Abstract
High mobility group (HMG) proteins of the HMGB family are chromatin-associated proteins that as architectural factors are involved in the regulation of transcription and other DNA-dependent processes. HMGB proteins are generally considered nuclear proteins, although mammalian HMGB1 can also be detected in the cytoplasm and outside of cells. Plant HMGB proteins studied so far were found exclusively in the cell nucleus. Using immunofluorescence and fluorescence microscopy of HMGB proteins fused to the green fluorescent protein, we have examined the subcellular localization of the Arabidopsis (Arabidopsis thaliana) HMGB2/3 and HMGB4 proteins, revealing that, in addition to a prominent nuclear localization, they can be detected also in the cytoplasm. The nucleocytoplasmic distribution appears to depend on the cell type. By time-lapse fluorescence microscopy, it was observed that the HMGB2 and HMGB4 proteins tagged with photoactivatable green fluorescent protein can shuttle between the nucleus and the cytoplasm, while HMGB1 remains nuclear. The balance between the basic amino-terminal and the acidic carboxyl-terminal domains flanking the central HMG box DNA-binding domain critically influences the nucleocytoplasmic distribution of the HMGB proteins. Moreover, protein kinase CK2-mediated phosphorylation of the acidic tail modulates the intranuclear distribution of HMGB2. Collectively, our results show that, in contrast to other Arabidopsis HMGB proteins such as HMGB1 and HMGB5, the HMGB2/3 and HMGB4 proteins occur preferentially in the cell nucleus, but to various extents also in the cytoplasm.
Within the cell nucleus, the genomic DNA is organized with histones and other proteins into a nucleoprotein complex termed chromatin. This packaging of the DNA has crucial consequences for DNA-dependent processes, including the transcription of genes. The chromatin structure is highly dynamic and is modulated by a variety of chromatin-associated proteins. Among these proteins are the high mobility group (HMG) proteins that represent a heterogeneous class of proteins that, after the histones, are the second most abundant family of chromosomal proteins (Bustin and Reeves, 1996; Thomas and Travers, 2001; Reeves, 2010). It has been estimated that they bind to 10% or fewer of the nucleosomes (Johns, 1982). Because of their abundance, HMG proteins are thought to serve a global structural function in the nucleus, and they act as architectural factors, facilitating various DNA-dependent processes, including transcription, recombination, and DNA repair (Bustin and Reeves, 1996; Thomas and Travers, 2001; Agresti and Bianchi, 2003).
In monocotyledonous and dicotyledonous plants, members of the HMGA and HMGB families have been identified that are expressed throughout the plant at different levels (Grasser et al., 2007). Plant HMGB proteins (approximately 13–27 kD) have a distinctive three-domain structure with a central HMG box DNA-binding domain that is flanked by a basic N-terminal domain and an acidic C-terminal domain. Plant HMGB proteins are structurally more diverse than their animal counterparts, in particular within the domains flanking the HMG box domain (Pedersen and Grasser, 2010). In addition, plants encode around six different HMGB proteins, whereas fewer HMGB variants are found in animals and yeast (Stros et al., 2007). Mediated by the HMG box domain, the plant HMGB proteins interact nonsequence specifically with linear DNA (Pedersen et al., 1991; Webster et al., 1997; Ritt et al., 1998; Wu et al., 2003), but they bind with high affinity certain DNA structures, including four-way junctions and DNA minicircles (Ritt et al., 1998; Wu et al., 2003; Zhang et al., 2003), and they interact with nucleosomes (Arwood and Spiker, 1990; Lichota and Grasser, 2001). Moreover, by functional interaction with certain transcription factors, members of the HMGB family contribute to the regulation of gene transcription (Grasser et al., 2007).
Studies addressing the biological function of HMGB proteins suggest that HMGB proteins have important cellular roles. In yeast, inactivation of one of the two NHP6A/B genes (encoding HMGB proteins) did not result in a phenotype distinct from the wild type, but the inactivation of both genes led to growth aberrations such as temperature-sensitive growth and morphological defects (Costigan et al., 1994). Knockout of the HMGB1 gene caused pleiotropic defects in mice, and they die soon after birth, but cell lines could grow normally without HMGB1 (Calogero et al., 1999). Mice lacking HMGB2 (which is approximately 80% identical to HMGB1) were viable, but male mice had reduced fertility, as HMGB2 seems to play a role in germ cell differentiation (Ronfani et al., 2001). Mice deficient in HMGB3 were also viable but had an altered rate of generation and differentiation of primitive hematopoietic progenitor cells, and HMGB3 appears to be required for the proper balance between hematopoietic stem cell self-renewal and differentiation (Nemeth et al., 2005, 2006). Ectopic expression of maize (Zea mays) HMGB1 in tobacco (Nicotiana tabacum) seedlings caused reduced length of the primary root, whereas HMGB4 did not affect root development (Lichota et al., 2004). In Arabidopsis (Arabidopsis thaliana), overexpression of HMGB2 reduced seed germination under salt and drought stress (Kwak et al., 2007). Altering the expression of HMGB1 influenced plant growth, stress response, and transcriptome in Arabidopsis (Lildballe et al., 2008).
Chromosomal HMGB proteins are generally considered nuclear proteins (Grasser et al., 2007; Reeves, 2010), although it is well documented that mammalian HMGB1 can be detected also outside the nucleus, acting as a kind of cytokine (Müller et al., 2004; Yang and Tracey, 2010). The Arabidopsis genome encodes seven HMGB-type proteins that differ in structure, expression pattern, and DNA-binding properties (Moehs et al., 1988; Stemmer et al., 1997; Grasser et al., 2004, 2006; Kwak et al., 2007; Launholt et al., 2007), suggesting (partially) specific functions of the different family members. The Arabidopsis HMGB1, HMGB5, and HMGB6 proteins as well as the HMGB-type protein encoded by the locus At2g34450 were found to localize to the nucleus (Grasser et al., 2004, 2006; Launholt et al., 2006). Within the cell nucleus, Arabidopsis HMGB1 and HMGB5 are highly dynamic, binding DNA/chromatin only transiently before moving on to the next binding site (Launholt et al., 2006). Here, we have examined the subcellular localization of the closely related Arabidopsis HMGB2/3 proteins and of HMGB4. Our experiments revealed that these three HMGB proteins occur predominantly in the nucleus, but variable amounts of the proteins are also detected in the cytoplasm, while HMGB1 and HMGB5 are exclusively nuclear proteins.
RESULTS
Subcellular Localization of Arabidopsis HMGB2/3 and HMGB4
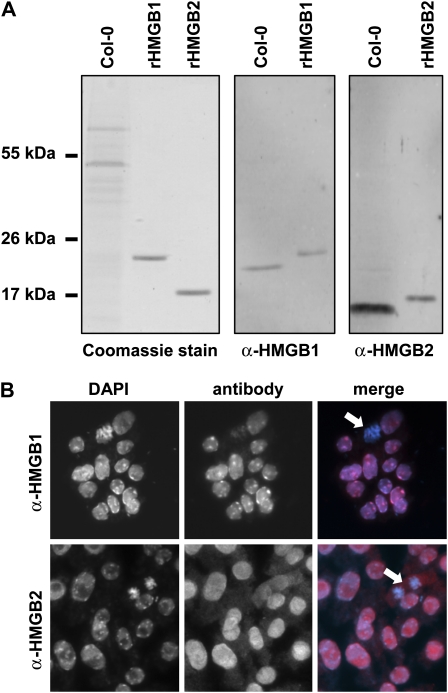
Since the subcellular localization of plant HMGB proteins has not been examined systematically, we raised an antiserum against Arabidopsis HMGB2, whose distribution has not been studied in plant cells. The antibody was tested in immunoblots, revealing that it reacted specifically with a protein band of the expected size of approximately 15 kD, when Arabidopsis ecotype Columbia (Col-0) leaf protein extracts were probed (Fig. 1A). When compared with an antiserum directed against Arabidopsis HMGB1 (Launholt et al., 2006), relative to the bands of the recombinant proteins the HMGB2 band appeared markedly more prominent, consistent with the higher expression level of HMGB2 (http://www.arabidopsis.org/portals/expression/index.jsp). In addition, the HMGB2 antiserum reacted equally well with recombinant HMGB2 and HMGB3 (data not shown), as both proteins share approximately 89% amino acid sequence identity and comigrate on SDS-PAGE. The HMGB2 antiserum was used for indirect immunofluorescence microscopic analyses of Arabidopsis root tip cells (Fig. 1B), while the HMGB1 antiserum served as a reference. In line with previous experiments (Launholt et al., 2006; Lildballe et al., 2008), the HMGB1 antiserum reacted specifically with interphase nuclei that were stained in parallel with 4′,6-diamino-phenylindole. Anti-HMGB1 signals were enhanced in heterochromatic interphase regions. Conversely, anti-HMGB2-labeled cells displayed uniformly stained interphase nuclei, and additional weak immunofluorescence signals were found in the cytoplasm. Dividing chromosomes were hardly stained with either anti-HMGB1 or anti-HMB2. This finding suggests that HMGB2 (and/or HMGB3) to some extent also occurs outside the cell nucleus.
Figure 1.
Detection of Arabidopsis HMGB2 by immunoblotting and immunofluorescence. A, Coomassie Brilliant Blue stain of a protein extract of Col-0 seedlings and recombinant HMGB1 and HMGB2 proteins separated by SDS-PAGE on 18% polyacrylamide gels (left). Immunoblot analysis of Col-0 protein extract and recombinant HMGB1/HMGB2 using an HMGB1 antiserum (middle) and an HMGB2 antiserum (right) are shown. Comparable amounts of Col-0 extract and of the recombinant proteins were used for the two immunoblots. Migration positions of marker proteins are given. Due to the 6× His tag, the recombinant HMGB1 and HMGB2 proteins displayed a slightly reduced electrophoretic mobility when compared with the corresponding proteins of the Col-0 extract. B, Indirect immunofluorescence analysis of Col-0 root tip cells using HMGB1 (top) and HMGB2 (bottom) antisera. For comparison, a 4′,6-diamino-phenylindole (DAPI) stain is shown, staining the nuclear DNA (left). The immunofluorescence images (middle) and the merge with the 4′,6-diamino-phenylindole stain (right) are shown. Dividing cells are indicated by arrows.
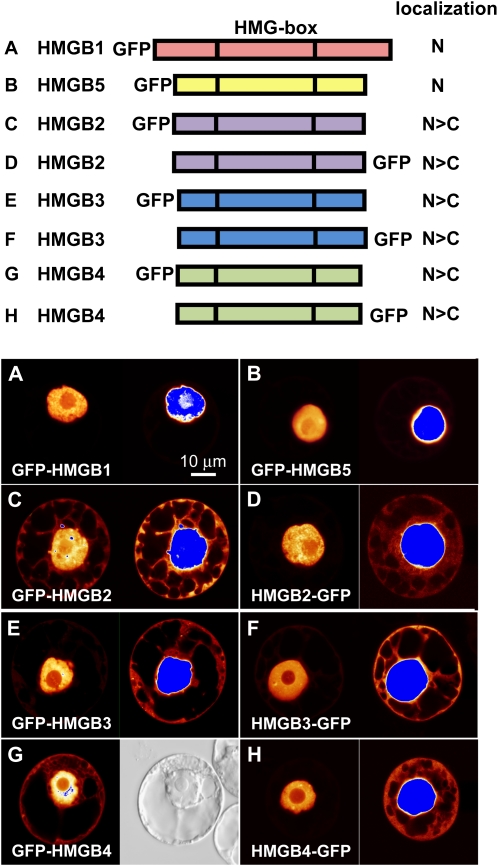
To further analyze the subcellular localization of HMGB2/3 and HMGB4, we generated plasmids that could direct the expression of GFP fusion proteins in plant cells. Tobacco protoplasts were transformed with these plasmids and examined by confocal laser scanning microscopy (CLSM; Fig. 2). The previously observed nuclear localization of HMGB1 and HMGB5 (Launholt et al., 2006; Lildballe et al., 2008) served as a reference. While GFP-HMGB1 and GFP-HMGB5 were found exclusively in the cell nucleus (Fig. 2, A and B), the fluorescence signals of HMGB2/3 and HMGB4 were detected primarily in the nucleus, but to various extents also in the cytoplasm (Fig. 2, C–H). Each fluorescent image (except G) is shown in normal exposure and overexposed to illustrate more clearly the cytoplasmic fluorescence seen with HMGB2/3 and HMGB4. The localization of HMGB2/3 and HMGB4 in both cytoplasm and nucleus was consistently observed for both the N- and C-terminal GFP fusion proteins, although quantitative differences were observed in the nucleocytoplasmic distribution with different protoplasts and with N- versus C-terminal GFP fusions (HMGB2 and HMGB4). These experiments showed that, in contrast to the strictly nuclear proteins HMGB1 and HMGB5, the HMGB2/3 and HMGB4 proteins are also found in the cytoplasm.
Figure 2.
Detection of HMGB1 and HMGB5 in the nucleus, while HMGB2/3 and HMGB4 are observed in both the nucleus and the cytoplasm. Protoplasts prepared from BY-2 tobacco cell suspension cultures were transformed with plasmids driving the expression of the indicated GFP fusion proteins (for overview, see top panel). The observed nucleocytoplasmic distribution is also summarized in the top panel (N, nuclear accumulation; N>C, indicating the tendency of higher fluorescence in the nucleus relative to the cytoplasm) based on inspecting 60 to 80 transformed protoplasts each. A, GFP-HMGB1. B, GFP-HMGB5. C, GFP-HMGB2. D, HMGB2-GFP. E, GFP-HMGB3. F, HMGB3-GFP. G, GFP-HMGB4. H, HMGB4-GFP. The left part of each panel represents an image of a protoplast at normal exposure, while the right part shows an overexposed image of the same protoplast (except in G, where instead of the overexposed image, the corresponding bright-field image is shown). Fluorescence intensities in CLSM images of transformed protoplasts are displayed using a false-color palette. Low-fluorescence signals are indicated by red color, and progressively stronger signals are indicated by orange over yellow to white. Blue indicates top-scale signals/overexposure. Bar = 10 μm.
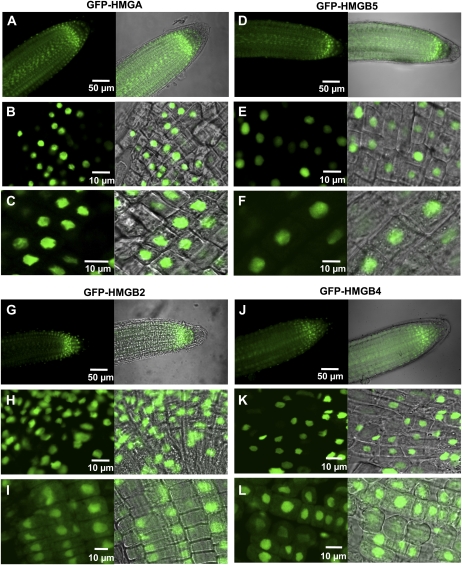
To examine the subcellular distribution of HMGB2 and HMGB4 in planta, we generated transgenic Arabidopsis plants expressing HMGB-GFP fusion proteins. Roots of the transgenic plants were analyzed by CLSM (Fig. 3) and compared with plants expressing HMGA and HMGB5 fused to GFP (which were constructed in the same way). The primary root tip was observed (Fig. 3, A, D, G, and J), and meristematic and cortical cells were inspected in more detail. Consistent with previous experiments (Launholt et al., 2006), HMGA and HMGB5 were detected exclusively in the nuclei of both the meristematic and cortical cells (Fig. 3, B, C, E, and F). For the HMGB2 and HMGB4 fusion proteins, in meristematic cells, GFP fluorescence was detected preferentially in nuclei (Fig. 3, H and K). In cortex cells, the GFP fusion proteins occurred predominantly in nuclei, but they were also clearly visible in the cytoplasm (Fig. 3, I and L). The cytoplasmic GFP signal in cortex cells was observed both in the GFP fluorescence images (left panels) and in the overlays of GFP fluorescence and the corresponding bright-field images (right panels). This experiment confirmed that HMGB2 and HMGB4 can occur in the cytoplasm and suggested that the nucleocytoplasmic distribution of HMGB2 and HMGB4 varies between cells.
Figure 3.
Nucleocytoplasmic distribution of HMGA, HMGB5, HMGB2, and HMGB4 fused to GFP in stably transformed Arabidopsis seedlings. The analyzed plants expressed GFP-HMGA (A–C), GFP-HMGB5 (D–F), GFP-HMGB2 (G–I), and GFP-HMGB4 (J–L). Detection of GFP fluorescence by CLSM is shown in root tips of 3-d-old transgenic seedlings expressing the indicated GFP constructs. The left images show the GFP fluorescence, and the right panels show an overlay with the corresponding bright-field image. The top images show overviews of the intact root tips, while closeup views of squeeze-preps of meristematic cells (middle images) and of cortical cells (bottom images) are shown. Bars = 10 or 50 μm.
Contribution of the Basic N-Terminal and Acidic C-Terminal Domains to the Subcellular Localization of HMGB2 and HMGB4
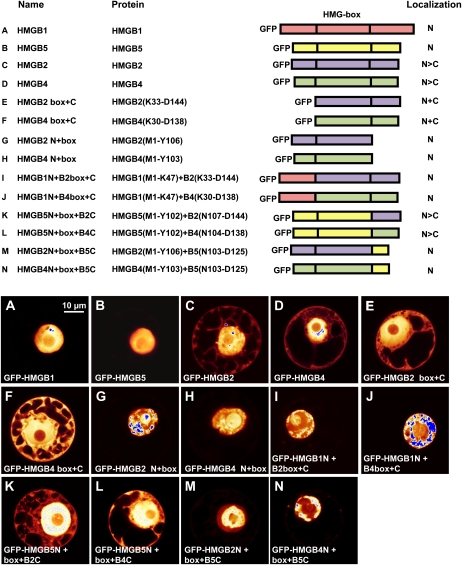
Since the Arabidopsis HMGB2/3 and HMGB4 proteins were detected both in nuclei and cytoplasm, while HMGB1 and HMGB5 were exclusively found in nuclei, the amino acid sequences of the HMGB proteins were searched for motifs that possibly are responsible for this difference in subcellular distribution. However, we were unable to identify differences in potential nuclear localization signals (NLSs) or nuclear export signals (Merkle, 2003) that could explain the observed differential localization. Moreover, incubation of protoplasts that expressed GFP fusion proteins with the inhibitor of the nuclear export receptor XPO1, leptomycin B, had no effect on the subcellular distribution of HMGB2 or HMGB4 (data not shown). To examine experimentally the contribution of HMGB protein domains on the subcellular localization, we constructed a variety of plasmids that direct the expression of truncated and chimeric HMGB proteins fused to GFP. Tobacco protoplasts transformed with these plasmids were examined by CLSM (Fig. 4). For comparison, the localization of full-length HMGB1 and HMGB5 (localized in the nucleus) and of HMGB2 and HMGB4 (localized in the nucleus and cytoplasm) are shown (Fig. 4, A–D). Deletion of the basic N-terminal domain of HMGB2 and HMGB4 had no marked influence on the localization of the proteins when compared with the full-length proteins (Fig. 4, E and F). However, deletion of the acidic C-terminal domain of HMGB2 and HMGB4 resulted in an increased accumulation of the proteins in the nucleus (Fig. 4, G and H). Replacement of the basic N-terminal domains of HMGB2 and HMGB4 by the basic N-terminal domain of HMGB1 (which contains a strong NLS; Launholt et al., 2006), caused efficient nuclear accumulation of the chimeric proteins (Fig. 4, I and J). When the acidic C-terminal domain of HMGB5 was replaced by the acidic C-terminal domains of HMGB2 or HMGB4, the chimeric proteins (unlike intact HMGB5) in addition to the nucleus were found in the cytoplasm (Fig. 4, K and L). Replacement of the acidic tails of HMGB2 and HMGB4 by the short acidic C-terminal domain of HMGB5 resulted in chimeric proteins that accumulated in the nucleus (Fig. 4, M and N). In summary, the comparative analyses of the subcellular localization of these full-length, truncated, and chimeric HMGB proteins indicated that the basic N-terminal and the acidic C-terminal domains are involved in directing the nucleocytoplasmic distribution of the Arabidopsis HMGB proteins, possibly by intramolecular interaction.
Figure 4.
Nucleocytoplasmic distribution of truncated and chimeric HMGB proteins in comparison with full-length proteins. BY-2 protoplasts were transformed with plasmids driving the expression of the indicated GFP fusion proteins (for overview, see top panel). For truncated and chimeric HMGB proteins, the origin of the domains (including amino acid positions) is represented in the scheme. The observed nucleocytoplasmic distribution is also summarized in the top panel (N, nuclear accumulation; N+C, localization in the nucleus and the cytoplasm; N>C, indicating the tendency of higher fluorescence in the nucleus relative to the cytoplasm) based on inspecting 60 to 80 transformed protoplasts each. A to N show representative images of the GFP fluorescence of the indicated GFP-HMGB proteins. Fluorescence intensities in CLSM images are displayed using a false-color palette (compare with Fig. 2). Bar = 10 μm.
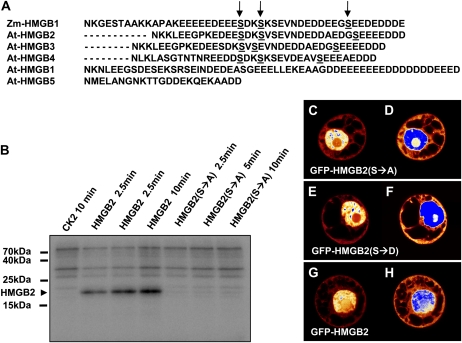
Since no amino acid sequence motif(s) could be detected that may confer the different subcellular localization of HMGB1 and HMGB5 on the one hand and of HMGB2/3 and HMGB4 on the other hand, the contribution of posttranslational modifications was considered. To date, the only posttranslational modification that has been identified for plant HMGB proteins is the protein kinase CK2-mediated phosphorylation of Ser/Thr residues within the acidic C-terminal domains of maize HMGB proteins (Stemmer et al., 2002). When Arabidopsis HMGB proteins were tested for CK2-mediated phosphorylation, HMGB2/3 and HMGB4 were readily phosphorylated, while HMGB1 was phosphorylated only weakly (and not within the C-terminal region that does not contain Ser/Thr residues) and HMGB5 was not phosphorylated at all (Stemmer et al., 2003). Thus, the three HMGB proteins that, in addition to the nucleus, are found in the cytoplasm are particularly good substrates for protein kinase CK2. In line with that, the C-terminal amino acid sequences of these proteins contain Ser residues within CK2 consensus sequences (Pinna and Ruzzene, 1996; Guerra et al., 1999) that resemble the sites known to be phosphorylated with maize HMGB proteins (Stemmer et al., 2002), while these sites do not occur in the C-terminal region of HMGB1 and HMGB5 (Fig. 5A). To test whether the CK2-mediated phosphorylation of these C-terminal Ser residues is involved in directing the subcellular localization of HMGB2, the three candidate Ser residues (Ser-120, Ser-123, and Ser-137) were changed to Ala residues. Phosphorylation assays using purified recombinant maize CK2α and [γ-32P]ATP were performed with wild-type HMGB2 and the S→A mutant version of HMGB2 lacking the three C-terminal Ser residues. The substrate proteins were reacted with CK2α for different times, and as expected, HMGB2 was vigorously phosphorylated by the enzyme (Fig. 5B). By contrast, the S→A mutations abolished the phosphorylation of HMGB2 by the protein kinase. The S→A mutant form of HMGB2, along with the corresponding phosphomimic mutant (S→D, with the three Ser residues changed to Asp) fused to GFP, were expressed in tobacco protoplasts and analyzed by CLSM in comparison with wild type GFP-HMGB2. The wild-type and mutant HMGB2 forms showed a similar nucleocytoplasmic distribution, but in contrast to GFP-HMGB2, which often showed GFP fluorescence in the nucleolus (Fig. 5, G and H), this effect was never seen with the mutant versions of GFP-HMGB2. In addition, the S→A and S→D mutant versions of HMGB2 displayed a more pronounced speckled pattern of fluorescence in the nucleus. Thus, CK2-mediated phosphorylation did not appear to influence the nucleocytoplasmic distribution of HMGB2, but it modulated the protein localization within the cell nucleus.
Figure 5.
Role of CK2-mediated phosphorylation on the subcellular localization of HMGB2. A, Alignment of the acidic C-terminal domains of Arabidopsis HMGB proteins in comparison with maize HMGB1. The Ser residues known to be phosphorylated by CK2 in maize HMGB1 (Stemmer et al., 2002) are indicated by arrows. These sites are essentially conserved in Arabidopsis HMGB2/3 and HMGB4 but not in HMGB1 and HMGB5. B, Phosphorylation assays using maize CK2α and radiolabeled ATP. Phosphorylation reactions were done without the addition of substrate protein or using HMGB2 or mutated (S→A) HMGB2 (in which the three Ser residues within the acidic tail were changed to Ala residues) as substrate proteins. The phosphorylation reactions were terminated at the indicated times, separated by SDS-PAGE on 18% polyacrylamide gels, and analyzed using a phosphor imager. The migration positions of marker proteins are given as well as the migration of HMGB2. C to H, Subcellular localization of wild-type and mutant HMGB2. BY-2 protoplasts were transformed with plasmids driving the expression of the indicated GFP fusion proteins. The above-mentioned S→A mutant (C and D) and the S→D phosphomimic mutant (E and F) of HMGB2 as well as wild-type HMGB2 (G and H) are shown. Normal exposures (C, E, and G) and overexposures (D, F, and H) of the GFP fluorescence are shown. Fluorescence intensities in CLSM images are displayed using a false-color palette (compare with Fig. 2).
HMGB2 and HMGB4 Shuttle between the Nucleus and the Cytoplasm
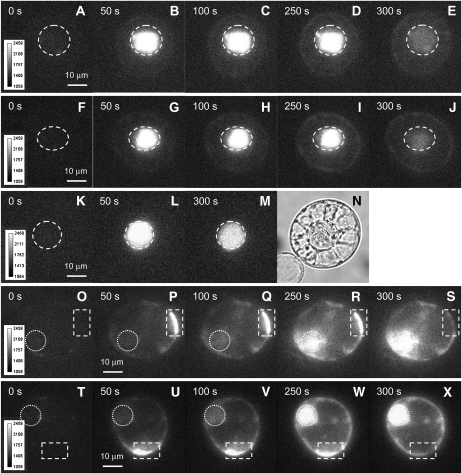
The occurrence of HMGB2/3 and HMGB4 in both nucleus and cytoplasm raised the possibility that these proteins can shuttle between the two locations. To test this possibility, HMGB2 and HMGB4 fused to photoactivatable GFP (paGFP) were expressed in tobacco protoplasts and examined by multifocal two-photon laser scanning microscopy (Martini et al., 2007). The dynamics of the diffusion of HMGB2 and HMGB4 fused to paGFP was recorded before (0 s) and at different times during and after activation of the paGFP in the nucleus (Fig. 6, A–E and F–J, respectively). For both paGFP fusion proteins, over time a decrease of nuclear fluorescence was observed. In parallel, an increase of fluorescence was seen in the cytoplasm. This demonstrated that significant amounts of both HMGB2 and HMGB4 fused to paGFP can migrate from the nucleus to the cytoplasm within the analyzed period. When HMGB1 fused to paGFP was examined in parallel, no cytoplasmic fluorescence was observed (Fig. 6, K–N), indicating that, in line with previous experiments (Launholt et al., 2006), HMGB1 remained in the nucleus and could not migrate to the cytoplasm. To study the migration from cytoplasm to the nucleus, paGFP fused to HMGB2 and HMGB4 was activated in a small area of the cytoplasm (indicated by a rectangle), and the fluorescence was recorded over time (Fig. 6, O–X). This experiment showed that HMGB2 and HMGB4 first spread from the activation site in the cytoplasm, and consistent with their preferential nuclear localization in the following, they accumulated in the cell nucleus. Monitoring the distribution of paGFP fused to HMGB2 and HMGB4 demonstrated that both proteins can shuttle between the nucleus and the cytoplasm, whereas HMGB1 remained nuclear localized.
Figure 6.
HMGB2 and HMGB4 show relocalization from the nucleus to the cytoplasm, while HMGB1 remains nuclear. Up to five selected fluorescence images of BY-2 protoplasts are shown that expressed paGFP-HMGB2 (A–E), paGFP-HMGB4 (F–J), or paGFP-HMGB1 (K–M). N shows a bright-field image of the protoplast shown in K to M. Images were taken before (0 s), during (50–250 s), and after (300 s) photoactivation of paGFP in the nuclei at the time points indicated. The regions of interest, where photoactivation was performed in A to M, were located within the nuclei that are indicated by dashed lines. Before photoactivation, the average fluorescence intensity was barely detectable. Shortly after the onset of photoactivation (performed with a two-photon laser scanning microscope during 250 s using femtosecond laser bursts covering an area of 7 × 8 μm with 64 parallel laser foci, at 4 mW at 800 nm per focus), strong fluorescent signals were detected, and the dynamics of photoactivated paGFP was monitored continuously in each case. Fluorescence intensity panels and scale bars are shown to the left of each row. In O to X, photoactivation was performed in the cytoplasm. Five selected fluorescence images of BY-2 protoplasts are shown that expressed paGFP-HMGB2 (O–S) or paGFP-HMGB4 (T–X). The region of interest is indicated by a dashed line, and the nucleus is indicated by a dotted line.
DISCUSSION
Chromatin-associated HMGB proteins occur in a wide variety of eukaryotes and are generally considered nuclear proteins (Thomas and Travers, 2001; Grasser et al., 2007; Reeves, 2010), but already early studies have revealed that HMGB1 can also be detected in the cytoplasm of mammalian cells (Bustin and Neihart, 1979; Rechsteiner and Kuehl, 1979). However, only in recent years was the extracellular presence of mammalian HMGB1 linked with its specific (additional) function as a cytokine during injury or inflammation (Müller et al., 2004; Yang and Tracey, 2010). Plant HMG proteins, which structurally differ from their mammalian counterparts (Stros et al., 2007), traditionally were purified from chromatin or isolated nuclei (Spiker, 1984; Grasser et al., 1991). All HMGB proteins, whose subcellular localization was studied by immunofluorescence and/or by fluorescence microscopic analyses of GFP-tagged proteins in plant cells, were found in the cell nucleus. Among these nuclear HMGB proteins were Arabidopsis HMGB1, HMGB5, and HMGB6 (Grasser et al., 2004; Launholt et al., 2006). While the (long) N-terminal basic domain was sufficient for nuclear accumulation of HMGB1, in the case of HMGB5, both the N-terminal and HMG box domains were required for efficient nuclear targeting (Launholt et al., 2006).
Here, we report that the Arabidopsis HMGB2 and HMGB4 proteins in addition to the nucleus can occur in the cytoplasm. Although the subcellular distribution of HMGB3 was assayed only in the protoplast system, due to its high degree of similarity to HMGB2 (89% amino acid sequence identity), it is likely that it behaves as HMGB2, which was studied in detail. Analysis of the amino acid sequences of the proteins did not reveal an explanation to the question of why some Arabidopsis HMGB proteins (HMGB1, HMGB5, and HMGB6) are exclusively nuclear while others (HMGB2/3 and HMGB4) are found in both the nucleus and the cytoplasm. However, examination of the subcellular localization of a variety of truncated and chimeric HMGB proteins (in comparison with the wild-type proteins) provided some insight. HMGB2 and HMGB4 lacking the basic N-terminal domain were also found in the nucleus and in the cytoplasm, but they showed a more prominent cytoplasmic localization when compared with full-length HMGB2 and HMGB4. In line with this finding, deletion of the acidic tails of HMGB2 and HMGB4 resulted in proteins that were exclusively nuclear, and these proteins were also found in the nucleoli. This demonstrated that the acidic C-terminal domain is critical for the occurrence of HMGB2 and HMGB4 in the cytoplasm. The nuclear accumulation of the chimeric proteins, in which the natural N-terminal domain was replaced by the N-terminal domain of HMGB1, showed that the strong NLS in the N-terminal domain of HMGB1 (Launholt et al., 2006) can “overrule” the effect of the acidic tails that in the natural context direct some of the protein to the cytoplasm. Replacement of the acidic tail of HMGB5 with those of HMGB2 or HMGB3 resulted in some cytoplasmic localization of HMGB5 (which normally is a nuclear protein), confirming the role the HMGB2/HMGB4 tails play in cytoplasmic localization. In addition, replacement of the acidic tail of HMGB2 and HMGB4 by the acidic C-terminal domain of HMGB5 abolished the cytoplasmic localization of HMGB2 and HMGB4.
Thus, both the basic N-terminal and acidic C-terminal domains of HMGB2 and HMGB4 are critical for the subcellular localization of the proteins. DNA-binding experiments suggested that the basic N-terminal and acidic C-terminal domains of maize and rice HMGB1 interact to regulate the affinity for DNA (Ritt et al., 1998; Wu et al., 2003). An interaction of the two terminal domains was also observed in spectrometric and cross-linking experiments, which in addition revealed that the intramolecular interaction is modulated by CK2-mediated phosphorylation of the acidic tail in maize HMGB1 (Stemmer et al., 2002; Thomsen et al., 2004). Similarly, this intramolecular interaction of the terminal domains was observed for Arabidopsis HMGB1 and HMGB4 and appears to be a general feature of plant HMGB proteins (Thomsen et al., 2004). Therefore, the intramolecular interaction between the two terminal domains offers the most likely explanation for the observed subcellular distribution of the truncated and chimeric HMGB proteins in comparison with the wild-type proteins. The C-terminal domains of HMGB2 and HMGB4 play a critical role for the proteins to localize to both the nucleus and the cytoplasm, since deleting the acidic tail or replacing it by the acidic tail of HMGB5 essentially abolished the cytoplasmic localization, rendering the proteins nuclear. However, in cases where the C-terminal domains of HMGB2 and HMGB4 occurred in conjunction with the basic N-terminal domain of HMGB1 (containing an efficient NLS), the chimeric proteins accumulated in the nucleus. In these proteins, the acidic tail of HMGB2 and HMGB4 could not confer cytoplasmic localization. In conclusion, we propose that the basic N-terminal and acidic C-terminal domains of HMGB2 and HMG4 need to be in a delicate balance of interaction. This interaction allows preferential nuclear accumulation of the proteins that most likely is mediated by nuclear targeting information both within the N-terminal and HMG box domains. This is similar to the situation with Arabidopsis HMGB5 (Launholt et al., 2006) and mammalian HMGB1 (Bonaldi et al., 2003), where the sites contributing to nuclear accumulation are scattered over larger regions of the proteins. At the same time, this intramolecular interaction allows that, dependent on the cell type, some of the HMGB2 and HMGB4 proteins are present in the cytoplasm.
As observed in the Arabidopsis root, the partitioning of the HMGB2 and HMGB4 proteins into nucleus versus cytoplasm appears to be cell type dependent. In mammals, it was reported that the amount of HMGB1 (and HMGB2) found in the cytoplasm is elevated in tissues rich in differentiated cells, while preferential nuclear localization is characteristic of undifferentiated cells (Mosevitsky et al., 1989). This is in line with the preferential nuclear localization of HMGB2 and HMGB4 in Arabidopsis root meristem cells when compared with cortical cells. Nucleocytoplasmic partitioning of mammalian HMGB1 is regulated by acetylation of various Lys residues (Bonaldi et al., 2003), but mass spectrometric analyses did not reveal evidence for acetylation of maize HMGB proteins (Stemmer et al., 2002, 2003). The nuclear accumulation of insect HMGB proteins is reduced by protein kinase C-mediated phosphorylation of the basic part of the protein (Wiśniewski et al., 1994). For some plant HMGB proteins, CK2-mediated phosphorylation of the acidic tail was demonstrated (Stemmer et al., 2002, 2003), but in the case of HMGB2, mutation of the phosphorylation sites did not markedly affect the nucleocytoplasmic partitioning of the protein, although it modulated the distribution within the nucleus. Since the proportion of HMGB2 and HMGB4 found in the cytoplasm appears to differ between cell types, it is likely that the nuclear import/export rates are modulated, perhaps by a yet to be identified posttranslational modification.
The puzzling question that remains is, what is the biological role of the chromosomal HMGB2 and HMGB4 proteins that are found in the cytoplasm? The extranuclear/extracellular role of HMGB1 that acts as a specific mediator related to injury and inflammation in mammals (Müller et al., 2004; Yang and Tracey, 2010) appears unlikely for plants. For the high levels of cytoplasmic HMGB1/HMGB2 associated with normal mammalian differentiated cell types, an inverse correlation was found with increased levels of linker histone H10 in nuclei (Mosevitsky et al., 1989). Based on the possibility that HMGB proteins and linker histones play a shared role in chromatin and that HMGB proteins confer a more open chromatin structure than H1 (Zlatanova and van Holde, 1998), Mosevitsky et al. (1989) proposed that the higher levels of nuclear HMGB proteins in undifferentiated cells reflect the more pronounced chromatin flexibility required for transcriptional dynamics during differentiation processes. Although this may explain the various levels of HMGB proteins in nuclei, it does not really explain the function of some chromosomal HMGB proteins in the cytoplasm of plant cells, while other HMGB types appear to be exclusively nuclear. Since HMGB proteins are involved in plant responses to abiotic stress conditions (Pedersen and Grasser, 2010), the presence of HMGB proteins in the cytosol may allow reacting rapidly to altered environmental conditions by modulating the nucleocytoplasmic distribution of these proteins. Analysis of mutant plants lacking the various types of HMGB proteins may provide insight into the potentially different roles of the differentially localized HMGB types.
MATERIALS AND METHODS
Plasmid Construction
The sequences encoding the various full-length, truncated, and chimeric HMG proteins were generated by amplifying the corresponding DNA fragments with Pfu DNA polymerase using an Arabidopsis (Arabidopsis thaliana) cDNA library as template and the primers listed in Supplemental Table S1. For expression of the mutant form of HMGB2, in which three Ser codons were replaced by Ala or Asp codons, the coding sequence was generated by overlap extension PCR (Ho et al., 1989). The PCR fragments were cloned into suitable plasmids as detailed below, and plasmid constructs were checked by DNA sequencing. For transient expression of GFP-HMGB fusions in protoplasts, the coding sequences were inserted into plasmid p5′GFP or p3′GFP (Haasen et al., 1999), providing the expressed protein with an N- or C-terminal GFP fusion, respectively. For expression of GFP-HMG fusions in stably transformed Arabidopsis plants, the GFP-HMG coding sequences were inserted into plasmid pGII0179-35S (Launholt et al., 2006). For analysis of protein dynamics, the HMGB coding sequences were inserted with paGFP in plasmid p5′paGFP (Martini et al., 2007). For expression of the wild-type and mutant forms of HMGB2 in Escherichia coli, the coding sequences were inserted into plasmid pQE9cm (Grasser et al., 1996), providing an N-terminal 6× His tag. Details of the plasmids generated in this work are summarized in Supplemental Table S1.
Production of Recombinant HMGB2 Proteins
The E. coli M15 strain was transformed with the pQE9 expression vectors, and the recombinant wild-type and mutant HMGB2 proteins were purified by three-step column chromatography (metal chelate, cation exchange, and anion exchange) as described previously (Grasser et al., 1996). Purified proteins were checked by SDS-PAGE and matrix-assisted laser-desorption ionization time of flight mass spectrometry.
CK2-Mediated Phosphorylation of HMGB2
Recombinant wild-type and mutant HMGB2 were phosphorylated in vitro using recombinant maize CK2α in the presence of [γ-32P]ATP as described previously (Stemmer et al., 2002). The samples were separated by SDS-PAGE on 18% SDS-polyacrylamide gels, and phosphorylated proteins were visualized with a Cyclone storage phosphor imager (Canberra Packard).
Immunoblot Analysis
An antiserum against purified recombinant Arabidopsis HMGB2 was produced by commercial immunization and tested as described previously (Launholt et al., 2006). The antiserum against Arabidopsis HMGB1 was described previously (Launholt et al., 2006; Lildballe et al., 2008). For immunoblot analyses, proteins were extracted from approximately 3 g of green tissue using 2% (w/v) TCA as described previously (Grasser et al., 1996). The protein extracts were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining to check the quality of the extracts. Immunodetection of the HMGB proteins was performed as described previously (Launholt et al., 2006).
Indirect Immunofluorescence
Immunodetection of HMGB1 and HMGB2 in root tip cells of Arabidopsis seedlings was performed using HMGB1/HMGB2-specific antisera as described previously (Launholt et al., 2006).
Plant Growth and Agrobacterium-Mediated Transformation
Arabidopsis (Col-0) and transgenic plant lines were grown in soil in a phytochamber at 22°C and 16 h of light per day as described previously (Lolas et al., 2010). The pGII0179 plasmids and the plasmid pSOUP (Hellens et al., 2000) were used to cotransform the Agrobacterium tumefaciens strain pGV3101 by electroporation. Transformed Agrobacterium cells were used to transform Arabidopsis plants of ecotype Col-0 employing the floral dip method (Clough and Bent, 1998). Plants growing on the selective medium were transferred to soil, and isolated genomic DNA was tested by PCR for the presence of the transgene (Lolas et al., 2010).
Transient Protoplast Transformation Assays with GFP Fusion Constructs
Protoplasts of dark-grown tobacco (Nicotiana tabacum) BY-2 cells were transiently transformed using polyethylene glycol-mediated transformation as described previously (Merkle et al., 1996). Excitation of GFP was performed with a standard UV light source and fluorescein isothiocyanate filters. For CLSM, samples were directly examined under oil with a 63× objective and a DM RBE TCS4D microscope (Leica) equipped with an argon-krypton laser (excitation at 488 nm, beam splitter at 510 nm, filter at 515 nm) using Leica Scanware. Analysis of the localization of the different GFP fusion proteins was performed in three independent experiments, each representing approximately 60 to 80 transformed protoplasts.
Microscopic Analysis of Protein Localization in Arabidopsis Root
Transgenic seedlings expressing GFP-HMGA and GFP-HMGB fusions were grown on MS medium (Murashige and Skoog, 1962) under long-day conditions at approximately 21°C in a plant incubator (Percival Scientific). Four-day-old seedlings were fixed in fixing solution (4.5% [w/v] paraformaldehyde, 50 mm phosphate buffer, pH 7.2, and 150 mm NaCl) prior to analysis by CLSM using a 510 Meta instrument (Zeiss) with Zeiss LSM Image browser software using the argon laser (pinhole, 394 μm; filter, BP 505–550).
Monitoring of Intracellular Protein Dynamics Using paGFP
The analysis of real-time protein mobility in transiently transformed tobacco BY-2 protoplasts expressing HMGB proteins fused to paGFP using multifocal two-photon laser scanning microscopy was performed as described previously (Martini et al., 2007).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Y14072 to Y14074.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. The plasmid constructs and primers used in this work.
Supplementary Material
Acknowledgments
We thank Andreas Houben (IPK Gatersleben) for performing the immunofluorescence experiment.
References
- Agresti A, Bianchi ME. (2003) HMGB proteins and gene expression. Curr Opin Genet Dev 13: 170–178 [DOI] [PubMed] [Google Scholar]
- Arwood LJ, Spiker S. (1990) Binding of wheat and chicken high mobility group chromosomal proteins to DNA and to wheat and chicken mononucleosomes. J Biol Chem 265: 9771–9777 [PubMed] [Google Scholar]
- Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. (2003) Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J 22: 5551–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M, Neihart NK. (1979) Antibodies against chromosomal HMG proteins stain the cytoplasm of mammalian cells. Cell 16: 181–189 [DOI] [PubMed] [Google Scholar]
- Bustin M, Reeves R. (1996) High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol 54: 35–100 [DOI] [PubMed] [Google Scholar]
- Calogero S, Grassi F, Aguzzi A, Voigtländer T, Ferrier P, Ferrari S, Bianchi ME. (1999) The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet 22: 276–280 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Costigan C, Kolodrubetz D, Snyder M. (1994) NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol Cell Biol 14: 2391–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser KD, Grill S, Duroux M, Launholt D, Thomsen MS, Nielsen BV, Nielsen HK, Merkle T. (2004) HMGB6 from Arabidopsis thaliana specifies a novel type of plant chromosomal HMGB protein. Biochemistry 43: 1309–1314 [DOI] [PubMed] [Google Scholar]
- Grasser KD, Grimm R, Ritt C. (1996) Maize chromosomal HMGc: two closely related structure-specific DNA-binding proteins specify a second type of plant high mobility group box protein. J Biol Chem 271: 32900–32906 [DOI] [PubMed] [Google Scholar]
- Grasser KD, Launholt D, Grasser M. (2007) High mobility group proteins of the plant HMGB family: dynamic chromatin modulators. Biochim Biophys Acta 1769: 346–357 [DOI] [PubMed] [Google Scholar]
- Grasser KD, Wurz A, Feix G. (1991) Isolation and characterisation of high-mobility-group proteins from maize. Planta 185: 350–355 [DOI] [PubMed] [Google Scholar]
- Grasser M, Lentz A, Lichota J, Merkle T, Grasser KD. (2006) The Arabidopsis genome encodes structurally and functionally diverse HMGB-type proteins. J Mol Biol 358: 654–664 [DOI] [PubMed] [Google Scholar]
- Guerra B, Boldyreff B, Sarno S, Cesaro L, Issinger OG, Pinna LA. (1999) CK2: a protein kinase in need of control. Pharmacol Ther 82: 303–313 [DOI] [PubMed] [Google Scholar]
- Haasen D, Köhler C, Neuhaus G, Merkle T. (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J 20: 695–705 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Johns EW, editor (1982) The HMG Chromosomal Proteins. Academic Press, London [Google Scholar]
- Kwak KJ, Kim JY, Kim YO, Kang H. (2007) Characterization of transgenic Arabidopsis plants overexpressing high mobility group B proteins under high salinity, drought or cold stress. Plant Cell Physiol 48: 221–231 [DOI] [PubMed] [Google Scholar]
- Launholt D, Grønlund JT, Nielsen HK, Grasser KD. (2007) Overlapping expression patterns among the genes encoding Arabidopsis chromosomal high mobility group (HMG) proteins. FEBS Lett 581: 1114–1118 [DOI] [PubMed] [Google Scholar]
- Launholt D, Merkle T, Houben A, Schulz A, Grasser KD. (2006) Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus. Plant Cell 18: 2904–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichota J, Grasser KD. (2001) Differential chromatin association and nucleosome binding of the maize HMGA, HMGB, and SSRP1 proteins. Biochemistry 40: 7860–7867 [DOI] [PubMed] [Google Scholar]
- Lichota J, Ritt C, Grasser KD. (2004) Ectopic expression of the maize chromosomal HMGB1 protein causes defects in root development of tobacco seedlings. Biochem Biophys Res Commun 318: 317–322 [DOI] [PubMed] [Google Scholar]
- Lildballe DL, Pedersen DS, Kalamajka R, Emmersen J, Houben A, Grasser KD. (2008) The expression level of the chromatin-associated HMGB1 protein influences growth, stress tolerance, and transcriptome in Arabidopsis. J Mol Biol 384: 9–21 [DOI] [PubMed] [Google Scholar]
- Lolas IB, Himanen K, Grønlund JT, Lynggaard C, Houben A, Melzer M, Van Lijsebettens M, Grasser KD. (2010) The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J 61: 686–697 [DOI] [PubMed] [Google Scholar]
- Martini J, Schmied K, Palmisano R, Toensing K, Anselmetti D, Merkle T. (2007) Multifocal two-photon laser scanning microscopy combined with photo-activatable GFP for in vivo monitoring of intracellular protein dynamics in real time. J Struct Biol 158: 401–409 [DOI] [PubMed] [Google Scholar]
- Merkle T. (2003) Nucleo-cytoplasmic partitioning of proteins in plants: implications for the regulation of environmental and developmental signalling. Curr Genet 44: 231–260 [DOI] [PubMed] [Google Scholar]
- Merkle T, Leclerc D, Marshallsay C, Nagy F. (1996) A plant in vitro system for the nuclear import of proteins. Plant J 10: 1177–1186 [DOI] [PubMed] [Google Scholar]
- Moehs CP, McElwain EF, Spiker S. (1988) Chromosomal proteins of Arabidopsis thaliana. Plant Mol Biol 11: 507–515 [DOI] [PubMed] [Google Scholar]
- Mosevitsky MI, Novitskaya VA, Iogannsen MG, Zabezhinsky MA. (1989) Tissue specificity of nucleo-cytoplasmic distribution of HMG1 and HMG2 proteins and their probable functions. Eur J Biochem 185: 303–310 [DOI] [PubMed] [Google Scholar]
- Müller S, Ronfani L, Bianchi ME. (2004) Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med 255: 332–343 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nemeth MJ, Cline AP, Anderson SM, Garrett-Beal LJ, Bodine DM. (2005) Hmgb3 deficiency deregulates proliferation and differentiation of common lymphoid and myeloid progenitors. Blood 105: 627–634 [DOI] [PubMed] [Google Scholar]
- Nemeth MJ, Kirby MR, Bodine DM. (2006) Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc Natl Acad Sci USA 103: 13783–13788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen DS, Grasser KD. (2010) The role of chromosomal HMGB proteins in plants. Biochim Biophys Acta 1799: 171–174 [DOI] [PubMed] [Google Scholar]
- Pedersen TJ, Arwood LJ, Spiker S, Guiltinan MJ, Thompson WF. (1991) High mobility group chromosomal proteins bind to AT-rich tracts flanking plant genes. Plant Mol Biol 16: 95–104 [DOI] [PubMed] [Google Scholar]
- Pinna LA, Ruzzene M. (1996) How do protein kinases recognize their substrates? Biochim Biophys Acta 1314: 191–225 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Kuehl L. (1979) Microinjection of the nonhistone chromosomal protein HMG1 into bovine fibroblasts and HeLa cells. Cell 16: 901–908 [DOI] [PubMed] [Google Scholar]
- Reeves R. (2010) Nuclear functions of the HMG proteins. Biochim Biophys Acta 1799: 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritt C, Grimm R, Fernández S, Alonso JC, Grasser KD. (1998) Basic and acidic regions flanking the HMG domain of maize HMGa modulate the interactions with DNA and the self-association of the protein. Biochemistry 37: 2673–2681 [DOI] [PubMed] [Google Scholar]
- Ronfani L, Ferraguti M, Croci L, Ovitt CE, Schöler HR, Consalez GG, Bianchi ME. (2001) Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development 128: 1265–1273 [DOI] [PubMed] [Google Scholar]
- Spiker S. (1984) High-mobility group chromosomal proteins of wheat. J Biol Chem 259: 12007–12013 [PubMed] [Google Scholar]
- Stemmer C, Leeming DJ, Franssen L, Grimm R, Grasser KD. (2003) Phosphorylation of maize and Arabidopsis HMGB proteins by protein kinase CK2α. Biochemistry 42: 3503–3508 [DOI] [PubMed] [Google Scholar]
- Stemmer C, Ritt C, Igloi GL, Grimm R, Grasser KD. (1997) Variability in Arabidopsis thaliana chromosomal high-mobility-group-1-like proteins. Eur J Biochem 250: 646–652 [DOI] [PubMed] [Google Scholar]
- Stemmer C, Schwander A, Bauw G, Fojan P, Grasser KD. (2002) Protein kinase CK2 differentially phosphorylates maize chromosomal high mobility group B (HMGB) proteins modulating their stability and DNA interactions. J Biol Chem 277: 1092–1098 [DOI] [PubMed] [Google Scholar]
- Stros M, Launholt D, Grasser KD. (2007) The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci 64: 2590–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO, Travers AA. (2001) HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci 26: 167–174 [DOI] [PubMed] [Google Scholar]
- Thomsen MS, Franssen L, Launholt D, Fojan P, Grasser KD. (2004) Interactions of the basic N-terminal and the acidic C-terminal domains of the maize chromosomal HMGB1 protein. Biochemistry 43: 8029–8037 [DOI] [PubMed] [Google Scholar]
- Webster CI, Packman LC, Pwee KH, Gray JC. (1997) High mobility group proteins HMG-1 and HMG-I/Y bind to a positive regulatory region of the pea plastocyanin gene promoter. Plant J 11: 703–715 [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Schulze E, Sapetto B. (1994) DNA binding and nuclear translocation of insect high-mobility-group protein-1 (HMG1) proteins are inhibited by phosphorylation. Eur J Biochem 225: 687–693 [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang W, Pwee KH, Kumar PP. (2003) Rice HMGB1 protein recognizes DNA structures and bends DNA efficiently. Arch Biochem Biophys 411: 105–111 [DOI] [PubMed] [Google Scholar]
- Yang H, Tracey KJ. (2010) Targeting HMGB1 in inflammation. Biochim Biophys Acta 1799: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu Q, Pwee KH, Manjunatha Kini R. (2003) Interaction of wheat high-mobility-group proteins with four-way-junction DNA and characterization of the structure and expression of HMGA gene. Arch Biochem Biophys 409: 357–366 [DOI] [PubMed] [Google Scholar]
- Zlatanova J, van Holde K. (1998) Linker histones versus HMG1/2: a struggle for dominance? Bioessays 20: 584–588 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.