Abstract
Huntington’s disease (HD) is a fatal autosomal dominant neurodegenerative disorder caused by an expansion of the polyglutamine (polyQ) repeat in exon-1 in the Huntingtin gene (HTT). This results in misfolding and accumulation of the huntingtin (htt) protein, forming nuclear and cytoplasmic inclusions. HD is associated with dysregulation of gene expression as well as mitochondrial dysfunction. We hypothesized that by improving transcriptional regulation of genes necessary for energy metabolism, the HD motor phenotype would also improve. We therefore examined the protective effects of nicotinamide (NAM), a well-characterized water-soluble B vitamin that is an inhibitor of sirtuin1/class III NAD+-dependent histone deacetylase (HDAC). In this study, both mini-osmotic pumps and drinking water deliveries were tested at 250mg NAM/kg/day, using the B6.HDR6/1 transgenic mouse model. Results were similar for both modes of delivery, and there was no evidence of toxicity. We found that NAM treatment increased mRNA levels of brain-derived neurotrophic factor (BDNF), and Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), the master regulator of mitochondrial biogenesis. Protein levels of BDNF were also significantly increased. In addition, NAM treatment increased PGC-1α activation in HD mice, pointing to a possible mode of action as a therapeutic. Critically, NAM treatment was able to improve motor deficits associated with the HD phenotype, tested as time courses of open field, rotarod, and balance beam activities. These improvements were substantial, despite the fact that NAM did not appear to reduce htt aggregation, or to prevent late-stage weight loss. Our study therefore concludes that NAM or similar drugs may be beneficial in clinical treatment of the motor dysfunctions of HD, while additional therapeutic approaches must be added to combat the aggregation phenotype and overall physiological decline.
Keywords: Huntington’s Disease, nicotinamide, vitamin, Brain-derived neurotrophic factor, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, histone deacetylase inhibitor
Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder characterized by motor dysfunction, emotional disturbances, cognitive dysfunction, weight loss, and premature death. HD is caused by an expansion of a trinucleotide (CAG)n repeat in the coding sequence of the huntingtin (HTT) gene on chromosome-4, resulting in an expanded polyglutamine (polyQ) tract in huntingtin (htt), with the age of onset inversely correlated with repeat number (Duyao et al., 1993). HD, along with Alzheimer’s disease, Parkinson’s disease (PD), spinal cerebellar ataxia, and prion-based dementia, is classified as a misfolded protein disorder, based on the characteristic intranuclear and cytoplasmic pathogenic aggregations of truncated and full-length htt protein observed in human patients and in mouse models of HD (Davies et al., 1997; DiFiglia et al., 1997). The HDR6/1 transgenic mouse model contains human huntingtin gene exon-1 under the control of the endogenous promoter, with approximately 115 CAG repeats. In this model, overt neurological symptoms similar to human HD become evident at 15–21 weeks of age, with premature death occurring at 32–40 weeks (Mangiarini et al., 1996). Behavioral abnormalities have been documented as early as 4 weeks of age (Bolivar et al., 2004).
Altered transcription of genes involved in cellular processes vital to neuronal function and survival has been demonstrated in mouse and human HD (Luthi-Carter et al., 2000; Zuccato et al., 2001; Zuccato et al., 2003) suggesting a direction for therapies. Acetylation and deacetylation of histones play an important role in regulation of chromatin condensation and gene transcription (Roth et al., 2001); therefore, increasing histone acetyltransferase activity, through inhibition of the histone deacetylases (HDACs), has been used as a strategy to slow the progression of many neurological disorders. The efficacy of HDAC inhibition in HD was first observed in a Drosophila model (Steffan et al., 2001), with suberoylanilide hydroxamic acid blocking photoreceptor neurodegeneration and increasing survival. In addition, HDAC inhibitors have provided phenotypic improvement in HD transgenic mouse models (Ferrante et al., 2003; Hockly et al., 2003).
The superfamily of HDACs consists of five main subtypes (Butler and Bates, 2006), including the structurally distinct class III which contains the family of sirtuins. Sirtuins comprise a unique class of nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases (Imai et al., 2000) that are involved in diverse biological functions such as metabolism, cell division and aging (Taylor et al., 2008). To date, seven mammalian homologues have been identified, with mammalian SIRT1 closest evolutionarily to yeast Sir2 (silent information regulator 2), the founding member of the sirtuin family (Rine et al., 1979).
SIRT1 is a multifunctional protein that regulates diverse cellular functions through deacetylation of important transcription factors, such as p53, forkhead subgroup O (FOXO) proteins and the DNA repair factor KU (Guarente, 2006). The observation that sirtuin activity requires NAD+ suggests a mechanistic link between sirtuin activity and intracellular energetics (Imai et al., 2000). The peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) is another well-known target of SIRT1 and its activation requires SIRT1 deacetylation (Nemoto et al., 2005). PGC-1α is a key regulator of mitochondrial biogenesis and respiration, modulating expression of several transcription factors (McGill and Beal, 2006).
Although the actions of sirtuins in the nervous system are only beginning to be explored, it has been reported that pharmacological modulation of SIRT1 can have significance in managing neurodegenerative diseases (Green et al., 2008; Lagouge et al., 2006). An early in vivo study using nicotinamide (NAM) also known as vitamin B3 or niacin, identified it as an effective suppressor of polyglutamine toxicity in a model of spinocerebellar ataxia (Ghosh and Feany, 2004). In addition to being a precursor for ATP, NAM is classified as a class III HDAC inhibitor, because of its role as a SIRT1 inhibitor (Bitterman et al., 2002). More recently, researchers have shown that high doses of NAM improved cognition in a mouse model of Alzheimer’s disease (Green et al., 2008), reduced motor deficits in a rodent PD model (Jia et al., 2008) and reduced both neuronal loss and motor impairment in the Drosophila model of HD (Pallos et al., 2008). Thus, we hypothesized that NAM will be effective in alleviating HD pathology in the B6.HDR6/1 mouse model.
NAM has been evaluated for its potential as a therapeutic for a range of disorders, from heart disease to juvenile diabetes; with doses as high as 6 grams/day with no major signs of toxicity (Mandrup-Poulsen et al., 1993). Most important to this study, NAM injected sub-cutaneously into rats has been shown to increase NAM levels in plasma, CSF, and brain (Erb and Klein, 1998). NAM is therefore pharmacologically feasible as an HD therapeutic.
Materials and Methods
Animals
B6.HD6/1 mice were generated and maintained by breeding transgenic males to C57BL/6J females (Jax # 000664). Offspring were genotyped by polymerase chain reaction (PCR) from tail biopsy. HDR6/1 mice, a moderate fragment model of HD, have human HTT Exon 1 with 1kB of the endogenous promoter (Mangiarini et al., 1996). This model recapitulates several phenotypes found in human HD patients, including neuropathology with nuclear and cytoplasmic aberrations in the corpus striatum and hippocampus, behavioral deficits, gene expression changes and premature death (Davies et al., 1997; Desplats et al., 2006; Iannicola et al., 2000; Turmaine et al., 2000). Mice were maintained on a 12h light–dark cycle under standard housing conditions, with LabDiet 5PO4 Autoclavable Prolab RMH 3500. All procedures were approved by the Wadsworth Center Institutional Animal Care and Use Committee.
Animal treatment
Male and female mice were administered 250 mg/kg NAM (Fluka). In separate experiments, either via Alzet® mini-osmotic pump (Model 2004; 0.9% saline controls, subcutaneous implants replaced after 5 weeks); or drinking water (50ml bottles). Mice were housed three animals per cage at 8 weeks of age, randomized for genotype to reduce symptomatic effects seen in our colony when animals from one genotype are housed together. Treatment was started at 8 weeks of age (early symptomatic), and ended at 20 weeks (Late symptomatic). Animals were monitored daily for signs of toxicity, such as wasting or inactivity, and sacrificed 2 weeks after the completion of behavioral testing. CAG sizing was done to maintain consistency among animals used in this study. CAG lengths were between 122–127 glutamines.
Behavioral assays
Behavioral experiments were conducted during the light cycle by a researcher blinded to genotype. Repeated tests were carried out in the same room, under the same climate conditions, at approximately the same time of day. Repeated behavioral tests were compared by repeated-measures ANOVA for all behavioral tasks. Tukey’s HSD post hoc test was used to determine significance. Effects of treatment were analyzed using Stat View 5.0 software (SAS Institute Inc.).
Open field testing
For evaluation of exploratory activity, each mouse was tested for 15 min in a completely dark, open field chamber (65×55×55 cm; Med Associates). Approximately 1 hr before testing the mice were placed in the testing room to permit acclimation. Each mouse was placed in the center of the dark chamber and allowed to move freely. Open field testing was performed biweekly, starting at 10 weeks of age, until 20 weeks. Data from the session were acquired using Digiscan 16-beam automated activity monitors (42×42×30 cm; AccuScan).
Rotarod performance
A rotarod apparatus (AccuScan) was used to measure motor coordination and balance. Each animal was tested bi-weekly, starting at 12 weeks and ending at 20 weeks of age. A constant-acceleration protocol was used (4–40 rpm for a max of 300 sec). The mice were trained for 3 consecutive days using the same parameters at 10 weeks. At 12 weeks each mice underwent 2 consecutive days of testing, with 3 trials/day. To avoid fatigue, sessions were separated by a 15-min rest period.
Balance Beam
Mice were assessed on time to cross the center region (80 cm) of a 1-m long beam using a protocol modified from (Carter et al., 2001). Six square or circular beams of different widths were used. The beams were mounted atop poles, with a narrow platform at the starting end and a dark box containing the animal's home-cage nesting material at the far end. A container filled with bedding was used to prevent injury to mice falling from the beam. At the start of the experiment, mice were placed at the start end of the beam, and the length of time between when the entire body of the mouse entered and when the nose of the mouse exited the 80-cm central region was measured manually. (Testing distance of the center 80 cm was used because the mice sometimes hesitate before starting and before entering the dark box). Mice initially were trained for 3 consecutive days on the 28-mm circle beam; training trials were repeated until each animal crossed the beam twice without stopping or turning around. On testing days, two trials were performed per mouse for each beam, with a 15-min rest period between beams. Testing trials in which the animal took longer than 60s to cross, or fell off the beam, were scored as 60s. Trials in which the animal stopped or turned around were repeated. The average of the trials was scored.
Quantitative Polymerase Chain Reaction (qPCR)
Animals were sacrificed using C02, brains removed, the left hemisphere placed in 4% paraformaldehyde for fixation, and the right hemisphere placed in RNAlater (Invitrogen). RNAlater-preserved tissue was placed in Trizol Reagent (Invitrogen), homogenized and RNA extracted using the manufacturer’s procedure. The acquired RNA pellet was resuspended in nuclease-free water (Ambion) containing RNase inhibitor (Invitrogen). Total RNA was confirmed with an ND-1000 spectrophotometer (NanoDrop Technologies). Genomic DNA was removed by treating the sample with RNase-free DNase (Promega). Using a high-capacity cDNA archiving kit (Applied Biosystems), total RNA was reverse-transcribed into single-stranded cDNA. To quantify mRNA levels, TaqMan® Gene Expression Assays (Applied Biosystems) were used: PGC-1α Mm00447180_m1 exon boundary 2–3; BDNF Mm00432069_m1 exon boundary 1–2.
Immunohistochemistry
After 20 hr in fixative at 4°C, the left hemisphere was placed in 15% sucrose and stored at 4°C. Thirty-micrometer (μm) frozen coronal brain sections were cut using a sliding microtome. Free-floating sections were processed as previously described (Pabello et al., 2005). In brief, sections were incubated in primary antibody (EM48, Chemicon 1:500) for 40 hr at 4°C; followed by horse anti-mouse biotin-conjugated secondary antibody (1:200) and ABC-peroxidase (Vector Labs). Immunostaining was visualized with a final incubation in 0.05% diaminobenzidine containing 0.0015% H2O2, and 0.25% nickel ammonium sulfate.
Immunoblotting
Mouse brains were homogenized in RIPA lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 0.1% SDS) supplemented with 1X
Complete protease inhibitor cocktail (Roche) and centrifuged for 15 min at 15,000g. The supernatants were separated by 4–16% gradient Tris–Hepes–SDS-PAGE (Pierce, Rockford, IL) and blotted onto polyvinylidene difluoride (PVDF) transfer membranes (PerkinElmer LifeSciences, Inc., Boston, MA). Membranes were blocked overnight at 4°C in 5% milk TBS-T buffer. On day two the membranes were incubated at room temperature in diluted primary antibodies. Immunoreactivity was detected after 1 h of incubation with goat anti-mouse (or anti-rabbit) IgG–horseradish peroxidase (1:2000; Santa Cruz Biotechnology) conjugate in 5% milk TBS-T buffer, using the Western Lightning Chemiluminescence kit (PerkinElmer LAS, Inc., Boston, MA). The primary antibodies that were used were rabbit polyclonal anti-PGC-1α (1:500; Santa Cruz Biotechnology), mouse monoclonal anti-acetyl-lysine (1:500; Millipore), SIRT1 (Cell Signaling; 1:1000), mouse monoclonal actin (Sigma; 1:2000), EM-48 (1:1000; Millipore).
Immunoprecipitation (IP)
Mouse brains were prepared in RIPA lysis buffer supplemented with 1X Complete protease inhibitor cocktail (Roche) and centrifuged for 10 min at 15,000g. The experiment was preformed using the manufacturer’s protocol (Invitrogen, Dynabeads G). However, specific to this experiment, beads were coated with 2μg primary antibody (PGC-1α Santa Cruz Biotechnology). Next, 500μg of lysates was added to beads and incubated at room temperature, with rotation for 45 min. After elution of sample, 25 μl was loaded onto a 4–16% gradient Tris–Hepes–SDS-PAGE. The remaining steps were as described in the immunoblotting section.
Enzyme-linked immunosorbent assay (ELISA)
The Promega BDNF Emax ImmunoAssay System (Promega Co., Madison, WI) was used to quantify the amount of BDNF in whole brain samples (cerebrum only) as described in (Szapacs et al., 2004). In brief, the left hemisphere was lysed in RIPA Buffer, supplemented with 1X Complete protease inhibitor cocktail (Roche), at 10x weight of the sample, centrifuged for 15 min at 15,000g. Supernatants were then removed and frozen at −80°C until analysis. Samples were not acid treated in order to measure mature BDNF (see manufacturer’s protocol for details). BDNF levels are reported in pg/ml ±S.E.M. which was extrapolated using the standard curve generated.
Agarose gel electrophoresis for resolving aggregates (AGERA)
For aggregation analysis, the right hemisphere of the brain was homogenized in 2% sodium dodecyl sulfate (SDS) lysis buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2% SDS) supplemented with 1X complete protease inhibitor cocktail (Roche). Protein was quantified using the Bio-Rad DC Protein Assay kit. 30μg of total protein per sample was processed and resolved on short 1.5% agarose gels as described (Weiss et al., 2008). Mouse monoclonal EM-48 (Chemicon; 1:1000), goat anti-mouse IgG–horseradish peroxidase (1:2000; Santa Cruz Biotechnology), and Western Lightning chemiluminescence kit (PerkinElmer LAS, Inc) were used.
Aggregate counting
Images of EM-48 immunostained striatum, captured using a 20X objective, were taken from 2 sections per animal. Sections were matched rostral-caudal for brain position to eliminate discrepancy in brain volume and structural differences Using ImageJ (NIH), particles/field were counted using the Threshold and Analyze Particles functions, to display and quantify particles representing aggregates.
Liver toxicity
Blood alanine aminotransferase (ALT) was measured in whole blood obtained at the time of sacrifice by cardiac puncture. ALT levels were quantified with a mammalian liver profile rotor (ABAXIS®). Samples were processed using the manufacturer’s protocol.
Results
NAM improves transcriptional dysregulation of BDNF and PGC-1α, restores BDNF protein levels and increases acetylated PGC-1α
Eight week old B6.HDR6/1 mice were treated with 250 mg/kg of NAM for 10 weeks via two consecutive minipumps, or for 12 weeks in drinking water. At the end of the treatment period, the animals were assessed for gene expression changes that we hypothesized would improve after NAM treatment. Importantly, neither mode of administration nor sex affected outcomes.
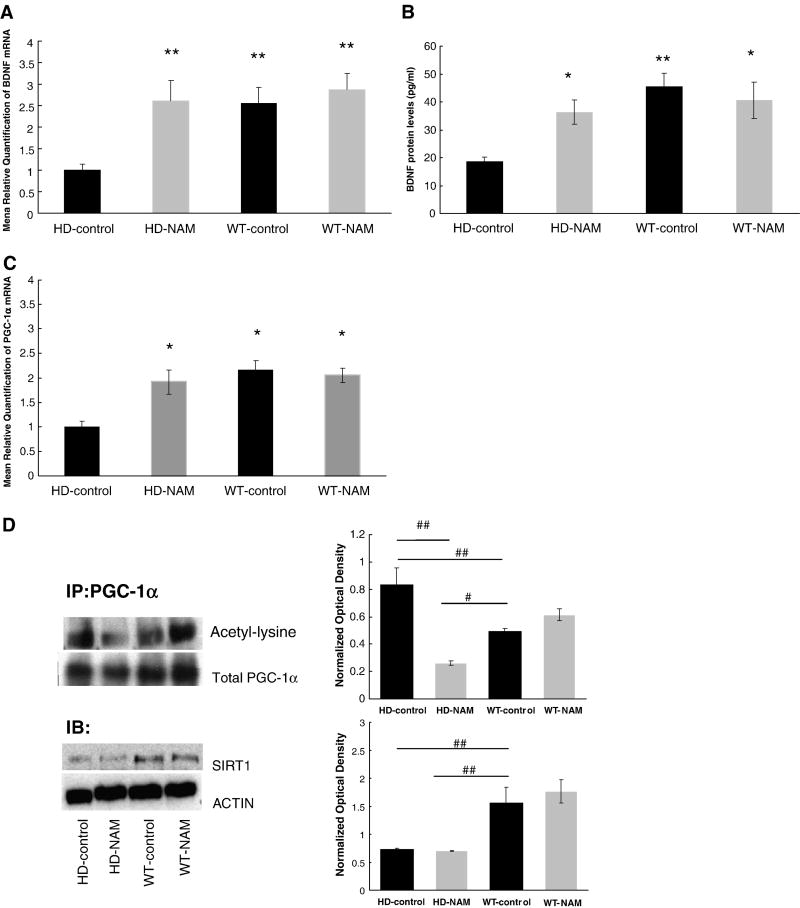
Brain-derived neurotrophic factor (BDNF) is critical to neuronal survival in the CNS, and has been shown to be down-regulated in HD patients and mice (Zuccato and Cattaneo, 2009). In this study, there was a 2.5-fold reduction in BDNF mRNA levels in HD-control mice when compared to wild-type (wt) mRNA levels [relative quantification (RQ) with NAM treatment as compared to animals receiving control treatment] (Fig. 1A). Importantly, NAM treatment did not affect the mRNA levels of BDNF in wt animals (Fig. 1A). In addition, protein levels of BDNF in HD mice were also improved with treatment (Fig. 1b). These findings specify a protective effect of NAM in the brains of HD mice.
Figure 1. Analysis of messenger RNA levels and protein levels.
Analysis of messenger RNA levels and protein levels of BDNF and PGC-1α. (A). Mice were treated for 12 weeks and sacrificed at 20 weeks of age. The right hemisphere was dissected and analyzed using a TaqMan® assay to quantify the levels of BDNF (A) and PGC-1α (C). Endogenous control is β-actin Expression shown in fold change. (B) ELISA data show BDNF protein levels in right hemisphere of the brain. (*=p<0.05 **=p<0.005 as compared to HD-control; ANOVA) n=6 per group (D) Immunoblot analysis of PGC-1α-activation and SIRT1 from right hemisphere. (#=p<0.05 ##=p<0.008; ANOVA) n= 3 per group. All values reported as mean ± SEM. NAM=nicotinamide IP=Immunoprecipation IB= Immunoblot.
NAM may exert its effects by improving mitochondrial function, either directly through ATP production, or through SIRT-1 inhibition. Since SIRT-1 regulates the activity of PGC-1α the effects of NAM on the expression of PGC-1α was evaluated (Handschin and Spiegelman, 2006). In HD mice, there was a ~2-fold lower expression of PGC-1α. NAM treatment restored the PGC-1α expression in transgenic mice to wt levels (Fig. 1c). We further analyzed the effects of NAM on PGC-1α by determining its acetylation state (level of activation) in response to NAM treatment. In HD, mice we show a decrease in PGC-1α acetylation, indicating increase activation of PGC-1α (Fig 1d). Finally, SIRT1 protein levels are reduced in HD mice relative to wt [as previously reported (Pallas et al., 2008)], and NAM treatment did not normalize these levels (Fig 1d).
NAM improves motor dysfunction in HD mice
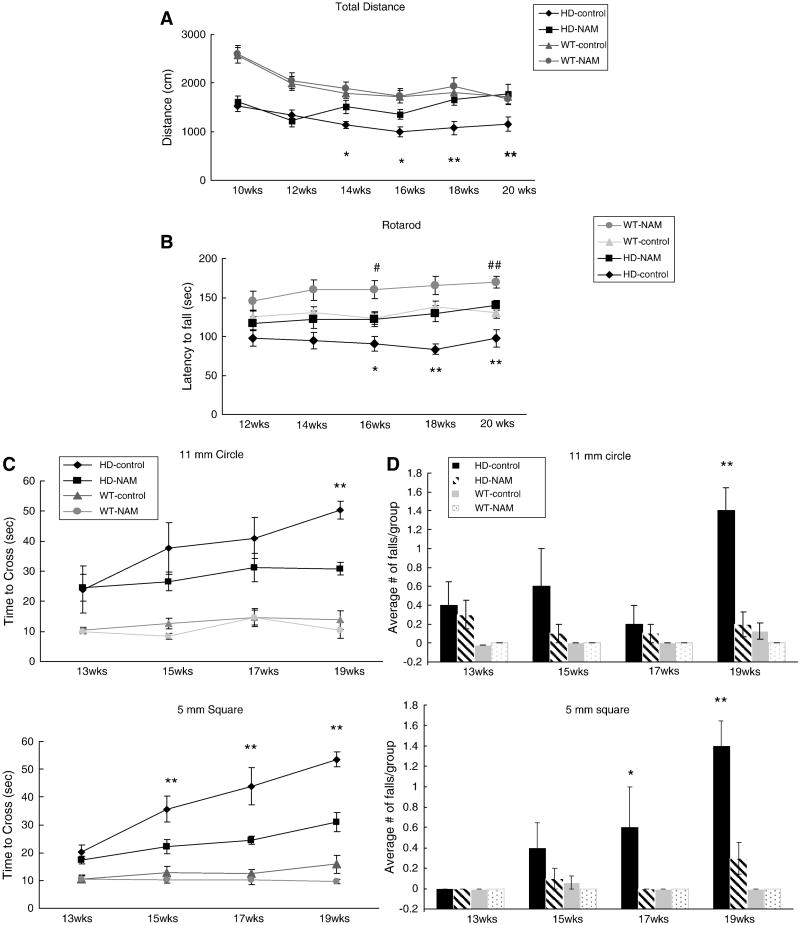
Correcting expression of genes involved in mitochondrial function may also ameliorate motor deficits associated with HD. We therefore tested three behaviors: openfield, rotarod, and balance beam. Female mice received 250 mg/kg NAM via their drinking water starting at 8 weeks, with motor testing throughout the treatment period, starting at 10 weeks of age. Decreased open field exploratory behavior of HD mice, compared with wt mice, was observed at every time point tested (Fig. 2A). Increased exploratory behavior, as measured by total distance traveled, was apparent by 14 weeks of age in NAM-treated HD animals; by the end of testing, their exploratory activity did not differ significantly from wt animals.
Figure 2. Multiple behavioral analyses of B6.HDR6/1 mice.
Multiple behavioral analyses of B6.HDR6/1 mice. (A) Openfield: mice were placed in an openfield chamber for 15 min every 2 weeks. Activity scored using Digiscan 16-beam automated activity monitors. (B) Rotarod: Each animal was tested at 2-week intervals on a constant acceleration protocol (4 to 40 rpm for a max of 5 min). (C) Balance beam: mice were assessed on the time to cross the center region (80 cm) of a 1 m beam, either 5 mm square or 11 mm circular. (D) Average number of falls from each group from each day of testing. Repeated behavior tests were compared using repeated-measures ANOVA for all behavioral tasks. Tukey’s HSD post hoc tests were used to determine significance. (*=p<0.05 **=p<0.01 HD-control vs. HD-NAM #=p<0.05 ## =p<0.01 with comparison of wt-control vs wt-NAM) (Average n=10 per HD group, 18 per wt group)
Animals receiving NAM also improved their rotarod performance, starting at 14 weeks of age (Fig. 2B). Increased performance was also seen in the wt mice, presumably due to the endurance-boosting nature of B vitamins. To further analyze the specificity and extent of motor improvement, we used a balance beam test, which tests balance and coordination and is not dependent on endurance. On the most difficult bars (5 mm square and 11mm circle) NAM-treated symptomatic animals crossed the beam faster (Fig. 2 C,D) and exhibited fewer falls (data not shown). Based on three behavioral tasks, we conclude that NAM significantly improves motor function of B6.R6/1 mice.
NAM has no effect on the aggregation phenotype
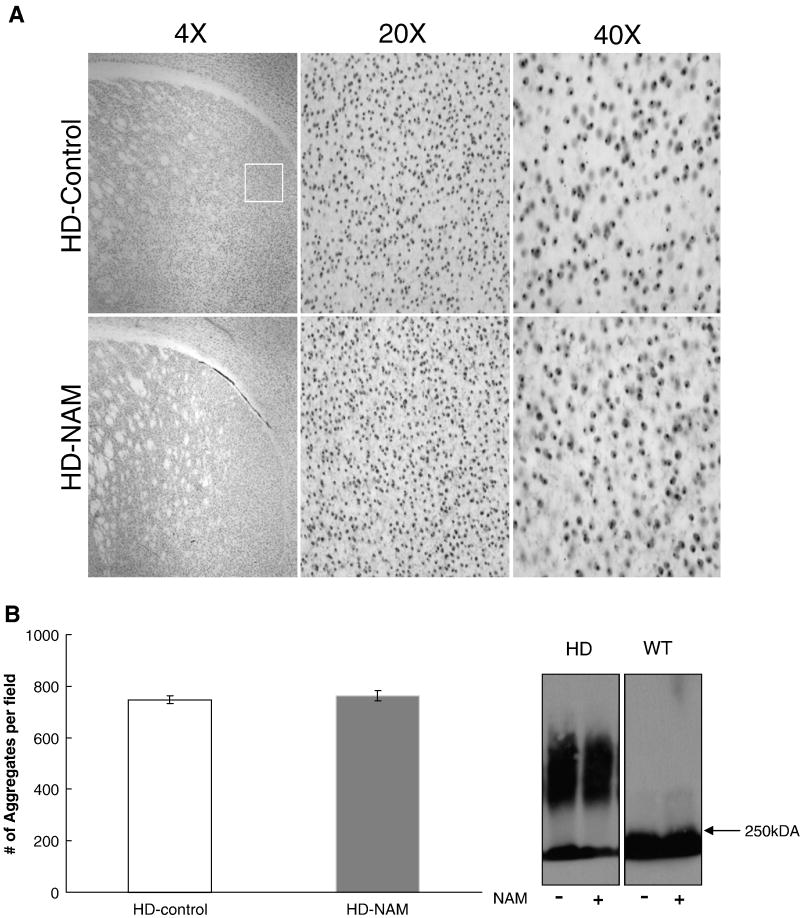
Aggregation of mHtt is central to HD pathology; therefore, brains were assessed for aggregate content after NAM treatment. The left hemisphere was fixed and probed using EM-48 antibody to visualize aggregates within the striatum (Fig. 3A). Counts revealed that the number of aggregates per field was unchanged with treatment (Fig. 3B). An additional experiment used SDS-AGE gels to assess size or conformational changes in aggregates in brain lysates. No change in migration of detergent-insoluble proteins (aggregates) was observed as a function of treatment (Fig. 3C). We therefore conclude that, despite the statistically significant improvement of motor behaviors and BDNF and PGC-1α mRNAs, NAM did not reduce observable aggregation.
Figure 3. Analysis of aggregates.
Analysis of aggregates. (A) Immunohistochemistry of aggregates. Representative images of sections probed with an anti-htt antibody (EM-48). White box represents the region that images were taken from for aggregate counts. (B) Analysis of number of aggregates in the striatum of B6.HDR6/1 mice. Aggregates were counted using ImageJ (NIH) using the Analyze Particles feature. N=5 per group (mean number from two 20X fields per animal). (C) AGERA, probed with EM-48 antibody, for analysis of aggregate load and conformation. Representative blot shown (3 replicates) (n=6 per group). 250kDA= highest band on protein ladder.
High dose NAM does not alter liver ALT enzyme levels or influence body weight
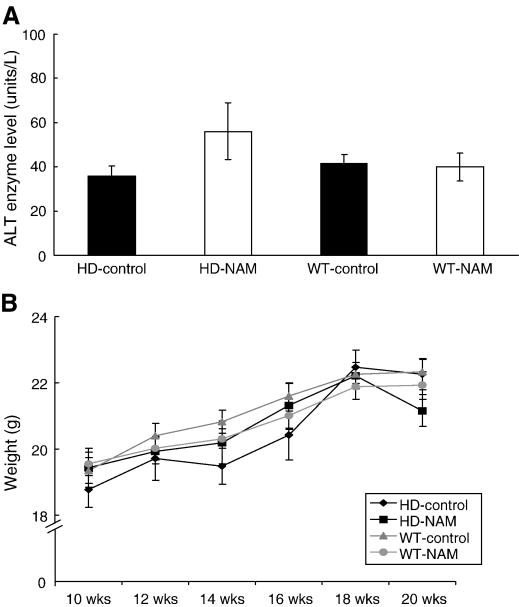
Two methods were used to assess potential drug-induced toxicity of the relatively high levels of NAM in this study. Alanine aminotransferase (ALT) is an enzyme present in healthy liver and heart cells that is released into blood when the liver or heart is damaged due to injury or toxicity. Analysis of ALT levels in the blood of HD mice receiving ad libitum NAM administration, as compared to levels in untreated mice, showed no significant differences in the amount of ALT (Fig. 4A). This strongly suggests that no major liver or heart toxicity was associated with administration of a high dose of NAM. During testing, animal weight was monitored to evaluate overall toxicity, and to determine whether NAM could affect HD-phenotypic weight loss. NAM neither exacerbated nor ameliorated weight loss associated with the HD phenotype (Fig. 4B).
Figure 4. Analysis of drug-induced toxicity.
Analysis of drug-induced toxicity. (A) Analysis of Alanine Aminotransferase (ALT). Upon sacrifice of female mice, blood was removed from each animal via a cardiac puncture. ALT levels were determined using a mammalian liver profile (Abaxis®). N=6 per group. (B) Animals were weighed weekly.
Discussion
The use of HDAC inhibitors to improve HD-mediated gene dysregulation is an appealing strategy, although it may result in non-specific targeting effects (Shao and Diamond, 2007). In addition, the long-term use of HDAC inhibitors in humans may be limited by high toxicity. In this study we use a class III HDAC (sirtuin) inhibitor that is in long-term human clinical use for other disorders, and that produces no apparent toxic effects here in the HD mice. Our results compare favorably with those seen with the use of other HDAC inhibitors. First, we showed that NAM treatment increases gene expression of BDNF and PGC-1α, correlated with improvement in BDNF protein levels and in PGC-1α activation. Secondly, NAM treatment improved HD-associated motor deficits on three behavioral tasks. Lastly, we did not see a reduction in mhtt aggregation nor HD-mediated weight-loss, suggesting that additional therapeutics will need to be added. Aggregation can be counteracted using engineered intrabodies in this B6.R6/1 mouse model (Snyder-Keller et. al. 2010), and in other HD mouse models (Southwell et al., 2009) and in Drosophila htt-exon1 models (Wolfgang et al., 2005). The Drosophila models also show enhanced protection with combinatorial therapies (Bortvedt et al., 2010; McLear et al., 2008).
Improving transcription of genes necessary for mitochondrial biogenesis as strategy for the development of therapy for HD was recently reviewed in article which focuses mostly on PGC-1α but discusses a possible interaction of BDNF and energy metabolism (Ross and Thompson, 2006). BDNF was shown to rapidly regulate energy metabolism through increased sympathetic nervous system activity in obese mice, attributed in part to induction of Ucp1 gene expression (Nonomura et al., 2001). Our data also suggest that BDNF may have actions linked to PGC-1α. However, the interplay of these different mechanisms in HD pathogenesis still needs to be elucidated.
There are several mechanisms by which the mhtt can result in mitochondrial dysfunction, including by directly interacting with mitochondria (Orr et al., 2008) or by altering gene expression (Cha, 2000). Mitochondrial impairment in HD results from interference of mhtt with PGC-1α activity and may increase cellular vulnerability to stress (Cui et al., 2006). In support of this, PGC-1α and its target genes were found to be reduced in HD patients and in HD mouse striatum, and the rate of mitochondrial oxygen consumption was found to be reduced (Weydt et al., 2006). The initial hypothesis that PGC-1α could play a role in HD pathogenesis came from studies on PGC-1α knockout mice (Leone et al., 2005; Lin et al., 2004), Although the parallels have limitations, the production of a phenotype that is reminiscent of HD in response to decreased PGC-1α raises the interesting possibility that this protein is involved in HD pathogenesis, and a potential target for HD therapy. Here we show that activated PGC-1α is decreased in the brains of B6.HDR6/1 mice, and that NAM treatment partially restores regulation. We postulate that NAM is being converted to NAD+ to compensate for the altered energy state associated with HD, and is acting as an activator rather than an inhibitor of SIRT 1. We do not rule out the possibilities of NAM acting via other Sirtuins or even via increased ATP. However, our data suggest that NAM can act as activator or inhibitor of SIRT1 depending upon cellular requirements. This could explain the inconsistent nature of SIRT 1 findings in the literature (Green et al., 2008; Lagouge et al., 2006). SIRT1 had been described as having a “yin-yang” effect because of findings that show both inhibition and activation can be beneficial (Dillin and Kelly, 2007). Nonetheless our results validate NAM as a therapeutic, as well as SIRT1 and PGC-1α as relevant targets.
Conclusions
This study demonstrates that NAM can increase gene expression of both BDNF and PGC-1α, indicating a beneficial effect. We hypothesized that this correction would alleviate the motor phenotype seen in HD mice. Indeed, we found that this small molecule can alleviate motor deficits in B6.HDR6/1 mice. NAM administration via either of two modes was able to increase exploratory behavior in B6.HDR6/1 mice. More extensive investigation of behavioral changes demonstrated improvements in rotarod performance and beam walking. Due to the lack of reduction in aggregation, we propose that NAM is having an effect downstream from mhtt aggregation, which is possibly through modulation of PGC-1α activation. These findings are exciting because for the first time, NAM treatment has been shown to be effective in improving motor deficits in a model of HD, at doses that are within a human clinical range. Our data therefore suggest that NAM is a potential therapeutic for HD symptoms, and should be strongly considered for clinical trials.
Acknowledgments
We thank Kevin Manley for expert assistance with the statistical analyses, Lizbeth Day for HD mouse assistance, Drs. David Butler for valuable consults on the SDS-AGE and RT-PCR, Valerie Bolivar for discussions of the behavioral testing, and Erik Kvam for valuable discussions and suggestions. This work was supported by NIH/NINDS to A. M. (NS053912), and a graduate student diversity supplement of NS053912 to T.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bitterman KJ, et al. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Bolivar VJ, et al. Early exploratory behavior abnormalities in R6/1 Huntington's disease transgenic mice. Brain Res. 2004;1005:29–35. doi: 10.1016/j.brainres.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Bortvedt SF, et al. Cystamine and intrabody co-treatment confers additional benefits in a fly model of Huntington's disease. Neurobiol Dis. 2010 doi: 10.1016/j.nbd.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler R, Bates GP. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat Rev Neurosci. 2006;7:784–96. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- Carter RJ, et al. Motor coordination and balance in rodents. Curr Protoc Neurosci. 2001;Chapter 8(Unit 8):12. doi: 10.1002/0471142301.ns0812s15. [DOI] [PubMed] [Google Scholar]
- Cha JH. Transcriptional dysregulation in Huntington's disease. Trends Neurosci. 2000;23:387–92. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–48. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Desplats PA, et al. Selective deficits in the expression of striatal-enriched mRNAs in Huntington's disease. J Neurochem. 2006;96:743–57. doi: 10.1111/j.1471-4159.2005.03588.x. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–3. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Dillin A, Kelly JW. Medicine. The yin-yang of sirtuins. Science. 2007;317:461–2. doi: 10.1126/science.1146585. [DOI] [PubMed] [Google Scholar]
- Duyao M, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet. 1993;4:387–92. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- Erb C, Klein J. Enhancement of brain choline levels by nicotinamide: mechanism of action. Neurosci Lett. 1998;249:111–4. doi: 10.1016/s0304-3940(98)00418-2. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J Neurosci. 2003;23:9418–27. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Feany MB. Comparison of pathways controlling toxicity in the eye and brain in Drosophila models of human neurodegenerative diseases. Hum Mol Genet. 2004;13:2011–8. doi: 10.1093/hmg/ddh214. [DOI] [PubMed] [Google Scholar]
- Green KN, et al. Nicotinamide restores cognition in Alzheimer's disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci. 2008;28:11500–10. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–74. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–35. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100:2041–6. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannicola C, et al. Early alterations in gene expression and cell morphology in a mouse model of Huntington's disease. J Neurochem. 2000;75:830–9. doi: 10.1046/j.1471-4159.2000.0750830.x. [DOI] [PubMed] [Google Scholar]
- Imai S, et al. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jia H, et al. High doses of nicotinamide prevent oxidative mitochondrial dysfunction in a cellular model and improve motor deficit in a Drosophila model of Parkinson's disease. J Neurosci Res. 2008;86:2083–90. doi: 10.1002/jnr.21650. [DOI] [PubMed] [Google Scholar]
- Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–35. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum Mol Genet. 2000;9:1259–71. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- Mandrup-Poulsen T, et al. Nicotinamide treatment in the prevention of insulin-dependent diabetes mellitus. Diabetes Metab Rev. 1993;9:295–309. doi: 10.1002/dmr.5610090410. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- McGill JK, Beal MF. PGC-1alpha, a new therapeutic target in Huntington's disease? Cell. 2006;127:465–8. doi: 10.1016/j.cell.2006.10.023. [DOI] [PubMed] [Google Scholar]
- McLear JA, et al. Combinational approach of intrabody with enhanced Hsp70 expression addresses multiple pathologies in a fly model of Huntington's disease. FASEB J. 2008 doi: 10.1096/fj.07-099689. [DOI] [PubMed] [Google Scholar]
- Nemoto S, et al. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Nonomura T, et al. Brain-derived neurotrophic factor regulates energy expenditure through the central nervous system in obese diabetic mice. Int J Exp Diabetes Res. 2001;2:201–9. doi: 10.1155/EDR.2001.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr AL, et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–92. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabello NG, et al. Regional expression of constitutive and inducible transcription factors following transient focal ischemia in the neonatal rat: influence of hypothermia. Brain Res. 2005;1038:11–21. doi: 10.1016/j.brainres.2004.12.047. [DOI] [PubMed] [Google Scholar]
- Pallas M, et al. Modulation of SIRT1 expression in different neurodegenerative models and human pathologies. Neuroscience. 2008;154:1388–97. doi: 10.1016/j.neuroscience.2008.04.065. [DOI] [PubMed] [Google Scholar]
- Pallos J, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease. Hum Mol Genet. 2008;17:3767–75. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, et al. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics. 1979;93:877–901. doi: 10.1093/genetics/93.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Thompson LM. Transcription meets metabolism in neurodegeneration. Nat Med. 2006;12:1239–1241. doi: 10.1038/nm1106-1239. [DOI] [PubMed] [Google Scholar]
- Roth SY, et al. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Shao J, Diamond MI. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum Mol Genet. 2007;16(Spec No 2):R115–23. doi: 10.1093/hmg/ddm213. [DOI] [PubMed] [Google Scholar]
- Southwell AL, et al. Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington's disease. J Neurosci. 2009;29:13589–602. doi: 10.1523/JNEUROSCI.4286-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–43. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Szapacs ME, et al. Exploring the relationship between serotonin and brain-derived neurotrophic factor: analysis of BDNF protein and extraneuronal 5-HT in mice with reduced serotonin transporter or BDNF expression. J Neurosci Methods. 2004;140:81–92. doi: 10.1016/j.jneumeth.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Taylor DM, et al. Biological and potential therapeutic roles of sirtuin deacetylases. Cell Mol Life Sci. 2008;65:4000–18. doi: 10.1007/s00018-008-8357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmaine M, et al. Nonapoptotic neurodegeneration in a transgenic mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2000;97:8093–7. doi: 10.1073/pnas.110078997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, et al. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington's disease. J Neurochem. 2008;104:846–58. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- Weydt P, et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–62. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Wolfgang WJ, et al. Suppression of Huntington's disease pathology in Drosophila by human single-chain Fv antibodies. Proc Natl Acad Sci USA. 2005;102:11563–11568. doi: 10.1073/pnas.0505321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–22. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
- Zuccato C, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–8. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]