This recombinant delineates the area responsible for the differential editing abilities of the product encoded by the two parental accessions, Col (high editor) and Ler (low editor). Seven nonsynonymous SNPs between Col and Ler lie in this area (Fig. 7) and thus are implicated either individually or in any possible combination in the differential editing ability of the accessions. However, the complementation analysis showed that the level of expression of REME1 also has an impact on the editing extent of the target sites. Three transgenic plants showed a level of editing extent almost twice the one found in Col, the accession providing the transgene. This observation is not surprising given the common low level of PPR proteins (Lurin et al., 2004) and the titration of editing factors when overexpressing a target site (Chateigner-Boutin and Hanson, 2002). Thus, the differential editing extent of nad2-558 and orfX-552 between Col and Ler could also come from a different level of REME1 expression in the two accessions. However, a differential REME1 expression level could not be detected between Col and Ler by our semiquantitative RT-PCR methodology (Fig. 10). This result does not rule out the involvement of REME1 expression in differential nad2-558 editing extent between the parental accessions, as a difference too small to be detected between Col and Ler might still have an impact on their nad2-558 editing ability.
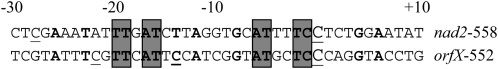
Among the 27 C residues edited in nad2 transcripts, REME1 silencing only affects one site, nad2-558. Similarly, orfX-552 and orfX-586 are the only C residues among 34 editing sites found on the orfX transcript whose editing extent appears to be reduced in REME1-silenced plants (orfX-586 was identified by bulk sequencing but not tested further by PPE; Table I). Like MEF1 (Zehrmann et al., 2009), MEF11 (Verbitskiy et al., 2010), OGR1 (Kim et al., 2009), and SLO1 (Sung et al., 2010), REME1 affects editing at multiple mitochondrial sites, thus reconciling the high number of mitochondrial targeted C residues in angiosperms (e.g. more than 500 in Arabidopsis) with the more limited number of PPRs belonging to the DYW or the E subclasses and predicted to be targeted to mitochondria. How a single PPR protein can recognize multiple C targets of editing is unknown but could be related to some sequence similarity in the cis-elements commonly found 5′ to C targets of editing. High-level transgenic expression of some individual chloroplast transcripts results in reduced editing of multiple C targets, a phenomenon thought to be due to saturation of a limiting amount of a trans-factor responsible for specifying editing at more than one site (Chateigner-Boutin and Hanson, 2002). The chloroplast sites that are coinhibited following transcript overexpression exhibit limited sequence similarity, sometimes requiring introduction of gaps into the alignment (Chateigner-Boutin and Hanson, 2002). Similarly, the number of conserved nucleotides is limited to seven in the 15 nucleotides upstream of the edited C in nad2-558 and orfX-552 (Fig. 12). However, despite the limited nucleotide conservation, when purine or pyrimidine conservation was considered in the 15-nucleotide sequence upstream of the targeted C in chloroplasts, the targeting of multiple editing sites by single factors could be rationalized (Hammani et al., 2009). On the other hand, some mitochondrial editing sites sharing the same editing factor show a high sequence similarity in the 15 nucleotides upstream of the edited C; for instance 60% of the nucleotides from −1 to −15 relative to the edited C are identical between nad4-449 and nad9-328, both of which are under the control of SLO1 (Sung et al., 2010). The occurrence of high sequence similarity in the cis-element upstream of the edited C was also reported for cox3-422 and nad4-124, the targets sites of MEF11, and allowed the identification of a third target site by an in silico search for similar upstream sequences (Verbitskiy et al., 2010). No sequence similarity was found in the sequences surrounding the edited C residues whose editing extent is negatively regulated by REME1.
Figure 12.
Alignment of sequences around editable C residues and putative cis-elements in the sites controlled by REME1. Thirty nucleotides upstream and 10 nucleotides downstream of the target C are shown. The target C is shown in larger font, and the edited C residues are underlined. Letters in boldface represent conserved nucleotides, and blocks of conserved nucleotides are shaded.
Multiple Factors May Control Editing Efficiency of Mitochondrial C Targets
Several lines of evidence indicate that single site-specificity factors do not completely control editing of individual mitochondrial C targets. Earlier, we reported that differential editing extent in Col and Ler at some sites was under the control of at least two QTLs located at different positions on the genetic map (Bentolila et al., 2008). Zehrmann et al. (2009) reported a total lack of editing in mef1 mutants for only two of the three mitochondrial sites under the control of MEF1. In mef1 mutants, the editing extent of nad2-1160 is reduced to 20%. Perhaps another factor is able to provide the function needed for low-level editing of nad2-1160. In addition, the level of editing extent in the study by Zehrmann et al. (2009) was assayed by bulk sequencing, which is rather inaccurate, particularly in evaluating low editing extent. For instance, no edited peak for orfX-552 is detectable in the electrophoretogram of the REME1-silenced pool RT-PCR product (Supplemental Fig. S2), even though the editing extent was measured to be 26% by PPE. Similarly, editing extent of nad4-C433 in the rice ogr1 mutant, a null mutant, is only reduced to 17%, but in that study, a reliable measurement was achieved by sequencing cDNA clones (Kim et al., 2009). In the REME1 insertional mutant we studied, the location of the insertion results in a predicted truncated product of 384 amino acids that lacks the E and DYW domains and three of the nine PPRs. Even if this truncated protein accumulates, it is very unlikely to be functional; the E motif has been shown to be essential for several of the chloroplast RNA editing factors (Okuda et al., 2007, 2009). Thus, the residual editing activity in REME1 homozygous mutant plants for both nad2-558 and orfX-552 may be explained by the existence of another factor active at these sites (Fig. 8).
In contrast to some of the mitochondrial mutants in PPR editing factors, all known null mutant alleles in chloroplast editing factors result in a complete lack of editing of the chloroplast sites they control (Hammani et al., 2009; Okuda et al., 2010). AccD-C794 is the only reported example of a chloroplast editing site under the control of two different PPR-DYW proteins, RARE1 and AtECB2 (Robbins et al., 2009; Yu et al., 2009). Nevertheless, RARE1 and AtECB2 do not exhibit redundant function; null mutation in either one completely abolishes the editing of the targeted C independently of the other.
REME1 Is a Positive Regulator and Might Be a Negative Regulator of Mitochondrial RNA Editing
All the approaches used in this study, positional cloning, VIGS, T-DNA insertional mutant, and complementation analysis, prove that REME1 is a positive regulator of mitochondrial editing for both nad2-558 and orfX-552. Unexpectedly, we observed that silencing of REME1 results in increased editing at multiple C targets in Arabidopsis mitochondria. Thirteen targets were verified by the precise PPE assay as significantly increased in editing efficiency in mitochondrial coding regions in plants subjected to VIGS of REME1. However, repetition of the VIGS experiment incorporating an empty vector control showed that about half of the increase in editing extent for matR-1771 was due to the virus inoculation, while the remaining half was due to specific REME1 silencing. This significant increase in editing extent upon REME1-specific silencing was further supported by an increase observed in T-DNA insertional mutants. While these two lines of evidence support an inhibitory role for REME1 in mitochondrial editing, the complementation analysis did not. Even transgenic plants outperforming the Col accession (high editor) with regard to nad2-558 and orfX-552 editing extent did not show any editing extent reduction in the expected sites, matR-1771 and rpl5-92. The observed absence of any decrease in matR-1771 and rpl5-92 editing extent in plants overexpressing REME1 would be expected if more than one editing factor positively controls these sites. This hypothesis is consistent with the fact that the REME1 knockout mutant still exhibits residual editing extent for nad2-558 and orfX-552, implying that another editing factor can operate on these sites. According to this theory of multiple factors, only one of the editing factors that increases matR-1771 and rpl5-92 editing extent is subjected to the REME1 inhibiting effect; thus, a reduction in REME1 by VIGS would release only the factor susceptible to the REME1 inhibitor effect, hence increasing editing extent of matR-1771 and rpl5-92 in REME1-silenced tissues. On the other hand, overexpressing REME1 would have no further inhibitory effect on editing, because the first factor susceptible to REME1 is already inhibited in the normal physiological state and the second, REME1-independent factor would still be free to function; hence, there is no change in matR-1771 and rpl5-92 editing extent.
Site-specific inhibition of editing by protein factors has previously been reported only once in plant organelles. PPR591, the only plant editing factor that belongs to the P subfamily of PPR-containing proteins, is a negative regulator of several mitochondrial sites that lie mostly on the rps3 transcript (Doniwa et al., 2010). There is also precedence for editing inhibition in the apolipoprotein B (apoB) RNA editing system in mammals. Two RNA-binding proteins, GRY-RBP and CUGBP2, have been shown to inhibit apoB mRNA editing in vitro (Anant et al., 2001; Blanc et al., 2001). In addition, antisense inhibition of expression of GRY-RBP or CUGBP2 in McA cells led to a 2- to 3-fold increase in endogenous apoB RNA editing, suggesting that both these factors may participate in the apoB editing complex as negative regulators in vivo. In contrast to GRY-RBP and CUGBP2, REME1 is able to promote editing at some sites while inhibiting editing at other sites. Future work may identify additional organelle editing factors with dual inhibitory and stimulatory activities. In fact, such activity could have been overlooked in some of the studies reported so far. For example, editing was assayed only by bulk sequencing and only for 120 mitochondrial C targets in mef1 mutants (Zehrmann et al., 2009), so that C targets with enhanced editing may not have been detected.
The existence of PPR proteins that inhibit editing efficiency may explain a previously unexpected result we obtained when studying the editing QTL that affects ccb206-406 in Col and Ler (Bentolila et al., 2005). We observed that the less edited phenotype was dominant: the F1 exhibited the same editing extent at ccb206-406 as the parent with the lower editing efficiency. We suggested that editing QTLs might encode inhibitory factors, which has now been demonstrated by REME1 silencing by VIGS. The mechanistic process by which REME1 is able to inhibit editing requires further investigation. REME1 might exert its inhibitory effect through binding to and sequestering of editing factors, as has been shown for GRY-RBP (Blanc et al., 2001). Alternatively, REME1 might directly compete with editing factors for binding to the cis-elements upstream of the C targets. We favor the first model, as the absence of sequence similarity in the surrounding sequences around the edited C residues suggests that REME1 does not exert its inhibitory effect by a direct interaction with the target RNAs.
Our finding raises the question of what functional roles in gene expression may be played by factors that inhibit plant mitochondrial RNA editing. Is the inhibitory effect on editing of particular C residues merely an accidental by-product of a complex system that edits over 500 C residues? Or does inhibition of editing of particular transcripts sometimes serve to increase the expression of mitochondrial protein variants? Although incompletely edited transcripts are known to be translated in plant mitochondria, no useful protein variation has ever been detected as a result (Lu and Hanson, 1994; Lu et al., 1996; Phreaner et al., 1996; Williams et al., 1998). Proteins derived from incompletely edited transcripts are usually thought to be degraded or nonfunctional. However, the presence of variants derived from incompletely edited transcripts has been tested rigorously for only a limited number of plant mitochondrial proteins. Further studies will be needed to discover whether reduced editing at particular C residues may play a role in the generation of protein diversity in plant mitochondria.
MATERIALS AND METHODS
Plant Materials and Growing Conditions
The population of Arabidopsis (Arabidopsis thaliana) RILs used to map editing QTLs and their parental accessions, Col and Ler, have been described in a previous work (Bentolila et al., 2008). One of the RILs, CS1985, which is Ler/Ler at the REME1 locus, was crossed with Col to generate the fine-mapping population. The F1 hybrid was allowed to self-pollinate to produce the F2 progeny. The F2 recombinants between 1113K and 1220K were grown in 16 h of light/8 h of dark under full-spectrum fluorescent lights in a growth room at 26°C. These growing conditions were also used for the Col line expressing GFP, kindly donated by Dominique Robertson (North Carolina State University), the F3 progeny from F2 recombinants, and the T-DNA insertional mutants. Line GT_5_23135 was obtained from the John Innes Center SM lines collection (Tissier et al., 1999).
Screening of Fine-Mapping Recombinants
Genomic DNA extraction of the 691 F2 plants followed the same protocols as described by Bentolila et al. (2008) but was scaled for the use of the Genogrinder 2000 apparatus (Spex CertiPrep) that allowed high-throughput DNA extraction in microtiter plate format. Screening for recombinants was done with the cleaved-amplified polymorphism markers 1027K and 1276K (Supplemental Table S1). PCR was performed with the Taq PCR Master Mix Kit (Qiagen). After amplification, the PCR products were digested by the appropriate restriction enzyme (Supplemental Table S1), and the digestion products were separated on a 1% agarose gel. All the markers used to genotype the recombinants are listed in Supplemental Table S1. For the sequencing markers, the PCR products after purification by using Exosap (USB) underwent bulk sequencing at the Cornell University Life Sciences Core Laboratory Center. Evaluation of the editing extent of the 11 recombinants for nad2-558 was done on three F3 progeny selected to be homozygous recombinant.
Identification of T-DNA Insertional Mutants
The T-DNA population was segregating for the presence of the T-DNA. Plants grown in soil were genotyped with the following primers: the wild-type allele was amplified with GT_5_23135-F1 (5′-AGGAGGGGTTGGAGTCTGAT-3′) and GT_5_23135-R1 (5′-GAACCCTACAAGCACCAAGC-3′). The mutant allele was amplified with GT_5_23135-F1 and Ds5-1 (5′-ACGGTCGGGAAACTAGCTCTAC-3′) or with GT_5_23135-R1 and Ds3-1 (5′-ACCCGACCGGATCGTATCGGT-3′). Both mutant amplicons were sequenced in order to precisely determine the location of the T-DNA insertion in REME1. Genotyping the T-DNA populations was performed by amplifying the mutant and wild-type alleles with BioMix Red (Bioline). The PCR products were then visualized on a 1% agarose gel.
VIGS
The protocol and materials used for VIGS are detailed by Robbins et al. (2009). The silencing fragment for REME1 was amplified by PCR using the following gene sequence tag primers designed by the CATMA database (Crowe et al., 2003): REME1-F1 (5′-TGGATTCATTGCAAAGTCATGG-3′) and REME-R1 (5′-CATTAACCAGAAACATCATTGGTCG-3′). The PCR product was cloned into the pCR8-GW-TOPO vector (Invitrogen), its sequence was verified, and then it was transferred to the silencing vector by using Gateway technology (Invitrogen). The empty vector control was built by first cutting the pCR8-GW-TOPO vector with EcoRI, which releases the amplicon, and then self-ligating the vector with a T4 DNA ligase (Fermentas). The minimal and only vector component of the self-ligated pCR8-GW-TOPO vector was transferred via Gateway technology to the silencing vector, which after validating its sequence was designated the empty vector in silencing experiments.
Screening of Mitochondrial Transcripts for Editing Sites Affected by REME1 Silencing
The screening of the mitochondrial transcripts in REME1-silenced and control plants relied on the production of cDNA for the entire set of mitochondrial genes. The methods and primers used for RT-PCR have been described previously (Bentolila et al., 2008); however, we had to design new primers for some of the genes in order to cover the entirety of the published sites (Supplemental Table S2). New primers were also developed to amplify UTRs and introns (Supplemental Table S2). Bulk sequencing of the RT-PCR products with the same primers used for the amplification of the cDNAs was performed at the Cornell University Life Sciences Core Laboratory Center. Whenever the amplicon was too long to obtain the full sequence with the primers used to amplify it, internal primers were designed (Supplemental Table S2).
Measurement of Editing Values
All the techniques and primers used to measure editing values have been reported in a previous work (Bentolila et al., 2008). The only difference concerns the use of PPE oligonucleotides with a fluorescent tag, 5′-hexachlorofluorescein, purchased from Integrated DNA Technologies to measure the editing extent of nad2-558, orfX-144, and rpl2-212. 5′-Hexachlorofluorescein-tagged extension products were detected by a Typhoon 9400 imager (GE Healthcare). Editing extent at other sites was detected by radiolabeled oligonucleotides as described previously (Bentolila et al., 2008). PPE primers are listed in Supplemental Table S3.
Intracellular Localization of REME1
The first 300 nucleotides of REME1 starting from the AUG codon were amplified and cloned into the pCR8-GW-TOPO vector (Invitrogen). After validating the sequence, the amplicon was transferred to the pEarleygate103 vector (35S-GW-GFP; Earley et al., 2006) by Gateway technology. Plasmid DNA was transiently introduced into onion (Allium cepa) epidermal cells by using a helium-driven particle accelerator (PDS-1000; Bio-Rad) according to the manufacturer’s instructions. After bombardment, the onion peels were kept for 36 h on Murashige and Skoog plates in the dark. The localization of the expressed proteins was visualized with a laser scanning confocal microscope (Leica SP2). The mitochondrial marker was developed by Nelson et al. (2007).
Plant Transformation
The coding sequence of the REME1 allele from Col, including the stop codon, was amplified and cloned into a Gateway-converted pBI121 binary vector (Wade Heller, personal communication). This construct was transformed into the Ler accession via Agrobacterium tumefaciens through the standard floral dip protocol.
Semiquantitative RT-PCR Conditions
RNA extracted by a combination of TRIzol and the PureLink RNA minikit (Invitrogen) was cleared from contaminating trace amounts of DNA by using the Turbo DNA-free kit (Ambion). Quantification of RNA was performed with a nanodrop spectrophotometer (Nanodrop Technologies). cDNA was produced with SuperScript III reverse transcriptase (Invitrogen) and gene-specific primers (Supplemental Table S4). Semiquantitative PCR was performed using the primers listed in Supplemental Table S4.
Statistical Analysis
Comparison of the ratio of cis-spliced and trans-spliced cDNAs and editing extent averages between REME1-silenced and control plants was performed by Student’s t test (two-tail t test with equal variances; Microsoft Excel).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Bulk sequencing assay can detect differential editing extent in REME1-silenced tissues.
Supplemental Figure S2. An example of differential editing between pools of control and silenced plants assayed by bulk sequencing.
Supplemental Figure S3. VIGS of REME1 results in increased editing of sites found in rpl5.
Supplemental Figure S4. Editing extent of nad2-558 is decreased in REME1-silenced plants.
Supplemental Figure S5. REME1 is targeted to mitochondria.
Supplemental Figure S6. Semiquantitative RT-PCR of a reference gene (At4g26410) shows no difference in expression level between transgenic lines and parental accessions.
Supplemental Figure S7. Semiquantitative RT-PCR is able to detect a 3- to 4-fold reduction in nad2 second intron-spliced or intron-adjoining product.
Supplemental Table S1. Markers used in REME1 fine mapping.
Supplemental Table S2. New primers used in mitochondrial gene screening.
Supplemental Table S3. New PPE primers used in this report.
Supplemental Table S4. Primers used for semiquantitative RT-PCR.
Supplementary Material
Acknowledgments
Some of this work comprised an undergraduate biology honors thesis submitted by W.K. We are grateful to Wade Heller for excellent technical advice on VIGS and to John Robbins for the identification of a REME1 insertional mutant. We acknowledge valuable technical help from Huijun Yang for use of the biolistic particle delivery system and from Amir Sattarzadeh for confocal microscopy. We thank Catherine Colas des Francs-Small for providing us with the primer sequences used in the semiquantitative RT-PCR.
References
- Anant S, Henderson JO, Mukhopadhyay D, Navaratnam N, Kennedy S, Min J, Davidson NO. (2001) Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing: CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J Biol Chem 276: 47338–47351 [DOI] [PubMed] [Google Scholar]
- Bentolila S, Chateigner-Boutin AL, Hanson MR. (2005) Ecotype allelic variation in C-to-U editing extent of a mitochondrial transcript identifies RNA-editing quantitative trait loci in Arabidopsis. Plant Physiol 139: 2006–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Elliott LE, Hanson MR. (2008) Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics 178: 1693–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V, Navaratnam N, Henderson JO, Anant S, Kennedy S, Jarmuz A, Scott J, Davidson NO. (2001) Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing. J Biol Chem 276: 10272–10283 [DOI] [PubMed] [Google Scholar]
- Cai W, Ji D, Peng L, Guo J, Ma J, Zou M, Lu C, Zhang L. (2009) LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol 150: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Hanson MR. (2002) Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol Cell Biol 22: 8448–8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, et al. (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Covello PS, Gray MW. (1990) Differences in editing at homologous sites in messenger RNAs from angiosperm mitochondria. Nucleic Acids Res 18: 5189–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe ML, Serizet C, Thareau V, Aubourg S, Rouzé P, Hilson P, Beynon J, Weisbeek P, van Hummelen P, Reymond P, et al. (2003) CATMA: a complete Arabidopsis GST database. Nucleic Acids Res 31: 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34: W362–W365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniwa Y, Ueda M, Ueta M, Wada A, Kadowaki K, Tsutsumi N. (2010) The involvement of a PPR protein of the P subfamily in partial RNA editing of an Arabidopsis mitochondrial transcript. Gene 454: 39–46 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Giegé P, Brennicke A. (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci USA 96: 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. (2000) Functions and mechanisms of RNA editing. Annu Rev Genet 34: 499–531 [DOI] [PubMed] [Google Scholar]
- Gray MW, Covello PS. (1993) RNA editing in plant mitochondria and chloroplasts. FASEB J 7: 64–71 [DOI] [PubMed] [Google Scholar]
- Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. (2001) TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res 29: 41–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. (2009) A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa H. (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res 31: 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernould M, Suharsono S, Litvak S, Araya A, Mouras A. (1993) Male-sterility induction in transgenic tobacco plants with an unedited atp9 mitochondrial gene from wheat. Proc Natl Acad Sci USA 90: 2370–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Yang JI, Moon S, Ryu CH, An K, Kim KM, Yim J, An G. (2009) Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J 59: 738–749 [DOI] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T. (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Lu B, Hanson MR. (1994) A single homogeneous form of ATP6 protein accumulates in petunia mitochondria despite the presence of differentially edited atp6 transcripts. Plant Cell 6: 1955–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Wilson RK, Phreaner CG, Mulligan RM, Hanson MR. (1996) Protein polymorphism generated by differential RNA editing of a plant mitochondrial rps12 gene. Mol Cell Biol 16: 1543–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM, Neckermann K, Igloi GL, Kössel H. (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol 251: 614–628 [DOI] [PubMed] [Google Scholar]
- Mower JP, Palmer JD. (2006) Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol Genet Genomics 276: 285–293 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K. (2002) The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics 268: 434–445 [DOI] [PubMed] [Google Scholar]
- Okuda K, Chateigner-Boutin AL, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. (2009) Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Hammani K, Tanz SK, Peng L, Fukao Y, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. (2010) The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J 61: 339–349 [DOI] [PubMed] [Google Scholar]
- Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. (2007) Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci USA 104: 8178–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. (2008) On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol 25: 1120–1128 [DOI] [PubMed] [Google Scholar]
- Phreaner CG, Williams MA, Mulligan RM. (1996) Incomplete editing of rps12 transcripts results in the synthesis of polymorphic polypeptides in plant mitochondria. Plant Cell 8: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JC, Heller WP, Hanson MR. (2009) A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15: 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Small ID, Peeters N. (2000) The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Sung TY, Tseng CC, Hsieh MH. (2010) The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J 63: 499–511 [DOI] [PubMed] [Google Scholar]
- Takenaka M. (2010) MEF9, an E-subclass pentatricopeptide repeat protein, is required for an RNA editing event in the nad7 transcript in mitochondria of Arabidopsis. Plant Physiol 152: 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Verbitskiy D, Zehrmann A, Brennicke A. (2010) Reverse genetic screening identifies five E-class PPR proteins involved in RNA editing in mitochondria of Arabidopsis thaliana. J Biol Chem 285: 27122–27129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki E, Hattori M, Sugita M. (2010) The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFc transcript. Plant J 62: 560–570 [DOI] [PubMed] [Google Scholar]
- Tillich M, Funk HT, Schmitz-Linneweber C, Poltnigg P, Sabater B, Martin M, Maier RM. (2005) Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J 43: 708–715 [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JD. (1999) Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsudzuki T, Wakasugi T, Sugiura M. (2001) Comparative analysis of RNA editing sites in higher plant chloroplasts. J Mol Evol 53: 327–332 [DOI] [PubMed] [Google Scholar]
- Verbitskiy D, Zehrmann A, van der Merwe JA, Brennicke A, Takenaka M. (2010) The PPR protein encoded by the LOVASTATIN INSENSITIVE 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. Plant J 61: 446–455 [DOI] [PubMed] [Google Scholar]
- Williams MA, Tallakson WA, Phreaner CG, Mulligan RM. (1998) Editing and translation of ribosomal protein S13 transcripts: unedited translation products are not detectable in maize mitochondria. Curr Genet 34: 221–226 [DOI] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN. (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Zehrmann A, van der Merwe JA, Verbitskiy D, Brennicke A, Takenaka M. (2008) Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion 8: 319–327 [DOI] [PubMed] [Google Scholar]
- Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. (2009) A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Cheng Y, Yap A, Chateigner-Boutin AL, Delannoy E, Hammani K, Small I, Huang J. (2008) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J 58: 82–96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.