Abstract
Seeds of grain legumes are important energy and food sources for humans and animals. However, the yield and quality of legume seeds are limited by the amount of sulfur (S) partitioned to the seeds. The amino acid S-methylmethionine (SMM), a methionine derivative, has been proposed to be an important long-distance transport form of reduced S, and we analyzed whether SMM phloem loading and source-sink translocation are important for the metabolism and growth of pea (Pisum sativum) plants. Transgenic plants were produced in which the expression of a yeast SMM transporter, S-Methylmethionine Permease1 (MMP1, YLL061W), was targeted to the phloem and seeds. Phloem exudate analysis showed that concentrations of SMM are elevated in MMP1 plants, suggesting increased phloem loading. Furthermore, expression studies of genes involved in S transport and metabolism in source organs, as well as xylem sap analyses, support that S uptake and assimilation are positively affected in MMP1 roots. Concomitantly, nitrogen (N) assimilation in root and leaf and xylem amino acid profiles were changed, resulting in increased phloem loading of amino acids. When investigating the effects of increased S and N phloem transport on seed metabolism, we found that protein levels were improved in MMP1 seeds. In addition, changes in SMM phloem loading affected plant growth and seed number, leading to an overall increase in seed S, N, and protein content in MMP1 plants. Together, these results suggest that phloem loading and source-sink partitioning of SMM are important for plant S and N metabolism and transport as well as seed set.
Sulfur (S) is a critical macronutrient for plant growth, and its assimilation is important for the production of the essential amino acids Cys and Met as well as the antioxidant glutathione (GSH; Saito, 2004; Jander and Joshi, 2010). Besides being a component of functional and structural molecules, S is crucial to the development of sink tissue, such as flowers and fruits, and its availability affects protein synthesis and the quality of seeds (Higgins et al., 1986; Ufaz and Galili, 2008). Acquisition of S occurs predominantly via the uptake of sulfate (SO42−) from the soil, which is mediated by sulfate transporters (Smith et al., 1995; Davidian and Kopriva, 2010). Following uptake, sulfate is either transiently stored in vacuoles or assimilated into reduced S compounds. The S reduction pathway is well characterized and conserved among different plant species. However, the major site of S reduction varies and might be dependent on environmental conditions and plant species (Rennenberg et al., 1979; Brunold and Suter, 1989; Hopkins et al., 2005; Hawkesford and De Kok, 2006; Hell et al., 2010). Following translocation of sulfate in the xylem transpiration stream, S reduction predominantly occurs in mature source leaves (Rennenberg et al., 1979; Bourgis et al., 1999; Lappartient et al., 1999; Hopkins et al., 2005), although in legumes, S reduction also takes place in other organs. For example, in pea (Pisum sativum), S assimilation has been localized to roots, as all the enzymes involved in sulfate reduction to Cys are present (Brunold and Suter, 1989). Regardless of the location of primary S assimilation, sink translocation of reduced S predominantly occurs via the phloem. It might be loaded into the phloem of source leaf minor veins following synthesis, or it may derive from xylem-phloem transfer along the translocation path when S assimilation takes place in roots (Buchner et al., 2004).
GSH, Cys, and Met have been suggested to be long-distance transport forms of organic S (Rennenberg et al., 1979; Bonas et al., 1982; Lappartient et al., 1999; Davidian and Kopriva, 2010), but in a study by Bourgis et al. (1999), it was shown that the amino acid S-methyl-Met (SMM) is a key contributor to phloem S transport in wheat (Triticum aestivum), canola (Brassica napus), legumes, and other plant species. In these plants, it is suggested that SMM is synthesized from Met by the enzyme Met methyltransferase (MMT) in source leaves and loaded into the phloem for transport to seeds (Bourgis et al., 1999; Lee et al., 2008). In the seed coat, SMM is recycled back to Met via homo-Cys methyltransferase (HMT; Gallardo et al., 2007) and amino acids are finally released into the seed apoplast, from where they are taken up by the embryo/cotyledons for development and storage product accumulation.
This work addresses the importance of phloem loading and source-sink partitioning of SMM for plant metabolism and growth. In recent years, much work has been invested in elucidating the role of Suc transporters in phloem loading and carbon movement to sinks (Tegeder et al., 2011). However, no information is available on the function of transporters in long-distance movement of organic nitrogen (N) and S, although a variety of amino acid, GSH, GSH derivative, and GSH conjugate transporters have been identified (Jasinski et al., 2003; Zhang et al., 2004; Bouvier et al., 2006; Rentsch et al., 2007; Tegeder et al., 2007, 2011; Tan et al., 2008). Since there are no known plant SMM transporters, in this study, a high-affinity yeast SMM transporter, S-Methylmethionine Permease1 (MMP1; Rouillon et al., 1999), was expressed in the pea phloem and seeds to determine the importance of SMM partitioning in plant physiology. We demonstrate that increasing phloem loading of SMM affects S metabolism in leaves as well as S uptake and assimilation in roots. In addition, N assimilation and translocation to sinks were altered in MMP1 plants. Changes in S and N metabolism and partitioning resulted in higher seed numbers and influenced the S-N ratio and storage protein profile in MMP1 seeds. Together, the data suggest that increasing long-distance transport of organic S positively affects plant metabolism and productivity. Our study raises the possibility that SMM might play a key role in balancing plant S and N status.
RESULTS
Molecular and Phenotypic Analyses of MMP1 Plants
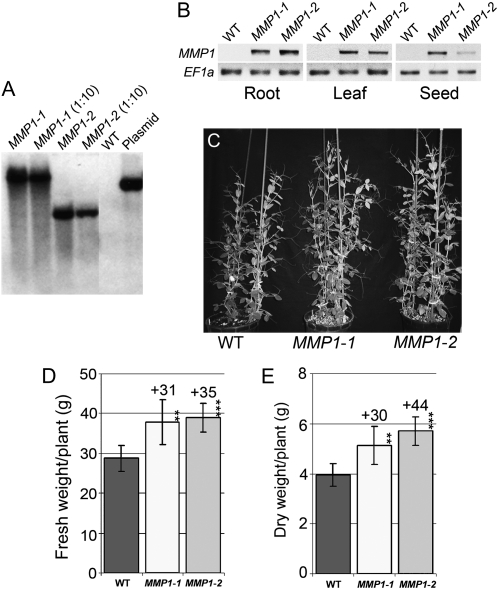
In general, transformation of pea plants is extremely difficult and has been successful in very few laboratories worldwide (Grant et al., 1995; McPhee et al., 2004; Rolletschek et al., 2005). Nonetheless, we were able to produce transgenic pea lines, MMP1-1 and MMP1-2, expressing yeast MMP1 (Rouillon et al., 1999) under the control of the Arabidopsis (Arabidopsis thaliana) Amino Acid Permease1 (AAP1) promoter (Hirner et al., 1998). In pea, this promoter targets gene expression to the phloem throughout the plant and to seeds (Tegeder et al., 2007). Southern-blot analysis resolved that both lines carry a single copy of the gene construct (Fig. 1A). RNA expression analysis established that MMP1 is expressed throughout the transgenic plants, including roots, leaves, and developing seeds (Fig. 1B), consistent with AAP1p-GUS studies in pea (Tegeder et al., 2007). Phenotypic analysis of vegetative growth illustrated that MMP1 plants were taller and displayed a bushier appearance compared with wild-type plants (Fig. 1C). When examining the shoot biomass of 5-week-old MMP1 plants, fresh and dry weight were increased by up to 35% and 44%, respectively, compared with the wild type (Fig. 1, D and E).
Figure 1.
Molecular and phenotypic analyses of MMP1 plants. A, Southern-blot analysis using HindIII-digested genomic DNA from MMP1-1, MMP1-2, and wild-type (WT) plants. Vector pTKan with the gene construct AAP1p-MMP1 (plasmid) was used as a positive control. Both transgenic lines carry a single copy of the gene construct. B, Analysis of MMP1 expression in MMP1-1 and MMP1-2 lines using RT-PCR. EF1α was amplified as a control for equivalent amounts of RNA used and even gel loading. C, Photograph of 5-week-old MMP1 and wild-type plants. Two plants were grown in each pot. D, Shoot fresh weight of 5-week-old MMP1 and wild-type plants (n = 6). E, Shoot dry weight of 5-week-old MMP1 and wild-type plants (n = 6). Results are presented as means ± sd. Asterisks indicate significant differences from the wild type detected by Student’s t test: ** P < 0.01, *** P < 0.001.
SMM Phloem Loading and S Metabolism Are Increased in MMP1 Plants
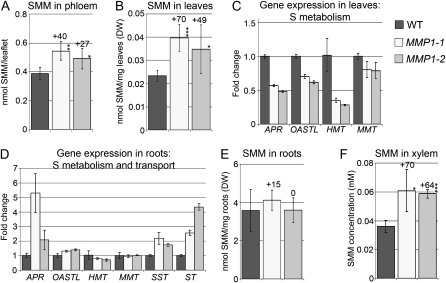
Phloem exudates from MMP1 leaflets were analyzed, and SMM concentrations were 27% to 40% higher in MMP1-1 and MMP1-2 plants compared with wild-type plants (Fig. 2A), suggesting an increase in SMM phloem loading and source-sink translocation in transgenic plants. Concurrently, whole leaf tissue SMM levels were increased by up to 70% (Fig. 2B). To decipher whether the positive changes in SMM content in leaves are due to increased SMM synthesis, expression levels of genes involved in S assimilation in leaves were analyzed using quantitative reverse transcription (RT)-PCR. Surprisingly, genes important for the synthesis of Cys from sulfate, such as adenosine 5′-phosphosulfate reductase (APR) and OAS (thiol)-lyase (OASTL), were down-regulated (Fig. 2C). Furthermore, transcript levels of MMT, the enzyme converting Met to SMM, were unchanged, whereas the expression of HMT, responsible for the synthesis of Met from SMM, was decreased (Fig. 2C). These findings indicate that reductive S assimilation in the leaf tissue does not account for the increased SMM content in MMP1 leaves and that the organic S compounds might be derived from roots.
Figure 2.
SMM levels and expression of genes involved in S metabolism and transport in MMP1 plants. A, SMM concentrations in phloem exudates of MMP1 and wild-type (WT) plants (n = 4). B, SMM levels in leaves (n = 4). DW, Dry weight. C and D, Fold change in gene expression of APR, OASTL, HMT, MMT, and sulfate transporters SST and ST in leaves (C) and roots (D). For OASTL, MMT, HMT, SST, and ST expression analyses, degenerated primers were used, since pea homologs are unknown. E, SMM levels in roots (n = 4). F, SMM concentrations in xylem sap (n = 3). Results are presented as means ± sd. Asterisks indicate significant differences from the wild type detected by Student’s t test: * P < 0.05, ** P < 0.01, *** P < 0.001.
Therefore, S assimilation in root tissue was examined. In fact, transcript levels of S assimilatory genes, especially APR, were increased in MMP1 versus wild-type roots (Fig. 2D). Furthermore, HMT expression was slightly down-regulated, while MMT1 RNA levels were unchanged. This suggests that de novo S assimilation is increased in MMP1 roots and that lower amounts of SMM are converted back to Met via HMT. When analyzing the expression levels of sulfate transporters, SST (Krusell et al., 2005) and ST (Smith et al., 1995), it was found that both transporters are up-regulated by up to 4-fold in MMP1 roots (Fig. 2D), indicating increased S uptake.
HPLC analysis was then performed to determine SMM levels in roots and in the xylem, and the results showed that SMM was unchanged in whole MMP1 root tissue (Fig. 2E). However, the SMM concentration in the xylem sap was increased by 64% to 70% in MMP1 compared with wild-type plants (Fig. 2F), indicating increased S uptake and assimilation in roots and translocation to the shoot.
Amino Acid Phloem Loading and Leaf N Metabolism Are Increased in MMP1 Plants
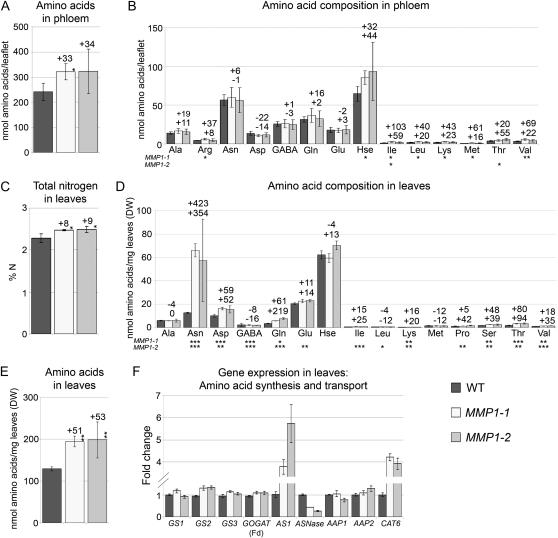
As S and N assimilation are tightly coordinated in plants (Wang et al., 2003; Dubousset et al., 2009) and S metabolism and partitioning in MMP1 plants are altered, N metabolism and transport are also likely to be affected. Consequently, analysis of phloem amino acid levels was performed and showed that the amounts of total amino acids in the MMP1 phloem were elevated by about 30% (Fig. 3A), with an increase in Arg, homo-Ser, Ile, Leu, Lys, Met, Thr, and Val (Fig. 3B). In addition, expression of Cationic Amino Acid Transporter6 (CAT6), a putative companion cell transporter (Brady et al., 2007) that uses most of the above-listed amino acids as substrates (Hammes et al., 2006), was strongly up-regulated in leaves (Fig. 3F), supporting increased phloem loading of amino acids.
Figure 3.
N levels in MMP1 phloem exudate and leaves, and expression of genes involved in amino acid synthesis and transport in leaves. A, Total free amino acid levels in the phloem of MMP1 compared with wild-type (WT) plants. B, Amino acid composition and concentration in the phloem exudates. Hse, Homo-Ser. C, Total N levels in leaves. D, Total free amino acid levels in leaves. DW, Dry weight. E, Amino acid composition and concentration in leaves. F, Fold change in gene expression of pea GS1, GS2, and GS3, AS1, and amino acid transporters AAP1, AAP2, and CAT6. Degenerated primers were used for ferredoxin-dependent Glu synthase [GOGAT (Fd)] and ASNase. Results are presented as means ± sd. Asterisks indicate significant differences from the wild type detected by Student’s t test (n = 4): * P < 0.05, ** P < 0.01, *** P < 0.001.
To resolve whether the increased amounts of phloem amino acids derive from elevated assimilation in the leaf, total N and amino acid levels and expression of genes involved in N assimilation in leaves were examined. The results showed that amounts of total N and free amino acids were increased in MMP1 leaves by up to 9% and 53%, respectively (Fig. 3, C and E). Analysis of single amino acid concentrations demonstrated that eight out of 20 amino acids were significantly elevated in both transgenic lines, with the strongest increase in Asn, by up to 400% (Fig. 3D). Along with this, there was a slightly increased expression of genes involved in the synthesis of Gln (GS) and Glu (GOGAT), which are the first products of N assimilation, an up to 6-fold up-regulation of AS1, important for Asn synthesis, and down-regulation of asparaginase (ASNase), involved in Asn catabolism (Fig. 3F).
Xylem But Not Root Amino Acid Levels Are Affected in MMP1 Plants
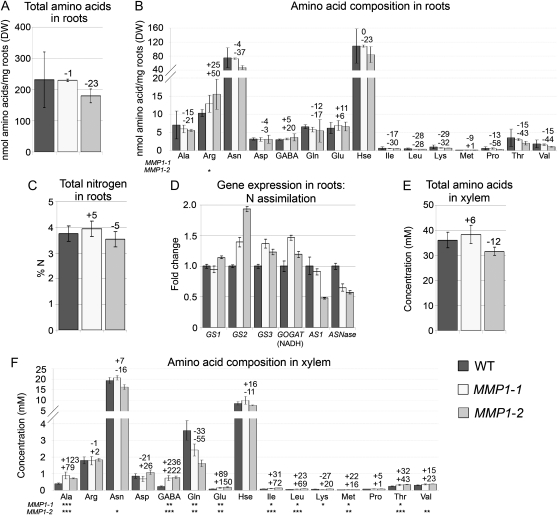
To determine whether alterations in root amino acid metabolism and export contribute to the observed increase in phloem and leaf amino acid contents in MMP1 plants, we analyzed total root N levels and amino acid concentrations in the root and xylem. It was found that amounts and profiles of free amino acids, as well as total N, were unchanged in MMP1 roots compared with the wild type (Fig. 4, A–C). Expression analysis demonstrated that genes involved in the initial assimilation of inorganic N, GS and GOGAT, were up-regulated in MMP1 roots, suggesting increased N reduction (Fig. 4D). Transcript levels of AS1 were unchanged or decreased depending on the MMP1 line, and ASNase expression was down-regulated in both lines (Fig. 4D).
Figure 4.
Analysis of N levels in root and xylem of MMP1 plants, and root gene expression studies. A, Total free amino acid levels in roots of MMP1 compared with wild-type (WT) plants (n = 4). DW, Dry weight. B, Amino acid composition and concentration in roots (n = 4). Hse, Homo-Ser. C, Total N levels in roots (n = 4). D, Fold change in gene expression of pea GS1, GS2, and GS3, and AS1. Degenerated primers were used for NADH-dependent Glu synthase [GOGAT (NADH)] and ASNase. E, Total free amino acid levels in xylem sap (n = 3). F, Amino acid composition and concentration in xylem sap (n = 3). Results are presented as means ± sd. Asterisks indicate significant differences from the wild type detected by Student’s t test: * P < 0.05, ** P < 0.01, *** P < 0.001.
Xylem sap analysis revealed that total amino acid levels were similar in overexpressors versus the wild type, but concentrations of single amino acids were altered (Fig. 4, E and F). Gln was decreased in both overexpressors, and Asn and Lys were reduced or not changed dependent on the MMP1 line. Glu, γ-aminobutyric acid (GABA), Ala, Leu, Val, Ile, Met, and Thr levels were up-regulated in MMP1 xylem sap compared with the wild type (Fig. 4F), suggesting increased export of these amino acids out of the root.
N Levels Are Increased in MMP1 Seeds, While S Content Is Unchanged
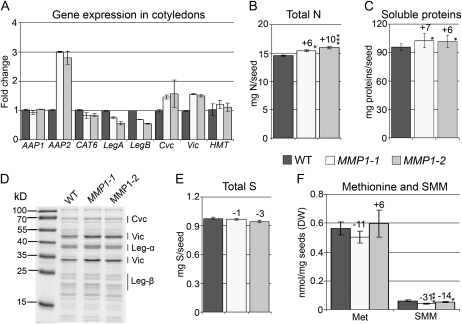
To investigate if increased phloem SMM and amino acid levels result in increased S and N distribution to individual pea seeds, MMP1 cotyledons were analyzed. Expression of AAP2, a transporter involved in the import of amino acids into pea cotyledons (Tegeder et al., 2000), was up-regulated in MMP1 seeds (Fig. 5A), indicating increased N uptake. This is consistent with changes in the total N content, which was elevated by up to 10% in MMP1 compared with wild-type seeds (Fig. 5B). In addition, total soluble protein levels were elevated by 6% to 7% in MMP1 seeds (Fig. 5C).
Figure 5.
Expression of genes involved in seed amino acid transport and seed storage protein synthesis, and total seed N, S, Met, SMM, and protein levels. A, Fold change in the expression of amino acid transporters AAP1, AAP2, and CAT6 and of the storage protein genes LegA, LegB, Cvc, and Vic as well as HMT in developing cotyledons of MMP1 compared with wild-type (WT) plants. B, Total N levels in dry seeds. C, Amounts of soluble seed proteins. D, Analysis of soluble seed proteins using a SDS-PAGE gel. Leg-α, Legumin α-subunits; Leg-β, legumin β-subunits. E, Total S levels in dry seeds. F, Concentrations of Met and SMM in developing MMP1 seeds. DW, Dry weight. Results are presented as means ± sd. Asterisks indicate significant differences from the wild type detected by Student’s t test (n = 4): * P < 0.05, *** P < 0.001.
In general, about 65% to 85% of pea seed proteins are present in the form of globulin storage proteins, with predominant localization to the seed cotyledons (Tzitzikas et al., 2006). Depending on the S content, globulins are further categorized into S-rich legumins and S-poor vicilins and convicilins, and increased availability of S positively affects seed levels of S-rich proteins (Higgins et al., 1986; Sexton et al., 1998; Tabe et al., 2002; Ufaz and Galili, 2008). To examine if S-poor and S-rich proteins were affected, soluble proteins including globulins were analyzed by SDS-PAGE. The stained gel showed that levels of convicilins and vicilins were increased in MMP1 seeds, while S-rich legumins were unaltered (Fig. 5D). For further confirmation, the expression of storage protein genes for legumins (LegA and LegB), convicilin (Cvc), and vicilin (Vic) was examined using RNA from developing cotyledons. While LegA and LegB expression was decreased, transcript levels of both Cvc and Vic were increased in MMP1 cotyledons (Fig. 5A), consistent with the changes in MMP1 convicilin and vicilin levels. When testing the total S content in seeds, no changes were observed (Fig. 5E), which concurs with unchanged levels of S-rich seed proteins in MMP1 plants. Analysis of Met and SMM in developing MMP1 seeds showed that Met concentrations were unchanged while SMM levels were decreased (Fig. 5F). However, expression of HMT, encoding for the enzyme converting SMM back to Met, was unchanged in developing MMP1 seeds (Fig. 5A).
Increased SMM Phloem Loading Positively Affects Seed Number
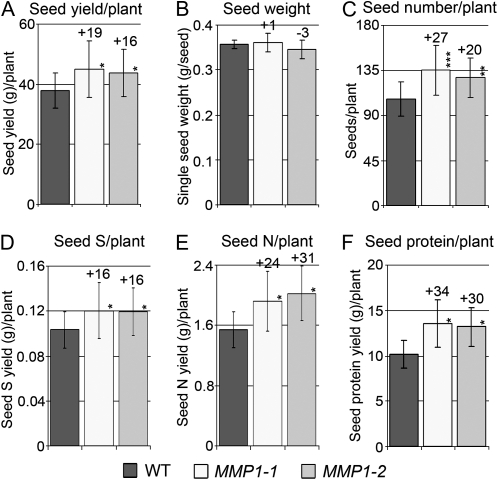
While SMM phloem levels were increased in MMP1 plants, seed S levels were unchanged. This raises the question of where the S is partitioned to. Analyses of yield-related parameters showed an increase in total seed yield of up to 19% in MMP1 plants (Fig. 6A). Seed weight was unchanged in transgenic compared with wild-type plants, but seed number per plant was positively affected by 20% to 27% (Fig. 6, B and C).
Figure 6.
Analysis of yield-related parameters in MMP1 and wild-type (WT) plants. A, Total seed yield per MMP1 or wild-type plant. B, Average single seed weight from eight plants. C, Total number of seeds per plant. D, Total seed S yield per plant. E, Total seed N yield per plant. F, Total seed protein yield per plant. Results are presented as means ± sd. Asterisks indicate significant differences from the wild type detected by Student’s t test (n = 8): * P < 0.05, ** P < 0.01, *** P < 0.001.
As described above, MMP1 seed S levels were unaltered, whereas N and protein levels were increased (Fig. 5, B–E). When calculating total seed S amount per plant, it was found that the overall S levels per MMP1 plant were increased by 16% compared with the wild type (Fig. 6D), whereas total seed N and protein yields per plant were elevated by up to 31% and 34%, respectively, dependent on the transgenic line (Fig. 6, E and F).
DISCUSSION
While the role of transporters in source-sink translocation of organic S in plants still needs to be addressed, it has recently been suggested that SMM is a major player in S phloem transport (Bourgis et al., 1999; Gallardo et al., 2007; Lee et al., 2008). This work links the function of organic S transporters with plant N and S metabolism and provides evidence for the importance of SMM phloem loading and source-sink distribution for plant growth and seed development.
Phloem Loading of SMM Affects Source S Metabolism
Targeted expression of the yeast SMM transporter MMP1 to the phloem throughout the pea plant led to an increase in SMM concentrations in the leaf phloem sap (Fig. 2A), which in turn affected S root uptake, SMM synthesis in root and leaves, and SMM translocation to the shoot. The concept that increased SMM phloem loading exhibits feedback is supported by the following: (1) up-regulation of gene expression of S uptake and assimilation in the root; (2) increased levels of SMM in the xylem; and (3) down-regulation of genes involved in leaf S assimilation (Fig. 2, C, D, and F). Most enzymes involved in S assimilation are transcriptionally regulated, with high S levels down-regulating gene expression (Lappartient et al., 1999; Vauclare et al., 2002; Davidian and Kopriva, 2010). This is consistent with the observed high levels of SMM in MMP1 leaves and the down-regulation of S assimilation genes (Fig. 2, B and C). On the other hand, it was shown that S deficiency leads to up-regulation of S transporter and biosynthesis genes (Vauclare et al., 2002; Hirai et al., 2003; Davidian and Kopriva, 2010). In MMP1 roots, the concentration of SMM was unchanged, suggesting a steady-state pool of the root amino acid. However, since xylem concentrations were strongly increased, the actual SMM levels in root cells might be decreased, triggering an up-regulation of uptake and synthesis to keep up with enhanced export rates (Fig. 2, D–F). Elevated levels of SMM in MMP1 xylem, leaves, and phloem (Fig. 2, A, B, and F) as well as the overall increase in seed S yield (Fig. 6D) further support an increase in S uptake, assimilation, and source-sink translocation in the transgenic plants. While it seems evident that improved SMM phloem loading or SMM phloem concentrations cause the changes in metabolism and transport in leaves and roots (see below), regulation of theses processes is probably highly complex. It most likely involves nutrient (SMM) sensing and signaling mechanisms in the different plant organs as well as shoot-root communication processes (Scheible et al., 2004; Walch-Liu et al., 2005; Nikiforova et al., 2006; Hirai and Saito, 2008; Miller et al., 2009; Ruffel et al., 2010; Yi et al., 2010). Future feeding experiments with radiolabeled SMM, as well as integrated “omics” studies, might help to identify some of the regulatory components leading to the observed changes.
SMM Phloem Levels Influence N Metabolism and Transport
It is well known that plant N and S metabolisms are tightly coordinated (Wang et al., 2003; Nikiforova et al., 2006). Our data demonstrate that increased SMM source-sink partitioning goes in hand with increased root S uptake and SMM synthesis and xylem transport to the leaf (Fig. 2, D and F). At the same time, substantial changes in leaf amino acid content and export were observed (Fig. 3). The changes in leaf and phloem N seem to be due to increased N assimilation in both MMP1 roots and leaves, as indicated by the up-regulation of N assimilation genes in root and leaf and the increased levels of specific amino acids in the xylem, leaf, and phloem (Figs. 3 and 4). Root N and amino acid steady-state levels are unchanged in MMP1 plants compared with the wild type, suggesting that, consistent with the expression data, increased synthesis is keeping up with enhanced export rates of amino acids (Fig. 4). Xylem levels of members of the Asp family, but only those of the homo-Ser branch, including Thr, Ile, and Met, were increased, while amounts of other amino acids of the Asp family, Asn and Lys, were not changed or decreased (Fig. 4F). This coincides with the observed up-regulation of SMM production and export in roots, which also requires the homo-Ser path for its synthesis (Fig. 2, D and F). In addition, export of Glu, GABA, and amino acids of the pyruvate family (Ala, Val, and Leu) that rely on Glu for synthesis was increased. On the other hand, xylem levels of Arg, Pro, Gln, and Asn were unchanged or decreased (Fig. 4F). Together, this suggests that in MMP1 plants, increased N assimilation in roots and export of amino acids is primarily focused on Glu, the pyruvate family, and the homo-Ser branch of the Asp family, including SMM (Fig. 2F), rather than on the synthesis and export of amides, Asp, Lys, and amino acids of the Glu family.
On the contrary, the synthesis of Asn, Asp, and Gln seems to be up-regulated in MMP1 leaves, as indicated by their increased levels and by gene expression studies (Fig. 3, D and F). In MMP1 plants, leaf N supply and content are increased (Fig. 3E; see below), and much of the N seems to be channeled into Asn synthesis. Asn might be transiently stored until usage (Genix et al., 1990; Tsai and Coruzzi, 1990; Lam et al., 1994) or it may be needed for improved synthesis of homo-Ser (Bauer et al., 1977; Murray and Cordova-Edwards, 1984) and other amino acids of the Asp family, including Lys for translocation to sinks (Fig. 3, B and D). Homo-Ser is one of the main phloem amino acids in pea (Urquhart and Joy, 1981; Rochat and Boutin, 1991; Fig. 3D). While homo-Ser levels in MMP1 compared with wild-type leaves (and xylem) were unchanged, phloem levels were increased, suggesting higher synthesis and export of this amino acid out of the leaves. Other amino acids that were increased in the MMP1 phloem are Met, Ile, Thr, Lys, Val, and Leu. With the exception of Lys, the accompanying increased levels of these amino acids in the xylem support the idea that they are root derived and probably transferred from the xylem to the phloem for sink supply (Pate et al., 1975; Atkins et al., 1979; for review, see Van Bel, 1990). However, it cannot be excluded that some improved assimilation of the Asp and pyruvate family amino acids in leaves might also contribute to their increased phloem content. In any case, increased loading of Val, Leu, Ile, Thr, and Met into the phloem is further consistent with the up-regulation of CAT6 (Fig. 3F), which in Arabidopsis is expressed in companion cells (Brady et al., 2007) and mediates the transport of the respective amino acids (Hammes et al., 2006). Elevated levels of amino acids in the phloem also suggest increased delivery of amino N to MMP1 seeds and uptake into the pea cotyledons. This is in agreement with the up-regulation of the N-inducible amino acid importer AAP2 (Tegeder et al., 2007) as well as the increased seed total N and protein levels (Fig. 5, A–C).
Taken together, these results support the notion that changes in SMM transport and metabolism affect N assimilation in root and leaves, probably by feedback regulation mechanisms, leading to improved source-sink translocation of N. Elevated levels of N in MMP1 leaves, phloem, and single seeds, as well as the enhanced overall seed N and protein yields (Fig. 6, E and F), further point to an increase in N uptake by MMP1 roots.
S and N Metabolism and Transport Are Important for Seed Development
Studies with Arabidopsis hmt2 mutants, in which the conversion of Met to SMM is inhibited, demonstrated that these plants overaccumulate SMM in source leaves and subsequently load more SMM into the phloem for transport to the seeds (Lee et al., 2008). In hmt2 seeds, the Met levels were increased due to almost normal enzymatic activity of HMTs. Medicago transcriptome and proteome studies provided further evidence for the importance of SMM long-distance transport for Met and protein synthesis in seeds (Gallardo et al., 2007). It was found that during the development of Medicago seed coats, de novo synthesis of Met was decreased while conversion from SMM to Met was up-regulated, suggesting that the Met required for protein synthesis during seed filling was mainly derived from SMM. In addition, the synthesis of S-adenosyl-Met, a precursor for SMM synthesis, was dramatically down-regulated during seed development, indicating that SMM synthesis might not occur in the seed coat at a later seed stage and that the SMM required for Met synthesis is mainly phloem derived (Gallardo et al., 2007). Surprisingly, in mature MMP1 seeds, the amounts of total S and S-rich proteins were unchanged, although SMM phloem levels were increased (Fig. 5, D and E). This suggests that S uptake into MMP1 seeds is not changed even though MMP1 is expressed in seeds (Fig. 1B; Tegeder et al., 2007). It further supports the idea that phloem SMM levels are a function of MMP1 transport activity in the leaf phloem rather than in the seeds (Tilsner et al., 2005). In addition, decreased SMM and unchanged Met levels were observed in developing MMP1 seeds (Fig. 5F), indicating that probably more SMM was converted to Met to obtain sufficient Met for the higher demand for storage protein synthesis in MMP1 seeds. Somewhat contradictory, gene expression of HMT, encoding the enzyme responsible for the conversion of SMM to Met, was unchanged in MMP1 seeds (Fig. 5A). However, near-normal HMT enzyme activity has been reported in Arabidopsis hmt2 mutant seeds, suggesting posttranscriptional control of HMT genes or alternative pathways for SMM conversion to Met (Lee et al., 2008).
While the mechanism is unclear, increased SMM phloem transport seems to affect seed number in MMP1 plants (Fig. 6, A–C). This is consistent with field experiments showing that fertilization of S alone increases seed yield (Malhi et al., 2007). In addition, responses to S application are greater when sufficient amounts of N are applied (Hocking et al., 1987; Zhao et al., 1999; Schonhof et al., 2007). Therefore, in MMP1 plants, the synergistic increase of S and N assimilates in the phloem or a change in the S-N ratio might trigger increased seed set (Fismes et al., 2000). However, it could also be caused by other factors, such as changes in carbon-N ratio, which has been hypothesized to be important for sink development (Lawlor, 2002). Furthermore, a number of studies indicate that SMM plays very diverse roles in plant physiology and might also function in the methylation of plant metabolites, maintenance of S-adenosyl-Met homeostasis, and cell membrane damage control under abiotic stress (Giovanelli et al., 1980; Kocsis et al., 2003; Rácz et al., 2008). Therefore, in addition to being an essential long-distance transport form of reduced S, SMM may act as a signal to regulate plant processes, including metabolism and growth. However, this is somewhat in disagreement with studies performed with Arabidopsis and maize (Zea mays) mmt mutants, in which the SMM cycle was abolished and no SMM was produced (Tagmount et al., 2002; Kocsis et al., 2003). The mmt plants grew and reproduced normally, with similar seed S content compared with the wild type. These findings suggest that SMM is not essential for plant growth and development and that other S compounds may substitute for SMM in S transport (Kocsis et al., 2003). Nevertheless, when SMM phloem levels are increased by either repressing HMT2 in the leaves or expressing SMM transporters in the phloem, changes in seed number and protein amounts (this study) or protein quality (higher Met, no growth effects; Lee et al., 2008) can potentially be made. The observed differences in alterations with respect to seeds in hmt2 mutants and MMP1 expressors might be species dependent or may be based on other factors that could potentially be identified by a detailed comparison of Arabidopsis lines (hmt2 versus MMP1) using combined transcriptome and metabolome studies.
CONCLUSION
Plant response to increased phloem loading and source-sink distribution of SMM seems to be managed by a number of strategically important processes. First, to accommodate the elevated demand for S caused by improved phloem loading, acquisition of sulfate by the root is increased and the S assimilatory machinery is up-regulated. Second, N metabolism in root and leaf is induced, probably to provide the precursors for the synthesis of organic S compounds and to deliver the required N for increased growth and sink development. Third, higher SMM phloem levels lead to increased seed/sink development and S partitioning to the increased number of seeds per plant, rather than to individual seeds. Whether altered SMM phloem transport positively affects seed numbers directly or indirectly is unknown. However, modifications in phloem metabolite levels might provide feed-forward regulation signals that finally lead to improved sink set. Fourth, elevated amino acid levels in the phloem lead to increased N uptake into seeds (and finally more protein), facilitated by amino acid importers that are up-regulated by the higher N levels.
Since the discovery of SMM as a long-distance transport form of reduced S (Bourgis et al., 1999), several studies have demonstrated that SMM is important for plant S metabolism, although it might not be essential for plant growth (Kocsis et al., 2003; Gallardo et al., 2007; Lee et al., 2008). Yet, the question remains: What advantage does SMM have over other S-containing compounds in terms of S transport? In this study, the SMM concentrations in the phloem and xylem were less than 0.2% of the total amino acids, and manipulation of SMM phloem levels promoted dramatic changes in S as well as N metabolism and transport in MMP1 pea plants. Perhaps one potential benefit of using SMM as a long-distance S transport compound lies in the fact that SMM might provide the signal for regulating plant S and N metabolism. The homeostasis of SMM, or a tightly coregulated key metabolite, might serve as a sensitive switch that, when tampered with, will trigger complex responses in whole plant S and N uptake, assimilation, and transport to regain a balanced optimum for plant growth and development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Pea (Pisum sativum ‘Bohatyr’) plants were grown in 2-gallon pots with a mixture of peat (60%), pumice (20%), and sand (20%; SunGro Horticulture) in a greenhouse or growth chamber under 14 h of light and with photosynthetically active radiation between 400 and 500 μmol photons m−2 s−1 at the soil surface. Greenhouse temperatures ranged from 18°C to 22°C during the day and from 15°C to 18°C at night. Growth chambers were at 22°C/18°C during the day/night. Plants were fertilized weekly with 20:20:20 (N:phosphate:potassium) fertilizer (J.R. Peters). Developing side branches were removed for a defined source-sink position and easy management of the plants. For confirmation of results, materials were harvested and analyzed from independently grown sets of plants. For biochemical analyses and gene expression studies, leaves, roots, and developing seeds (75%–80% relative water content) were collected from at least six 10-week-old plants. Plant samples were collected 6 h after the beginning of the light period and over a maximum of 3 h. Tissues were sampled concurrently for wild-type and transgenic plants. Dry seeds were harvested from eight desiccated plants.
Construct Preparation and Plant Transformation
The coding sequence of the MMP1 gene (YLL061W; Rouillon et al., 1999) from Saccharomyces cerevisiae was amplified using PCR with MMP1-specific primers (Supplemental Table S1) and cloned into pTKan (Hajdukiewicz et al., 1994) carrying the AAP1 promoter from Arabidopsis (Arabidopsis thaliana; Hirner et al., 1998). The construct was introduced into the Agrobacterium tumefaciens strain AGL1, and pea plants were transformed (Grant et al., 1995). Homozygous transgenic plants were identified by PCR using genomic DNA and the primers listed in Supplemental Table S1. For Southern-blot analysis, standard procedures were applied.
Gene Expression Analyses
RNA was isolated from root, leaf, and seed tissue of homozygous MMP1 plants (Pélissier and Tegeder, 2007) and used for RT-PCR with two independent sets of primers (Tan et al., 2008; Supplemental Table S1). Amplification products were analyzed by gel electrophoresis. Pea elongation factor 1α (EF1α; X96555) reverse transcription products were amplified (for primers, see Supplemental Table S1) as a control for even amounts of total cDNAs used for the different PCRs.
cDNA synthesis and quantitative RT-PCR were performed following Sanders et al. (2009). The expression of genes of S and N transport and metabolism and storage proteins was analyzed. Primers were designed based on published pea sequences (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/). In cases where specific pea gene sequences were unknown, degenerated primers were designed along known cDNAs from at least two legume species. Supplemental Table S2 lists the genes analyzed (including accession numbers), the primers used, and the gene accessions and legume species when degenerated primers were applied. For the pea homolog to Arabidopsis CAT6, a partial clone was produced using PCR with degenerated primers (Supplemental Table S1) performed on pea cotyledon cDNAs. The 921-bp partial clone was then sequenced, and gene-specific primers were designed and used for quantitative RT-PCR analysis (Supplemental Table S2). Fold changes in gene expression were calculated by comparison with expression of EF1α (X96555) and ubiquitin (PUB1-4; L81139, L81140, L81141, L81142). Representative results of three independent experiments using EF1α as the control gene are shown.
Collection and Analysis of EDTA-Phloem Exudates and Xylem Sap
Phloem exudates were obtained from a total of 16 plants using two leaflets per plant (Urquhart and Joy, 1981). The exudates of four plants were pooled and analyzed. Xylem sap was collected from nine 6-week-old plants (Bauer et al., 1977), and pools from three plants were analyzed.
Elemental Nutrient and Metabolite Analyses
Total C and N contents were analyzed according to Sanders et al. (2009). For total S measurements, seed samples were combusted with a Costech ECS 4010 elemental analyzer (Costech Analytical Technologies) with a dual reactor configuration (Fry et al., 1992). SO2 gases were separated with a 0.8-m gas chromatography column at 105°C and analyzed for total area (Brenna et al., 1997). Protein extractions and SDS-PAGE were done after Sanders et al. (2009). Protein concentrations were determined using the NanoOrange kit (Invitrogen).
Free amino acids were extracted from 3 mg of lyophilized tissue with 80% methanol at 70°C for 15 min of shaking at 1,000 rpm, with α-aminobutyric acid (7 nmol mg−1 tissue) added as an internal standard. After centrifugation and collection of supernatants, the pellets were reextracted using 20% methanol as described above and centrifuged. The supernatant was collected, combined with that from the previous step, and dried to a pellet under vacuum (SpeedVac concentrator; Savant Instruments). The resulting pellet was dissolved in HPLC-grade water. Samples for amino acid measurements were derivatized with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate using the AccQ-fluor kit (Waters) according to the manufacturer’s instructions. Derivatized samples were filtered (0.22 μm; SunSri) to remove debris and particles. An amino acid mixture (catalog no. A2407; Sigma) with added Gln, SMM, Orn, GABA, Arg, Lys, His, ammonium chloride, and homo-Ser was used as a standard.
HPLC analysis was performed using a Waters 2695 separation module with column heater, autosampler, and Empower2 software. Separation was performed on a Waters 4.6- × 150-mm, 3.5-μm SunFire C18 column equipped with a Sentry guard column at 37°C. Solvent A (acetonitrile), solvent B (HPLC-grade water), solvent C (AccQ-Taq Eluent A, diluted), and solvent D (100 mm sodium acetate, 783 μL L−1 triethylamine, and 250 μL L−1 phosphoric acid, pH 5.8) were used at 1.35 mL min−1 flow rate with the following gradient: 0 to 0.5 min, 100% C; 0.5 to 16.8 min, linear gradient to 96.5% C and 3.5% B; 16.8 to 29.85 min, to 96.5% D and 3.5% B; 29.85 to 34.2 min, to 95% C and 5% B; 34.2 to 35.68 min, to 91% C and 9% B; 35.68 to 49 min, to 83% C and 17% B; 49 to 57.35 min, immediate change to 60% B and 40% A; 57.35 to 64.5 min, immediate change to 100% C; 64.5 to 72 min, 100% C. Amino acids were detected using a Waters 2475 multi λ fluorescence detector with excitation at 280 nm and emission at 395 nm. Amino acid concentrations were calculated by comparing peak areas with amino acid standards.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used for molecular analyses other than quantitative RT-PCR.
Supplemental Table S2. Primers employed for quantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank Dr. Raymond Lee (Washington State University) and the Washington State University Stable Isotope Core for the isotope analyses and Dr. Sanja Roje (Washington State University) for advice about HPLC analyses. We are very grateful for the support from our greenhouse manager Chuck Cody.
References
- Atkins CA, Pate JS, Layzell DB. (1979) Assimilation and transport of nitrogen in nonnodulated (NO3-grown) Lupinus albus L. Plant Physiol 64: 1078–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Joy KW, Urquhart AA. (1977) Amino acid metabolism of pea leaves: labeling studies on utilization of amides. Plant Physiol 59: 920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas U, Schmitz K, Rennenberg H, Bergmann L. (1982) Phloem transport of sulfur in Ricinus. Planta 155: 82–88 [DOI] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL, et al. (1999) S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 11: 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, Linka N, Isner JC, Mutterer J, Weber AP, Camara B. (2006) Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18: 3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brenna JT, Corso TN, Tobias HJ, Caimi RJ. (1997) High-precision continuous-flow isotope ratio mass spectrometry. Mass Spectrom Rev 16: 227–258 [DOI] [PubMed] [Google Scholar]
- Brunold C, Suter M. (1989) Localization of enzymes of assimilatory sulfate reduction in pea roots. Planta 179: 228–234 [DOI] [PubMed] [Google Scholar]
- Buchner P, Takahashi H, Hawkesford MJ. (2004) Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J Exp Bot 55: 1765–1773 [DOI] [PubMed] [Google Scholar]
- Davidian JC, Kopriva S. (2010) Regulation of sulfate uptake and assimilation: the same or not the same? Mol Plant 3: 314–325 [DOI] [PubMed] [Google Scholar]
- Dubousset L, Abdallah M, Desfeux AS, Etienne P, Meuriot F, Hawkesford MJ, Gombert J, Ségura R, Bataillé MP, Rezé S, et al. (2009) Remobilization of leaf S compounds and senescence in response to restricted sulphate supply during the vegetative stage of oilseed rape are affected by mineral N availability. J Exp Bot 60: 3239–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fismes J, Vong PC, Guckert A, Frossard E. (2000) Influence of sulfur on apparent N-use efficiency, yield and quality of oilseed rape (Brassica napus L.) grown on a calcareous soil. Eur J Agron 12: 127–141 [Google Scholar]
- Fry B, Brand W, Mersch FJ, Tholke K, Garritt R. (1992) Automated analysis system for coupled δ13C and δ15N measurements. Anal Chem 64: 288–291 [Google Scholar]
- Gallardo K, Firnhaber C, Zuber H, Héricher D, Belghazi M, Henry C, Küster H, Thompson R. (2007) A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: evidence for metabolic specialization of maternal and filial tissues. Mol Cell Proteomics 6: 2165–2179 [DOI] [PubMed] [Google Scholar]
- Genix P, Bligny R, Martin JB, Douce R. (1990) Transient accumulation of asparagine in sycamore cells after a long period of sucrose starvation. Plant Physiol 94: 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanelli J, Mudd S, Datko A. (1980) Sulfur amino acids in plants. Miflin B, ed, The Biochemistry of Plants Academic Press, New York, pp 453–505 [Google Scholar]
- Grant JE, Cooper PA, McAra AE, Frew TJ. (1995) Transformation of peas (Pisum sativum L.) using immature cotyledons. Plant Cell Rep 15: 254–258 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hammes UZ, Nielsen E, Honaas LA, Taylor CG, Schachtman DP. (2006) AtCAT6, a sink-tissue-localized transporter for essential amino acids in Arabidopsis. Plant J 48: 414–426 [DOI] [PubMed] [Google Scholar]
- Hawkesford MJ, De Kok LJ. (2006) Managing sulphur metabolism in plants. Plant Cell Environ 29: 382–395 [DOI] [PubMed] [Google Scholar]
- Hell R, Khan M, Wirtz M. (2010) Cellular biology of sulfur and its functions in plants. Robinson DG, ed, Cell Biology of Metals and Nutrients Springer, Heidelberg, pp 243–279 [Google Scholar]
- Higgins TJ, Chandler PM, Randall PJ, Spencer D, Beach LR, Blagrove RJ, Kortt AA, Inglis AS. (1986) Gene structure, protein structure, and regulation of the synthesis of a sulfur-rich protein in pea seeds. J Biol Chem 261: 11124–11130 [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Saito K. (2008) Analysis of systemic sulfur metabolism in plants using integrated ‘-omics’ strategies. Mol Biosyst 4: 967–973 [DOI] [PubMed] [Google Scholar]
- Hirner B, Fischer WN, Rentsch D, Kwart M, Frommer WB. (1998) Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J 14: 535–544 [DOI] [PubMed] [Google Scholar]
- Hocking PJ, Randall PJ, Pinkerton A. (1987) Sulphur nutrition of sunflower (Helianthus annuus) as affected by nitrogen supply: effects on vegetative growth, the development of yield components, and seed yield and quality. Field Crops Res 16: 157–175 [Google Scholar]
- Hopkins L, Parmar S, Błaszczyk A, Hesse H, Hoefgen R, Hawkesford MJ. (2005) O-Acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiol 138: 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Joshi V. (2010) Recent progress in deciphering the biosynthesis of aspartate-derived amino acids in plants. Mol Plant 3: 54–65 [DOI] [PubMed] [Google Scholar]
- Jasinski M, Ducos E, Martinoia E, Boutry M. (2003) The ATP-binding cassette transporters: structure, function, and gene family comparison between rice and Arabidopsis. Plant Physiol 131: 1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocsis MG, Ranocha P, Gage DA, Simon ES, Rhodes D, Peel GJ, Mellema S, Saito K, Awazuhara M, Li C, et al. (2003) Insertional inactivation of the methionine S-methyltransferase gene eliminates the S-methylmethionine cycle and increases the methylation ratio. Plant Physiol 131: 1808–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Krämer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al. (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Peng SS, Coruzzi GM. (1994) Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol 106: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass AD, Touraine B. (1999) Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant J 18: 89–95 [DOI] [PubMed] [Google Scholar]
- Lawlor DW. (2002) Carbon and nitrogen assimilation in relation to yield: mechanisms are the key to understanding production systems. J Exp Bot 53: 773–787 [PubMed] [Google Scholar]
- Lee M, Huang T, Toro-Ramos T, Fraga M, Last RL, Jander G. (2008) Reduced activity of Arabidopsis thaliana HMT2, a methionine biosynthetic enzyme, increases seed methionine content. Plant J 54: 310–320 [DOI] [PubMed] [Google Scholar]
- Malhi SS, Gan Y, Raney JP. (2007) Yield, seed quality, and sulfur uptake of Brassica oilseed crops in response to sulfur fertilization. Agron J 99: 570–577 [Google Scholar]
- McPhee KE, Gollasch S, Schroeder HE, Higgins TJV. (2004) Gene technology in pea. Curtis IS, ed, Transgenic Crops of the World: Essential Protocols Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 351–359 [Google Scholar]
- Miller AJ, Shen Q, Xu G. (2009) Freeways in the plant: transporters for N, P and S and their regulation. Curr Opin Plant Biol 12: 284–290 [DOI] [PubMed] [Google Scholar]
- Murray DR, Cordova-Edwards M. (1984) Amino acid and amide metabolism in the hulls and seeds of developing fruits of garden pea, Pisum sativum. II. Asparagine. New Phytol 97: 253–260 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Bielecka M, Gakière B, Krueger S, Rinder J, Kempa S, Morcuende R, Scheible WR, Hesse H, Hoefgen R. (2006) Effect of sulfur availability on the integrity of amino acid biosynthesis in plants. Amino Acids 30: 173–183 [DOI] [PubMed] [Google Scholar]
- Pate JS, Sharkey PJ, Lewis OAM. (1975) Xylem to phloem transfer of solutes in fruiting shoots of legumes, studied by a phloem bleeding technique. Planta 122: 11–26 [DOI] [PubMed] [Google Scholar]
- Pélissier HC, Tegeder M. (2007) PvUPS1 plays a role in source-sink transport of allantoin in French bean (Phaseolus vulgaris). Funct Plant Biol 34: 282–291 [DOI] [PubMed] [Google Scholar]
- Rácz I, Páldi E, Szalai G, Janda T, Pál M, Lásztity D. (2008) S-Methylmethionine reduces cell membrane damage in higher plants exposed to low-temperature stress. J Plant Physiol 165: 1483–1490 [DOI] [PubMed] [Google Scholar]
- Rennenberg H, Schmitz K, Bergmann L. (1979) Long-distance transport of sulfur in Nicotiana tabacum. Planta 147: 57–62 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M. (2007) Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett 581: 2281–2289 [DOI] [PubMed] [Google Scholar]
- Rochat C, Boutin J. (1991) Metabolism of phloem-borne amino acids in maternal tissues of Pisum sativum L. J Exp Bot 42: 207–214 [Google Scholar]
- Rolletschek H, Hosein F, Miranda M, Heim U, Götz KP, Schlereth A, Borisjuk L, Saalbach I, Wobus U, Weber H. (2005) Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol 137: 1236–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon A, Surdin-Kerjan Y, Thomas D. (1999) Transport of sulfonium compounds: characterization of the S-adenosylmethionine and S-methylmethionine permeases from the yeast Saccharomyces cerevisiae. J Biol Chem 274: 28096–28105 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Coruzzi GM. (2010) A systems view of responses to nutritional cues in Arabidopsis: toward a paradigm shift for predictive network modeling. Plant Physiol 152: 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K. (2004) Sulfur assimilatory metabolism: the long and smelling road. Plant Physiol 136: 2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A, Collier R, Trethewy A, Gould G, Sieker R, Tegeder M. (2009) AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J 59: 540–552 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhof I, Blankenburg D, Müller S, Krumbein A. (2007) Sulfur and nitrogen supply influence growth, product appearance, and glucosinolate concentration of broccoli. J Plant Nutr Soil Sci 170: 65–72 [Google Scholar]
- Sexton PJ, Paek NC, Shibles RM. (1998) Effects of nitrogen source and timing of sulfur deficiency on seed yield and expression of 11S and 7S seed storage proteins of soybean. Field Crops Res 59: 1–8 [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT. (1995) Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe L, Hagan N, Higgins TJV. (2002) Plasticity of seed protein composition in response to nitrogen and sulfur availability. Curr Opin Plant Biol 5: 212–217 [DOI] [PubMed] [Google Scholar]
- Tagmount A, Berken A, Terry N. (2002) An essential role of S-adenosyl-l-methionine:l-methionine S-methyltransferase in selenium volatilization by plants: methylation of selenomethionine to selenium-methyl-l-selenium-methionine, the precursor of volatile selenium. Plant Physiol 130: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Grennan AK, Pélissier HC, Rentsch D, Tegeder M. (2008) Characterization and expression of French bean amino acid transporter PvAAP1. Plant Sci 174: 348–356 [Google Scholar]
- Tegeder M, Offler CE, Frommer WB, Patrick JW. (2000) Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol 122: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Rentsch D, Patrick JW. (2011) Organic carbon and nitrogen transporters. Murphy A, Peer W, Schulz B, eds, Plant Plasma Membrane: Plant Cell Monographs Springer, Berlin, pp 331–352 [Google Scholar]
- Tegeder M, Tan Q, Grennan AK, Patrick JW. (2007) Amino acid transporter expression and localisation studies in pea (Pisum sativum). Funct Plant Biol 34: 1019–1028 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Kassner N, Struck C, Lohaus G. (2005) Amino acid contents and transport in oilseed rape (Brassica napus L.) under different nitrogen conditions. Planta 221: 328–338 [DOI] [PubMed] [Google Scholar]
- Tsai FY, Coruzzi GM. (1990) Dark-induced and organ-specific expression of two asparagine synthetase genes in Pisum sativum. EMBO J 9: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzitzikas EN, Vincken JP, de Groot J, Gruppen H, Visser RG. (2006) Genetic variation in pea seed globulin composition. J Agric Food Chem 54: 425–433 [DOI] [PubMed] [Google Scholar]
- Ufaz S, Galili G. (2008) Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol 147: 954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart AA, Joy KW. (1981) Use of phloem exudate technique in the study of amino acid transport in pea plants. Plant Physiol 68: 750–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel AJE. (1990) Xylem-phloem exchange via the rays: the undervalued route of transport. J Exp Bot 41: 631–644 [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos P, Krähenbühl U, den Camp RO, Brunold C. (2002) Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J 31: 729–740 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Filleur S, Gan Y, Forde BG. (2005) Signaling mechanisms integrating root and shoot responses to changes in the nitrogen supply. Photosynth Res 83: 239–250 [DOI] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Galant A, Ravilious GE, Preuss ML, Jez JM. (2010) Sensing sulfur conditions: simple to complex protein regulatory mechanisms in plant thiol metabolism. Mol Plant 3: 269–279 [DOI] [PubMed] [Google Scholar]
- Zhang MY, Bourbouloux A, Cagnac O, Srikanth CV, Rentsch D, Bachhawat AK, Delrot S. (2004) A novel family of transporters mediating the transport of glutathione derivatives in plants. Plant Physiol 134: 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FJ, Hawkesford MJ, McGrath SP. (1999) Sulphur assimilation and effects on yield and quality of wheat. J Cereal Sci 30: 1–17 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.