Abstract
Precise targeting of various voltage-gated ion channels to proper membrane domains is crucial for their distinct roles in neuronal excitability and synaptic transmission. How each channel protein is transported within the cytoplasm is poorly understood. Here, we report that KIF5/kinesin I transports Kv3.1 voltage-gated K+ (Kv) channels through the axon initial segment (AIS) via direct binding. First, we have identified a novel interaction between Kv3.1 and KIF5, confirmed by immunoprecipitation from mouse brain lysates and by pull-down assays with exogenously expressed proteins. The interaction is mediated by a direct binding between the Kv3.1 N-terminal T1 domain and a conserved region in KIF5 tail domains, in which proper T1 tetramerization is crucial. Overexpression of this region of KIF5B markedly reduces axonal levels of Kv3.1bHA. In mature hippocampal neurons, endogenous Kv3.1b and KIF5 colocalize. Suppressing the endogenous KIF5B level by RNA interference significantly reduces the Kv3.1b axonal level. Furthermore, mutating the Zn2+-binding site within T1 markedly decreases channel axonal targeting and forward trafficking, likely through disrupting T1 tetramerization and hence eliminating the binding to KIF5 tail. The mutation also alters channel activity. Interestingly, coexpression of the YFP (yellow fluorescent protein)-tagged KIF5B assists dendritic Kv3.1a and even mutants with a faulty axonal targeting motif to penetrate the AIS. Finally, fluorescently tagged Kv3.1 channels colocalize and comove with KIF5B along axons revealed by two-color time-lapse imaging. Our findings suggest that the binding to KIF5 ensures properly assembled and functioning Kv3.1 channels to be transported into axons.
Introduction
Dendrites and axons are two prominent subcellular compartments for most neurons to receive synaptic inputs and to convey action potential outputs, respectively. Kv channels are differentially targeted on dendritic and axonal membranes (Lai and Jan, 2006; Vacher et al., 2008), allowing them to regulate neuronal excitability locally and thus to play distinct roles in synaptic transmission and input–output relationship. Whereas dendritic A-type Kv4.2 channels regulate action potential backpropagation, dendritic integration, and plasticity (Hoffman et al., 1997; Cai et al., 2004; Losonczy et al., 2008), axonal Kv1 channels regulate action potential initiation, waveform and propagation, and synaptic efficacy (Zhou et al., 1998; Hille, 2001; Kole et al., 2007; Goldberg et al., 2008). Kv3 channels display complex targeting patterns in dendrites and axons, correlating with their functional diversity in shaping large dendritic depolarization (Martina et al., 2003), regulating action potential duration and frequency, and regulating transmitter release (Rudy and McBain, 2001; Lien and Jonas, 2003; Goldberg et al., 2005).
Each Kv channel complex contains four voltage-sensing and pore-forming subunits. Each subunit consists of six membrane-spanning segments, and cytoplasmic N- and C-terminal domains (Jan and Jan, 1997; Long et al., 2005). N-terminal T1 domains form tetramers within a Kv subfamily, which are responsible for the proper assembly of channel tetramers (Li et al., 1992; Xu et al., 1995; Choe, 2002). T1 tetramers from Kv2, Kv3, and Kv4 but not Kv1 contain a conserved Zn2+-binding site at the interface, required for tetramerization (Bixby et al., 1999; Choe, 2002; Jahng et al., 2002). Our previous study has identified a conditional axonal targeting motif (ATM) (for polarized distribution on axonal membranes) in the C termini of the two Kv3.1 splice variants (Kv3.1a and Kv3.1b) (Xu et al., 2007). Ankyrin G, a critical adaptor protein at the axon initial segment (AIS) (Bennett and Chen, 2001; Jenkins and Bennett, 2001), interacts with the ATM, and may function as a conditional barrier differentially regulating Kv3.1a and Kv3.1b polarized targeting (Xu et al., 2007). However, it remains unknown how Kv3.1b channels are transported down the axon.
Conventional kinesin I, a major anterograde motor in axons, consists of a heavy chain (KIF5) dimer and two light chains [kinesin light chains (KLCs)]. The heavy chains (three isoforms: KIF5A, KIF5B, and KIF5C) have an N-terminal motor domain, followed by a stalk domain responsible for dimerization through coiled-coil regions, and a C-terminal tail domain containing cargo-binding sites (Goldstein, 2001; Asbury et al., 2003; Hirokawa and Noda, 2008; Gennerich and Vale, 2009). KLCs directly bind to KIF5 C termini, mediating the transport of many cargos in axons (Setou et al., 2002; Hirokawa and Takemura, 2005; Glater et al., 2006). However, there is no precedent yet for a direct binding between the pore-forming subunit of an ion channel and kinesin I.
In this study, we have identified KIF5 as a novel interacting protein of Kv3.1. We provide compelling evidence that Kv3.1 T1 tetramers, but not monomers, directly bind to an evolutionarily conserved region in KIF5 tail domains. The binding is most likely essential for axonal targeting and forward trafficking of properly assembled Kv3 channels.
Materials and Methods
cDNA constructs.
Kv1.2HA, Kv3.1aHA, Kv3.1bHA, Kv3.1aHAIPR, Kv3.1aHAPP, Kv3.1bHAKKK, and Kv3.1bHAKKH were described previously (Gu et al., 2003; Xu et al., 2007). Point mutations, Kv3.1aHAH77A, Kv3.1aHAC83A, Kv3.1aHAC2A2, and Kv3.1bHAC83A were made with the QuickChange strategy. GST-31N, GST-31aC, GST-31bC, GST-31sC, GST-Kvβ2, His-31T1, KIF5B-YFP, KIF3A-YFP, CFP-Kv1.2, YFP-Kvβ2, and YFP-Kvβ2K235E were described previously (Gu et al., 2006; Xu et al., 2007). GST-12N and GST-12C were made by fusing the N-terminal (amino acids 2–140) and C-terminal (amino acids 420–499) regions of Kv1.2 to the glutathione S-transferase (GST) C terminus in pGEX4T-2.
GST-Tail758–820, GST-Tail805–934, GST-Tail865–934, GST-Tail865–963, GST-Tail865–912, GST-Tail892–934, GST-Tail865–896, and GST-Stalk (amino acids 337–760) were made by fusing the regions (indicated by residue numbers) of KIF5B to the GST C terminus in pGEX4T-2. GST-ATail (amino acids 756–1027) and GST-ATail863–932 were constructed by fusing the regions of the mouse KIF5A tail domain to the GST C terminus. His-12T1 and His-42T1 were constructed by inserting the N termini of Kv1.2 (amino acids 2–140) and Kv4.2 (amino acids 2–140) into pRSETB between BglII and SalI. Point mutations in the Zn2+-binding site of the T1 domain, His-31T1C83A, His-31T1C2A2, and His-31T1H77A were made with QuickChange based on His-31T1; Kv3.1aHAH77A, Kv3.1aHAC83A, and Kv3.1aHAC2A2 were made with QuickChange based on Kv3.1aHA; Kv3.1bHAC83A was made with QuickChange based on Kv3.1bHA. KIF5B1–760-YFP (amino acids 1–760) and YFP-Tail (amino acids 758–963) were constructed by subcloning the corresponding regions of human KIF5B into pEYFP vectors. YFP-KLC was made by inserting the PCR fragment of the full-length kinesin light chain 1 (Open Biosystems) into pEYFP-C1. All constructs were confirmed by sequencing.
Antibodies and immunocytochemistry.
Antibodies used include rabbit polyclonal anti-green fluorescent protein (GFP) and mouse anti-6×His antibodies (Invitrogen), rabbit anti-Kv3.1b antibody (Alomone Labs), rabbit anti-microtubule-associated protein 2 (MAP2) and anti-Tau1 antibodies (Millipore Bioscience Research Reagents), mouse monoclonal anti-GFP and anti-Kv3.1b antibodies (Antibodies, Inc.), rat monoclonal anti-HA antibody (Roche), mouse anti-kinesin heavy chain (KIF5) H2 (Sigma-Aldrich), Cy2-, Cy3-, and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). The procedures of immunocytochemistry were described previously (Gu et al., 2006; Xu et al., 2007). In brief, the neurons were fixed with 4% formaldehyde (from 10% ultrapure EM grade and methonal free; Polysciences) and 4% sucrose in PBS for 20 min, and stained with specified antibodies under permeabilized conditions (in the presence of 0.2% Triton X-100) to label total proteins. To distinguish axons and dendrites of neurons, an anti-Tau1 (Tau1, axonal marker) antibody or an anti-MAP2 (MAP2, dendritic marker) antibody was used in costaining.
Immunoprecipitation and Western blotting.
Mouse brains were homogenized with Dounce tissue grinder in homogenization buffer [50 mm Tris buffer, pH 8.0, 2 mm MgCl2, 1 mm EDTA, and a Complete protease inhibitor tablet (Roche)], followed by low-speed centrifugation at 700 × g for 5 min at 4°C. Crude membranes were pelleted from the suspension by high-speed centrifugation at 50,000 × g for 15 min at 4°C, solubilized in immunoprecipitation (IP) buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1% Triton X-100, and a Complete protease inhibitor tablet) for 2 h at 4°C, and centrifuged at 50,000 × g for 30 min at 4°C. The supernatant (1 ml in each condition) was incubated (2–4 h, 4°C) with 4 μl of mouse anti-KIF5 H2 antibodies (or control mouse IgG) and 50 μl of protein G-agarose beads (Roche). The beads were washed six times with the IP buffer and eluted with 2× sample buffer. The immunoprecipitants were resolved by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and subjected to Western blotting with a mouse anti-Kv3.1b antibody (1:1000 dilution) or with the H2 antibody (1:1000 dilution).
Protein purification and in vitro binding assay.
Expression of GST- or 6×His-tagged fusion proteins was induced in BL21 Escherichia coli cells with 1 mm IPTG (isopropyl β-d-thiogalactoside) for 4 h at 37°C. Bacterial pellets were solubilized with sonication in the IP buffer at 4°C, and centrifuged at 50,000 × g for 30 min at 4°C. The supernatants were incubated either with glutathione beads (GE Healthcare Bio-Sciences) or with Co2+ beads (Clontech) at 4°C for 3 h. After extensive washing, the beads coated with purified fusion proteins were eluted with the IP buffer containing either 20 mm glutathione or 150 mm imidazole. The elution was further dialyzed with the IP buffer at 4°C overnight. In in vitro binding assays, purified His-31T1 (0.5 mg) and GST fusion protein (0.5 mg) were incubated in either the regular IP buffer or one containing 1 mm EDTA, at 4°C overnight, and then precipitated with glutathione beads.
In some experiments, glutathione beads coated with purified GST fusion proteins were further incubated with either bacterial lysate supernatant containing 6×His-tagged fusion proteins or the supernatant from HEK293 cells expressing yellow fluorescent protein (YFP) fusion proteins. The precipitants were eluted with 2× sample buffer, resolved in SDS-PAGE, and then subjected to Western blotting and Coomassie staining. Each immunoprecipitation or pull-down assay was performed at least three times.
Hippocampal neuron cultures and transfection.
Hippocampal neuron culture was prepared as previously described from embryonic day 18 (E18) embryos (Gu et al., 2006). In brief, 2 d after neuron plating, 1 μm cytosine arabinose (Sigma-Aldrich) was added to the neuronal culture medium to inhibit glial growth for the subsequent 2 d, and then replaced with the normal culture medium. The culture medium was replenished twice a week by replacing one-half of the volume. For transient transfection, neurons in culture at 5 d in vitro (DIV) were incubated in Opti-MEM containing 0.8 μg of cDNA plasmid and 1.5 μl of Lipofectamine 2000 (Invitrogen) for 20 min at 37°C.
To obtain a GABAergic interneuron enriched culture, we made hippocampal cultures from rat pups at postnatal day 8 (P8) using the same procedure as the E18 culture. Staining with anti-GABA antibody and nuclear dye showed that more than one-half of the neurons in culture were GABAergic (data not shown). Enrichment of GABAergic interneurons in P8 cultures may reflect differences in the birth and migration of GABAergic interneurons versus pyramidal neurons. In addition, pyramidal neurons may die more readily during dissociation for culture, since P8 pyramidal neurons already have lengthy and complex dendritic and axonal arbors. At least three independent transfections were performed for each condition.
Short interfering RNA knockdown of endogenous KIF5B.
Construction and validation of vector-based short interfering RNA (siRNA) strategy to suppress the levels of endogenous proteins in rat hippocampal neurons were previously described (Gu et al., 2006). We designed eight probes against rat KIF5B and subcloned them in pRNAT-H1.1/neo vector (GenScript). The siRNA vectors were transfected into neurons at 4 DIV and fixed and stained 6 d later. When transfected into E18 and P8 hippocampal neurons, one of them, KIF5B siR6 (TATCTGGTTGATTTAGCTGGA), significantly suppressed endogenous levels of KIF5. The GFP coding region in the vector was replaced with the coding sequence for mCherry between BamH1 and HindIII sites. Therefore, the neurons transfected with the siRNA plasmid expressed mCherry as the indicator for transfection.
Fluorescence microscopy and quantification.
Fluorescence images were captured with a Spot CCD camera RT slider (Diagnostic Instruments) in a Zeiss upright microscope, Axiophot, using Plan Apo objectives 20×/0.75 and 100×/1.4 oil, saved as 16 bit TIFF files, and analyzed with NIH ImageJ and SigmaPlot 10.0 for fluorescence intensity quantification. Exposure times were controlled so that the pixel intensities in dendrites and axons were below saturation, but the same exposure time was used within each group of an experiment. The quantification procedure was described previously (Gu et al., 2006). Only transfected neurons with clearly separated dendrites and axons, and isolated from other transfected cells, were chosen for analysis. Using NIH ImageJ, we laid a line along the major axon to acquire its average fluorescence intensity (in arbitrary unit) (Faxon), and laid lines along proximal dendrites that connect with soma to obtain the average fluorescence intensity of the somatodendritic region (Fsd) to represent the total level. Thus, the ratio (Faxon/Fsd) reflects the relative axonal level. The background fluorescence intensity was measured for each image and subtracted.
Live-cell imaging.
Neurons growing on 25 mm coverslips were loaded into the imaging chamber (Molecular Devices) and incubated with imaging buffer [HE-LF medium (Brainbits) plus 2% B-27, 0.5 mm glutamine, and 25 μm glutamate] at room temperature. The time-lapse imaging setup was built on a Nikon TE2000 inverted microscope. Images were captured with a CCD camera Coolsnap HQ (Photometrics) through cyan fluorescent protein (CFP) or YFP filter sets with 1 s exposure time. The filters were changed through filter wheels controlled through Lamda 10-3 (Sutter Instrument) by MetaMorph software (Molecular Devices). The time-lapse imaging was performed with 2 s interval for 100 frames.
Fluorescence resonance energy transfer imaging.
There are three major strategies to perform fluorescence resonance energy transfer (FRET) imaging, sensitized emission, donor dequenching, and measurements of fluorescence lifetime (Erickson et al., 2001). Among them, only sensitized emission is suitable for detecting FRET in living cells with a conventional fluorescence microscope. The strategy and protocol of FRET imaging were described previously (Sorkin et al., 2000; Gu et al., 2001, 2006). In brief, three images were acquired sequentially through (1) YFP filter (excitation, 500/20 nm; emission, 535/20 nm); (2) CFP filter (emission, 430/25 nm; emission, 470/30 nm); (3) FRET filter set (excitation, 430/25 nm; emission, 535/30 nm). A single dichroic mirror (86004BS; Chroma Technology) was used with all three filter channels. FRET between CFP and YFP was measured and calculated for the entire image on a pixel-by-pixel basis by a three-filter “microFRET” method. The raw FRET images consisted of both FRET and non-FRET components (the donor and acceptor fluorescence bleeding through the FRET filter). The extent of cross-bleeding was characteristic of the particular optical system and determined by the use of cells that express either CFP- or YFP-fusion protein alone. Background fluorescence values were subtracted from each image before calculation. In our system, 64.7 ± 0.5% (n = 18) of CFP and 1.47 ± 0.08% (n = 12) of YFP fluorescence bleed through the FRET channel. Therefore, FRETcorrected = FRETraw − (0.65 × CFP) − (0.015 × YFP). The calculation of FRETcorrected was performed with the MetaMorph software.
Patch-clamp whole-cell recording.
The transfected HEK293 cells were identified by fluorescence from cotransfected YFP. They were recorded in Hanks buffer (150 mm NaCl, 4 mm KCl, 1.2 mm MgCl2, 10 mg/ml glucose, 1 mm CaCl2, 20 mm HEPES, pH 7.4). The internal solution for electrical pipettes was composed of the following (in mm): 122 KMeSO4, 20 NaCl, 5 Mg-ATP, 0.3 GTP, and 10 HEPES, pH 7.2. The resistance of electrodes was between 2 and 5 MΩ. Isolated cells were voltage clamped in the whole-cell mode using an Axonpatch 200B amplifier (Molecular Devices), held at −80 mV, and given 250 ms voltage episodes from −60 to +60 mV with a 10 mV increment. Voltage commands were made from pCLAMP10 software through Digidata 1440A, and the currents were recorded at 5 kHz. Conduction–voltage relationships (G–V curves) for Kv3.1 channel constructs were G = I/(Vm − Vrev), Vrev = −95 mV, normalized to the maximal conduction. Curves were fitted with Boltzmann function, G/Gmax = 1/(1 + exp[−(V − V1/2)/k]), where V1/2 is the potential at which the value of the Boltzman function is 0.5, and k is the slope factor. To obtain τon, activation curves were fitted with a single exponential function raised to a power of 4, I(t) = A(1 − exp(−t/τon))4. In the studies of deactivation of Kv3.1 channels, the cells were held at −80 mV, given a 2 ms prepulse to 60 mV and 15 ms voltage episodes from −100 mV to −10 mV. To obtain τoff, tail currents were fitted with the following equation: I(t) = Aexp(−t/τoff). SigmaPlot10 was used for fitting.
Results
A novel interaction between Kv3.1 and KIF5
To identify conserved Kv3-interacting proteins that regulate channel trafficking and activity, we performed a pull-down assay using bacterially expressed and purified GST-31N (GST-fused to the Kv3.1 N-terminal region containing the conserved T1 domain) to precipitate rat brain lysates, followed by in-gel digestion and tandem mass spectrometry analysis. Among precipitants, we identified KIF5/kinesin I by the presence of three peptides, TGAEGAVLDEAK, LYLVDLAGSEK, and VSYFEIYLDK, all from its N-terminal motor domain. This result suggests a potential association between Kv3.1 channels and KIF5 motors (Fig. 1A).
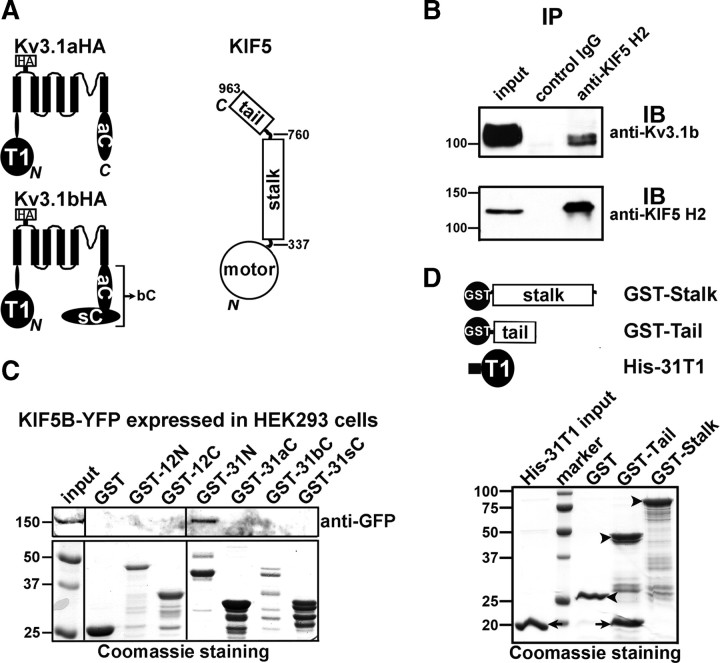
Figure 1.
Kv3.1 channels interact with KIF5/kinesin I. A, Structural diagrams of Kv3.1aHA, Kv3.1bHA, and KIF5 (residue numbers taken from KIF5B, a ubiquitous isoform). Each Kv3.1 subunit consists of six membrane-spanning segments (indicated by 6 black bars), and intracellular N (N) (including the T1 domain) and C termini (C). The HA tag is inserted in the first extracellular loop. The splice domain (sC) is at the C terminus of Kv3.1b. The N termini of Kv3.1a and Kv3.1b are identical. KIF5 consists of an N-terminal motor domain, a stalk domain for dimerization, and a tail domain at the C terminus for associating with cargos. B, Endogenous Kv3.1b binds to KIF5 shown by a tissue IP experiment. Crude membranes from mouse brains were solubilized in the IP buffer and then centrifuged at a high speed. The supernatants were precipitated by a monoclonal anti-KIF5(H2) antibody or a control IgG. Inputs and precipitants were blotted with anti-Kv3.1b (top) and anti-KIF5(H2) (bottom) antibodies. One percent of the inputs was loaded (left lanes). In the bottom panel, only 1.6% of the precipitants of the top panel was loaded for blotting with the anti-KIF5 H2 antibody. C, Purified GST-31N, but not GST, GST-31aC, GST-31bC, GST-31sC, GST-12N, or GST-12C, pulled down KIF5B-YFP expressed in HEK293 cells. Five percent of the inputs of HEK293 cell lysates were loaded. Fifty percent of the inputs of GST fusion proteins were loaded for Coomassie staining (bottom). D, Purified GST-Tail, but not GST or GST-Stalk efficiently precipitated purified His-31T1. Arrows, His-31T1 protein bands. Arrowheads, Bands of GST and GST fusion proteins. Molecular weights are indicated on the left in kilodaltons. All pull-down experiments were performed at least three times.
To assess the interaction between endogenous Kv3.1b (the more abundant form of the two splice variants of Kv3.1 in adult brains) and KIF5, we performed an immunoprecipitation experiment using an anti-KIF5 (H2) antibody to pull down the supernatants from solubilized crude membranes of mouse brains. A monoclonal anti-Kv3.1b antibody (Fig. 1B, top) and the anti-KIF5 antibody (Fig. 1B, bottom) were used to blot the precipitants. The anti-KIF5 antibody, but not an unrelated mouse IgG, precipitated endogenous Kv3.1b channels (Fig. 1B). This result confirmed that endogenous Kv3.1b channels indeed interact with KIF5 motors.
To determine whether KIF5 specifically binds to the N-terminal region of Kv3.1 channels, we used purified GST fusion proteins of Kv channel cytoplasmic domains to pull down KIF5B-YFP expressed in HEK293 cells. KIF5B-YFP was precipitated only by purified GST-31N but not by GST, GST-12N (GST fused to Kv1.2 N-terminal domain), GST-12C (GST fused to Kv1.2 C-terminal domain), GST-31aC, GST-31bC, or GST-31sC (Fig. 1C). Thus, consistent with the initial pull-down result, KIF5B binds to the Kv3.1 N terminus, but not Kv3.1 C terminus, nor the Kv1.2 cytoplasmic domains (Fig. 1C).
The Kv3.1 N terminus contains only one major domain, the T1 domain. To map the potential Kv3.1 T1-binding site within KIF5B, we used purified GST fusion proteins of KIF5B stalk and tail domains to pull down His-tagged Kv3.1 T1 (His-31T1). GST-Tail (amino acids 760–963) but not GST-Stalk (amino acids 337–760) precipitated His-31T1 revealed by Coomassie staining (Fig. 1D). Therefore, the interaction between Kv3.1 and KIF5 is most likely mediated by a direct binding between the Kv3.1 T1 domain and the KIF5 tail domain. Since Kv3.1a and Kv3.1b share the same T1 domain, both should bind to KIF5 motors.
Direct binding between the Kv3.1 T1 domain and a conserved region in KIF5 C termini
The KIF5 tail domain primarily mediates the motor-cargo association and regulation. KLCs, major adaptor proteins of KIF5, bind to a region near the N terminus of KIF5 tail and associate with approximately one-half of the KIF5 dimers in vivo (Diefenbach et al., 1998; Kanai et al., 2000; Gyoeva et al., 2004; Hirokawa et al., 2009). The tail domain of KIF5 can regulate the activity of KIF5 motors by binding to its own motor domain, myosin and microtubule (Coy et al., 1999; Huang et al., 1999; Seeger and Rice, 2010). Furthermore, various ion channel proteins and organelles associate with KIF5 tail through different adaptor proteins (Ong et al., 2000; Diefenbach et al., 2002; Setou et al., 2002; Gindhart et al., 2003; Macioce et al., 2003; Kanai et al., 2004; Cai et al., 2005; Glater et al., 2006; Twelvetrees et al., 2010). Although many proteins have been shown previously to bind to this region (supplemental Fig. S1, available at www.jneurosci.org as supplemental material), the present study is the first to suggest a direct binding between a pore-forming subunit of an ion channel and KIF5 tail. Therefore, it is important to identify the exact binding site for Kv3.1 T1 for two primary reasons. First, this is the critical initial step for understanding the relationship of Kv3.1 channels and other KIF5-binding proteins, and hence for further elucidating the regulation of Kv3.1 trafficking. For instance, does Kv3.1 T1 compete for the same binding site with KLCs? Second, to identify the specific binding site is helpful in the dominant-negative experiment. The overexpressed fragment is less likely to affect the trafficking of other binding proteins.
To identify the Kv3.1 T1-binding site within the KIF5B tail domain, we first made seven GST fusion constructs (Fig. 2A). Bacterially expressed His-31T1 was precipitated by purified GST-Tail, GST-Tail805–934, GST-Tail865–934, or GST-Tail865–963, but not by GST or GST-Tail758–820 (including the KLC-binding site) (Fig. 2B). The GST fusion with a 70 residue fragment (amino acids 865–934) in the tail domain displayed the strongest binding to His-31T1 (Fig. 2C). Additional deletion of ∼20 residues either at its N or C terminus reduced the pull-down efficiency (Fig. 2C).
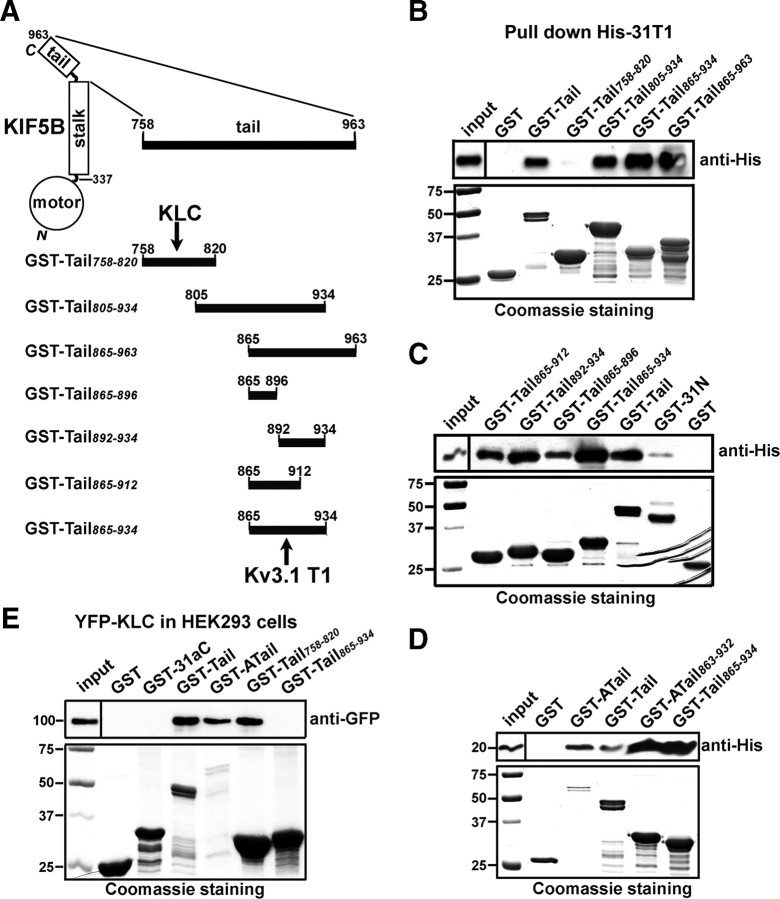
Figure 2.
Identifying the Kv3.1 T1-binding site in KIF5 tails. A, Different regions of the KIF5B tail domain were fused with GST. Binding sites for KLC and Kv3.1 T1 were indicated with black arrows. B, Bacterially expressed His-31T1 was precipitated by GST-Tail, GST-Tail805–934, GST-Tail865–934, and GST-Tail865–963, but not by GST or GST-Tail758–820 (it contains the binding site for KLC). Precipitated His-31T1 was blotted with an anti-His antibody (top). Five percent of the inputs of His-31T1-containing bacterial lysates were loaded (top left) and 50% of purified GST fusion proteins were loaded for Coomassie staining (bottom). C, His-31T1 was efficiently precipitated by a 70 residue region (865–934) in the KIF5B tail domain (GST-Tail865–934). D, His-31T1 was precipitated by GST fusions of the KIF5A tail domain (GST-ATail) and the corresponding 70 residue region (GST-ATail863–932). GST-ATail863–932 contains 10 extra residues from the multiple cloning site of the pGEX4T-2 vector. E, Purified GST-Tail, GST-ATail, and GST-Tail758–820, but not GST-Tail865–934, precipitated YFP-KLC expressed in HEK293 cells. YFP-KLC was transfected into HEK293 cells for 2 d. Molecular weights are indicated on the left in kilodaltons.
To determine whether Kv3.1 T1 specifically binds to KIF5B or binds to all three isoforms (KIF5A, KIF5B, and KIF5C), we examined the potential interaction between Kv3.1 T1 and KIF5A tail, since KIF5A is more divergent from KIF5B in the 70 residue region (amino acids 865–934) compared with KIF5C (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). We made two GST fusion proteins from KIF5A, its whole C-terminal tail (amino acids 765–1027) (GST-ATail) and the region corresponding to the 70 residue fragment of KIF5B (GST-ATail863–932). Similar to GST-Tail, purified GST-ATail and GST-ATail863–932 efficiently precipitated His-31T1 (Fig. 2D). Thus, the Kv3.1 T1-binding site is likely common within all KIF5 members. In fact, this region is evolutionarily conserved in fruit fly Drosophila (DKHC), Xenopus laevis (XKHC), mouse (mKIF5B), and human (hKIF5A-C) (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). In a control experiment, purified GST-Tail, GST-ATail, and GST-Tail758–820, but not GST-Tail865–934, GST-31aC, or GST precipitated YFP-KLC expressed in HEK293 cells (Fig. 2E). The Kv3.1 T1-binding site is ∼45 residues away from the KLC-binding site (Fig. 2A).
Binding to KIF5 motors is important for Kv3.1b channel trafficking into axons
To determine whether the binding to KIF5 motors is essential for the axonal transport of Kv3.1 channels, we performed the following experiments using two different approaches, the dominant-negative mutant and siRNA knockdown of endogenous KIF5B. First, we examined whether disrupting the binding between Kv3.1 T1 and KIF5 tail prevents Kv3.1b channels from trafficking into axons. YFP-tagged KIF5 tail (YFP-Tail), containing the Kv3.1 T1-binding site, and Kv3.1bHA were cotransfected into young hippocampal neurons at 5 DIV from the E18 culture. Two days after transfection, the neurons were fixed and stained. At this stage, there is no detectable endogenous Kv3.1 channel. YFP-Tail can function as a dominant-negative mutant via direct binding to Kv3.1 T1 domains to mask the binding site for endogenous KIF5 motors. Indeed, the axonal level of Kv3.1bHA markedly decreased in the presence of YFP-Tail (Faxon/Fsd, 0.31 ± 0.02; n = 40) compared with YFP (Faxon/Fsd, 0.72 ± 0.03; n = 40) (Fig. 3A,B). However, YFP-Tail was mainly concentrated in somatodendritic regions when expressed alone (supplemental Fig. S2A, available at www.jneurosci.org as supplemental material), maybe resulting from its many binding partners.
Figure 3.
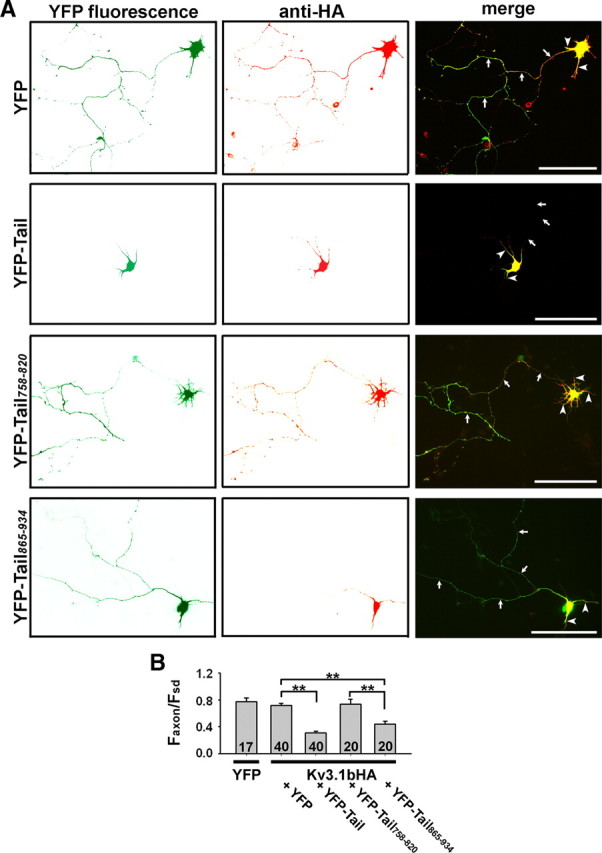
Coexpression of KIF5B tail domains suppresses the axonal level of Kv3.1bHA. A, Hippocampal neurons from the E18 culture cotransfected with YFP-fusion proteins and Kv3.1bHA at 5 DIV, were fixed 2 d later and stained with an anti-HA antibody under permeabilized conditions. Kv3.1bHA (red; total proteins) localized in axons as well as soma and proximal dendrites in the presence of YFP (green) (top). When coexpressed with YFP-Tail or YFP-Tail865–934 but not with YFP-Tail758–820, Kv3.1bHA (red) became highly restricted in somatodendritic regions. B, Summary of the effects of KIF5B tail domains on the relative axonal level (Faxon/Fsd) of Kv3.1bHA. The ratio (Faxon/Fsd) of soluble YFP used as a neutral cytoplasmic marker was used to normalize the volume difference between axons and dendrites. Arrows, Axons. Arrowheads, Dendrites. Scale bars, 100 μm. **p < 0.01, t test. Error bars indicate SEM.
Therefore, we made two smaller fragments of the KIF5B tail region, YFP-Tail758–820 and YFP-Tail865–934, and both expressed rather uniformly along dendrites and axons (supplemental Fig. S2B,C, available at www.jneurosci.org as supplemental material). YFP-Tail865–934 (containing the binding site of Kv3.1 T1, 70 residues; Faxon/Fsd, 0.44 ± 0.05; n = 20) but not YFP-Tail758–820 (containing the KLC-binding site, 63 residues; Faxon/Fsd, 0.74 ± 0.07; n = 20) markedly reduced the Kv3.1bHA axonal level (Fig. 3A,B). Here, we used the ratio Faxon/Fsd (the average axonal fluorescence intensity divided by the average fluorescence intensity of the somatodendritic region) to represent the relative axonal level of Kv3.1 channels, since the expression level of channel proteins varied in each neuron. Moreover, axons, dendrites, and soma differ in their thickness. Therefore, we calculated the ratio of soluble YFP as a neutral cytoplasmic marker (Faxon/Fsd, 0.77 ± 0.06; n = 17) to normalize the volume difference (Fig. 3B). Moreover, we show that coexpression of KIF5B tail constructs and Kv3.1bHA for 2 d in our assays did not affect the morphology of dendrites and axons of neurons (supplemental Fig. S3, available at www.jneurosci.org as supplemental material).
Our overexpression experiments suggest that KIF5 motors might play an essential role in axonal transport of Kv3.1 channels. Next, we examined whether endogenous Kv3.1 channels colocalize with KIF5 motors. Kv3.1 channels are expressed in parvalbumin-containing interneurons in hippocampus (Weiser et al., 1995; Du et al., 1996). In the E18 hippocampal neuron culture, some mature neurons (presumably interneurons) did express Kv3.1b channels. The onset of the Kv3.1b expression was ∼20 DIV. By costaining with endogenous voltage-gated sodium channels (localized along axons), we previously showed that endogenous Kv3.1b channels (total proteins) localized in axons, axonal terminals, soma, and proximal dendrites (Xu et al., 2007). In the present study, endogenous Kv3.1b channels coexpressed and colocalized with KIF5 motors (revealed by the staining using the anti-KIF5 H2 antibody, which recognize all three KIF5 isoforms) (Fig. 4A). As an important axonal ion channel, Kv3.1b should mainly localize on axonal membranes, and thus it might not be transported by KIF5 motors all the time within axons. The interaction between KIF5 and Kv3.1 may be highly dynamic, instead of being persistent. Despite that neither KIF5 nor Kv3.1b displayed highly clustered patterns along axons, we did observe some clusters containing both Kv3.1b and KIF5 along axons (Fig. 4A).
Figure 4.
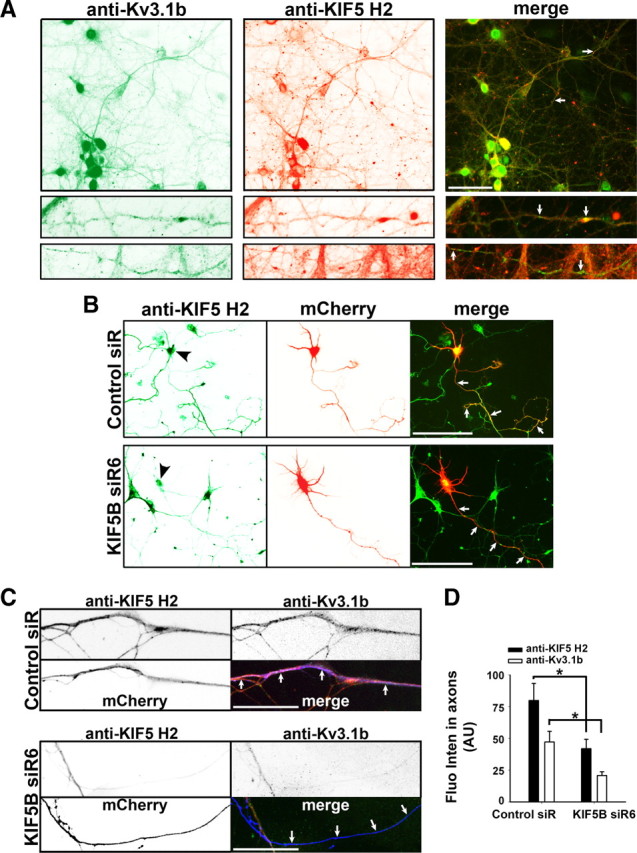
Knocking down endogenous KIF5B by siRNA reduces the axonal level of endogenous Kv3.1b. A, Colocalization of endogenous Kv3.1b and KIF5 in mature hippocampal neurons. The hippocampal neurons from the E18 culture were fixed at 24 DIV, and stained for endogenous Kv3.1b (with a rabbit anti-Kv3.1b antibody; in green) and KIF5 motors (with a mouse anti-KIF5 H2 antibody; in red). Colocalization of endogenous Kv3.1b and KIF5 along axons is shown in the bottom panels in fourfold higher magnification. Arrows, Axons. B, Vector-based KIF5B siR6 significantly suppressed the endogenous KIF5 level in E18 hippocampal neurons. The siRNA vector contains a mCherry (red) as the indicator for transfection. Hippocampal neurons from the E18 culture were transfected with the siRNA vector at 4 DIV, fixed at 10 DIV, and stained with the anti-KIF5 antibody (green). Black arrowheads, Transfected neurons. C, Suppressing endogenous KIF5B reduced the axonal level of endogenous Kv3.1b channels in hippocampal neurons from the P8 culture. Hippocampal neurons, prepared from the P8 culture and containing enriched interneurons expressing Kv3.1b channels, were transfected with siRNA vectors (mCherry in blue in merged) at 4 DIV, fixed at 10 DIV, and stained for endogenous Kv3.1b (green in merged) and KIF5 (revealed by the H2 antibody, red in merged). D, Summary of the siRNA knockdown effects on axonal levels of KIF5 (control siR, 79.9 ± 13.5, n = 9; KIF5B siR6, 41.9 ± 7.4, n = 12) and Kv3.1b (control siR, 47.3 ± 8.4, n = 9; KIF5B siR6, 20.8 ± 3.0, n = 12). Scale bars: A, B, 100 μm; C, 20 μm. *p < 0.05, t test. Error bars indicate SEM.
To determine whether KIF5 is the major kinesin motor to transport Kv3.1 channels along axons, we used the second approach, examining how knocking down endogenous KIF5B by siRNA may affect the axonal level of endogenous Kv3.1b channels. A vector-based siRNA plasmid (KIF5B siR6; H2 fluorescence intensity, 125.5 ± 7.2) significantly suppressed endogenous KIF5 levels compared with the control (control siR, 160.4 ± 11.5) in soma from the E18 culture, although there were some variations among cells (Fig. 4B). The siRNA plasmid contained mCherry as the expression indicator. This result suggests that KIF5B is likely the predominant isoform of KIF5 motors expressed in hippocampal neurons. The presence of other two isoforms (KIF5A and KIF5C) may contribute to the variation in our siRNA knockdown experiment. Endogenous Kv3.1b channels can be clearly detected only in mature neurons (>21 DIV) from the E18 culture. The neurons cultured for >14 d are no longer transfectable. In contrast, endogenous Kv3.1b channels can be clearly detected from the P8 neurons at ∼10 DIV. Therefore, we transfected siRNA plasmids into hippocampal neurons of the P8 culture at 4 DIV (this culture made from hippocampi of rat pups at postnatal day 8 had more enriched GABAergic interneurons that express Kv3.1 channels) (for details, see Materials and Methods). The neurons were costained at 10 DIV for endogenous Kv3.1b and KIF5. KIF5B siR6 significantly decreased the axonal levels of both KIF5 and Kv3.1b, compared with the control siRNA (Fig. 4C,D; supplemental Fig. S4, available at www.jneurosci.org as supplemental material). Together, our results suggest KIF5 plays a critical role in axonal transport of Kv3.1b channels.
The Zn2+-binding site essential for T1 tetramerization is critical for Kv3.1 T1 binding to KIF5B tail and Kv3.1bHA targeting to axons
Our results suggest that the direct binding between Kv3.1 and KIF5 may be essential for Kv3.1b transport in axons. We further analyzed the determinant(s) for Kv3.1 and KIF5 binding. Kv3 T1 domains share high sequence homology (>70%) within the subfamily, but share significantly less homology (≈40%) with the T1 domains of other Kv channels (Shen and Pfaffinger, 1995; Nanao et al., 2003). To determine whether the T1 domains from other Kv channel subfamilies also interact with KIF5B tail, we fused a 6×His tag to Kv1.2 (His-12T1) and Kv4.2 T1 domains (His-42T1) and expressed them in E. coli BL21 cells. Purified GST-31N and GST-Tail precipitated bacterially expressed His-31T1 (Fig. 5A), but not His-12T1 (Fig. 5B) or His-42T1 (Fig. 5C). As a positive control, despite the low protein level, GST-Kvβ2 efficiently precipitated His-12T1 (Fig. 5B). Therefore, our data suggest that the binding between KIF5 tail and Kv3 T1 might be specific within the Kv channel family.
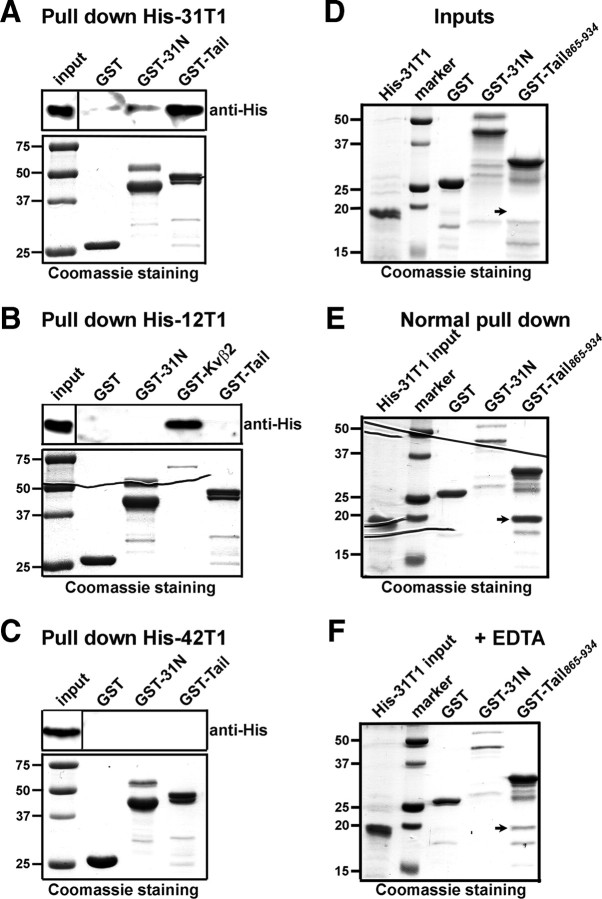
Figure 5.
Specific binding between KIF5B tail and tetramerized Kv3.1 T1 domains. A, His-31T1 in bacterial lysates was more efficiently precipitated by GST-Tail than by GST-31N. Precipitated His-31T1 was blotted with an anti-His antibody (top). Five percent of the inputs of His-31T1-containing bacterial lysates were loaded (top left) and 50% of purified GST fusion proteins were loaded for Coomassie staining (bottom). B, His-12T1 (the His tag fused to Kv1.2 T1 domain) was efficiently precipitated by GST-Kvβ2, but not by GST, GST-31N, or GST-Tail. C, His-42T1 (the His tag fused to Kv4.2 T1 domain) was not precipitated by GST, GST-31N, or GST-Tail. D, Purified His-31T1, GST, GST-31N, and GST-Tail865–934 revealed by Coomassie staining. E, Purified His-31T1 was efficiently precipitated by GST-Tail865–934 under normal conditions. F, In the presence of 1 mm EDTA, precipitation of His-31T1 by GST-Tail856–934 was markedly weakened. The arrows indicate the positions of His-31T1 bands. Molecular weights are indicated on the left in kilodaltons. All pull-down experiments were performed at least three times.
Since the majority of Kv3.1 T1 domains of GST-31N or His-31T1 expressed in bacteria already form stable tetramers even in the absence of specifically added Zn2+ (Bixby et al., 1999; Nanao et al., 2003), purified GST-31N did not efficiently pull down His-31T1 (Fig. 5A). Compared with GST-31N, purified GST-Tail pulled down His-31T1 much more efficiently (Figs. 2C, 5A). Therefore, we predicted that Kv3.1 T1 might bind to KIF5 tail as tetramers. To test this hypothesis, we first purified His-31T1, GST, GST-31N, and GST-Tail865–934 (Fig. 5D), and performed pull-down assays in the absence or presence of 1 mm EDTA (Fig. 5E,F). Since Zn2+ is required for tetramerization of Kv3 T1 domains, addition of EDTA that chelates Zn2+ disassembles Kv3.1 T1 tetramers into monomers (Bixby et al., 1999; Nanao et al., 2003). Indeed, in the presence of EDTA, the binding between purified GST-Tail865–934 and His-31T1 was significantly weakened (Fig. 5F).
Next, we made point mutations to disrupt the Zn2+-binding site in Kv3.1 T1, and assayed their binding to KIF5B tail. Four residues, H77, C83, C104, and C105, in the conserved Zn2+-binding motif (HX5CX20CC), are essential for T1 domains to coordinate Zn2+ at the tetramer interface (Bixby et al., 1999; Nanao et al., 2003). Mutating any one of them to Ala disrupts the Zn2+ binding and thereby yields monomeric T1 domains (Bixby et al., 1999; Nanao et al., 2003). GST-Tail865–934, His-31T1, His-31T1H77A, His-31T1C83A, and His-31T1C2A2 (a double mutant with both C104 and C105 mutated to Ala) were bacterially expressed and purified. His-31T1, but none of the three mutants, was precipitated by GST-Tail865–934, as revealed by Coomassie staining and confirmed by Western blotting (Fig. 6A,B). Therefore, both the EDTA and mutagenesis results suggest that the KIF5 tail domain preferentially binds to Kv3.1 T1 tetramers, but not monomers.
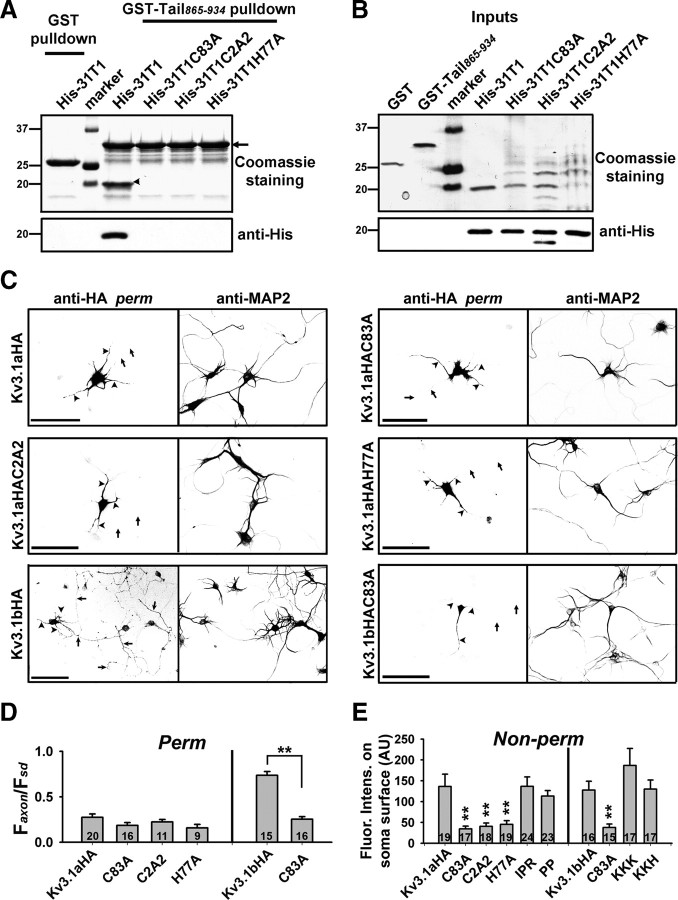
Figure 6.
The Zn2+-binding site required for Kv3.1 T1 tetramerization is critical for binding to KIF5B tail, and for Kv3.1 targeting to axons and plasma membranes. A, Mutations of the Zn2+-binding site within the Kv3.1 T1 domain eliminated direct binding between Kv3.1 T1 and KIF5B tail. Bacterially expressed and purified GST-Tail865–934 efficiently precipitated purified His-31T1, but not His-31T1C83A, His-31T1C2A2, or His-31T1H77A. Eluted proteins were shown by Coomassie staining (top) and Western blotting with an anti-His antibody (bottom). As a negative control, purified GST failed to pull down His-31T1 (left lane). Arrowhead, The His-31T1 band. B, Inputs of the pull-down experiment. Twenty-five percent of the inputs were loaded for Coomassie staining (top). Loading for Western blotting (bottom) was 10% of that for Coomassie staining (top). Molecular weights are indicated on the left in kilodaltons. C, Mutants of the Zn2+-binding site in the Kv3.1 T1 domain were all restricted in somatodendritic regions. Hippocampal neurons transfected with Kv3.1HA or mutants were stained with monoclonal anti-HA (left) and polyclonal anti-MAP (right) antibodies under permeabilized conditions. Signals are inverted. Arrows, Axons. Arrowheads, Dendrites. D, Summary of axonal levels (Faxon/Fsd) of Kv3.1 wild types and mutants under permeabilized conditions. **p < 0.001 in t test. E, Mutating the N-terminal Zn2+-binding site, but not the C-terminal axonal targeting motif, significantly decreased the surface levels of Kv3.1 channels in the soma. Hippocampal neurons transfected with Kv3.1HA or mutants were stained with an anti-HA antibody under the nonpermeabilized condition. The average immunofluorescence intensities on the soma surface were measured and the background was subtracted for Kv3.1aHA (Fsoma, 136.4 ± 29.3), Kv3.1aHAC83A (Fsoma, 34.6 ± 6.6), Kv3.1aHAC2A2 (Fsoma, 40.7 ± 7.9), Kv3.1aHAH77A (Fsoma, 45.3 ± 8.8; n = 19), Kv3.1aHAIPR (Fsoma, 136.5 ± 22.7), Kv3.1aHAPP (Fsoma, 113.4 ± 13.1), Kv3.1bHA (Fsoma, 127.7 ± 21.2), Kv3.1bHAC83A (Fsoma, 37.8 ± 8.4), Kv3.1bHAKKK (Fsoma, 186.8 ± 40.6), and Kv3.1bHAKKH (Fsoma, 130.1 ± 21.6). **p < 0.001, one-way ANOVA followed by Dunnett's test for p values. The number within each bar indicates “n.” Scale bars, 100 μm. Error bars indicate SEM.
To assess how mutating the Zn2+-binding site affects the axonal level of Kv3.1 channels, we examined the targeting of Kv3.1aHA and Kv3.1bHA constructs carrying mutated residues. When expressed in hippocampal neurons, Kv3.1aHAH77A (Faxon/Fsd, 0.18 ± 0.05; n = 9), Kv3.1aHAC83A (Faxon/Fsd, 0.19 ± 0.03; n = 16), and Kv3.1aHAC2A2 (Faxon/Fsd, 0.22 ± 0.03; n = 11) were restricted in somatodendritic regions similar to the wild-type Kv3.1aHA (Faxon/Fsd, 0.27 ± 0.04; n = 20) (Fig. 6C,D). In contrast, the axonal level of Kv3.1bHAC83A (Faxon/Fsd, 0.25 ± 0.03; n = 16) markedly decreased compared with that of the wild-type Kv3.1bHA (Faxon/Fsd, 0.70 ± 0.04; n = 15) (Fig. 6C,D). Therefore, mutating the Zn2+-binding site disrupts T1 tetramerization and thereby may prevent Kv3.1bHA from entering axons through interrupting the interaction between Kv3.1 T1 and KIF5 tail.
We further examined surface levels of these mutants in the soma by staining transfected neurons with the anti-HA antibody under nonpermeabilized conditions. Surface levels of T1 domain mutations (H77A, C83A, and C2A2), but not those of ATM mutations (IPR, PP, KKK, and KKH) (Xu et al., 2007), significantly decreased in the neuronal soma (Fig. 6E). Therefore, this result suggests that proper T1 tetramerization is important for the membrane targeting of Kv3.1 channels.
The Zn2+-binding site in T1 is essential for the proper activity of Kv3.1 channels
Kv channel T1 domains are involved in not only channel assembly but also channel gating (Cushman et al., 2000; Minor et al., 2000). Therefore, proper T1 tetramerization is critical for both channel assembly and activity. In fact, mutating the Zn2+-binding site in Kv4.2 channels disrupted the channel assembly and eliminated channel activity (Strang et al., 2003). Interestingly, coexpression of KCHIP3 rescued the function of Zn2+ site mutants by driving the mutant subunits to assemble to tetramers and to traffic to plasma membranes (Kunjilwar et al., 2004). How mutating the Zn2+-binding site affects Kv3.1 channel activity is still unknown. Moreover, it is of interest to determine whether the KIF5 binding affects channel activity. Therefore, we performed whole-cell voltage-clamp recording on HEK293 cells transfected with Zn2+ site mutants of Kv3.1. Channel currents for all mutants decreased by approximately one order of magnitude (Fig. 7A,B; scale bars are different between the wild type and mutants). Although the decrease may result from reduced channel number on the cell surface and/or decreased channel conductance, biophysical properties of the channel mutants were clearly altered.
Figure 7.
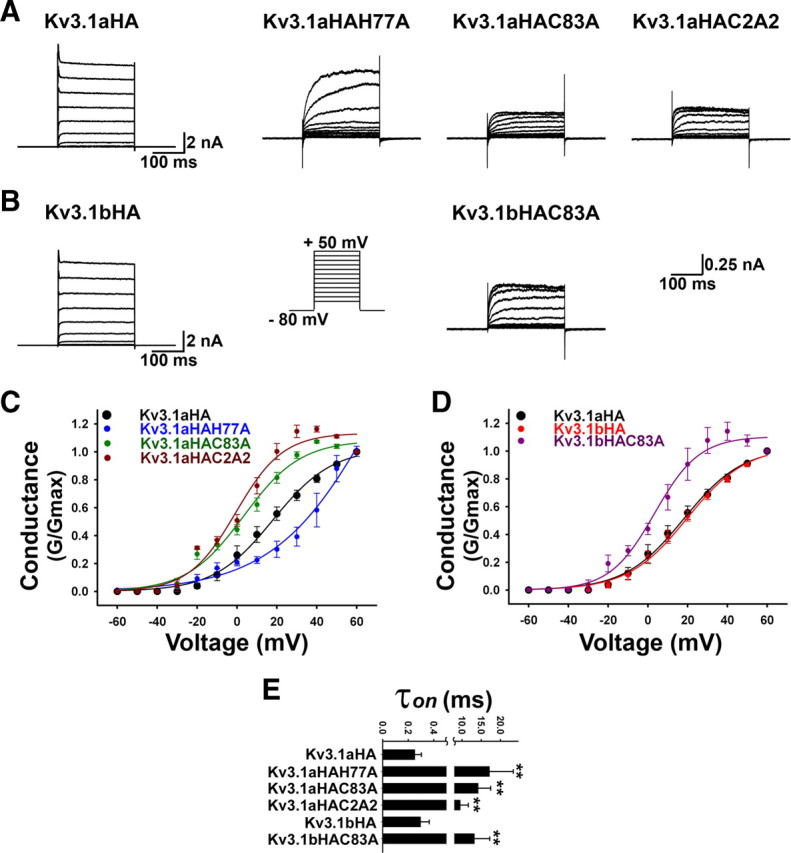
The Zn2+-binding site required for proper T1 tetramerization is critical for the proper Kv3.1 channel activity. Kv3.1 channel constructs were cotransfected with GFP into HEK293 cells and studied with whole-cell voltage-clamp recording. Transfected cells were identified with GFP fluorescence, held at −80 mV, and given 250 ms voltage episodes from −60 to +60 mV with a 10 mV increment. Examples of current traces of Kv3.1a constructs (A) and Kv3.1b constructs (B) are given in the range from −60 to +50 mV. The wild types and mutants have different scale bars. C, Conductance (G/Gmax)–voltage curves of Kv3.1a wild type and mutants. D, Conductance–voltage curves of Kv3.1 wild type and the C83A mutant of Kv3.1b. E, Activation constants (τon) of all the T1 mutants drastically increased. **p < 0.001, one-way ANOVA followed by Dunnett's test for p values. Error bars indicate SEM.
Whereas conduction–voltage curves for wild-type Kv3.1aHA and Kv3.1bHA completely overlapped, C83A and C2AC mutations shifted the curve to the left, and the H77A mutation shifted the curve to the right (Fig. 7C,D). The activation of all T1 mutants was drastically slower compared with that of the wild-type channels. The two splice variants were similar in both the activation constant (τon) (Kv3.1aHA τon, 0.25 ± 0.05, n = 12; Kv3.1bHA τon, 0.30 ± 0.07, n = 8) and deactivation constant (τoff) (Kv3.1aHA τoff, 0.66 ± 0.05, n = 12; Kv3.1bHA τoff, 0.59 ± 0.02, n = 8) (Fig. 7E). τon of the mutants increased by ∼50-fold (Kv3.1aHAH77A τon, 17.14 ± 6.21, n = 5; Kv3.1aHAC83A τon, 14.21 ± 3.24, n = 6; Kv3.1aHAC2A2 τon, 9.55 ± 2.09, n = 6; Kv3.1bHAC83A τon, 13.21 ± 3.98, n = 5) (Fig. 7E). Because of the small currents of mutants, it was difficult to unequivocally determine their deactivation constants.
Deleting the T1 domain completely abolished the channel activity for some Kv channels but not for others (VanDongen et al., 1990; Shen et al., 1993; Tu et al., 1996; Schulteis et al., 1998). It appears that the high expression of subunits lacking T1 is sufficient for some channel assembly. Interestingly, an artificial tetramerization domain restores efficient assembly of functional Shaker channels lacking T1 (Zerangue et al., 2000). It was hypothesized that T1 tetramerization promotes subfamily-specific assembly through kinetic partitioning of the assembly process but is not required for subsequent steps in channel assembly and folding (Zerangue et al., 2000). For Kv3.1 channels, mutating the Zn2+-binding site did not completely eliminate the channel activity. All mutants displayed some channel currents, indicating these proteins were still properly folded. Thus, mutating the Zn2+-binding site in Kv3.1 channels does not result in misfolded proteins.
To determine whether binding to KIF5 affects Kv3.1 channel activity, we performed voltage-clamp recording on HEK293 cells cotransfected with wild-type Kv3.1 channels and KIF5B-YFP. No major effect was observed (data not shown). This is consistent with the hypothesis that KIF5 binds to the external surface of the T1 tetramer of the Kv3.1 channel complex. Alternatively, KIF5 motors may bind to intracellular Kv3.1 channels, but not to the channel proteins already on plasma membranes.
Expressed KIF5B-YFP assists dendritic Kv3.1 to penetrate the AIS and enter distal axons
Our results suggest that, since KIF5 specifically binds to T1 tetramers, only properly assembled and hence functioning Kv3.1 channels can bind to KIF5 motors and be transported into axons. To further understand how Kv3.1 channels can be specifically transported by KIF5 motors into axons, we examined the effects of overexpressed KIF5 constructs on Kv3.1 channel targeting. Interestingly, the axonal level of dendritic Kv3.1aHA significantly increased in the presence of KIF5B-YFP (Faxon/Fsd, 0.87 ± 0.07; n = 20), but not KIF3A-YFP (Faxon/Fsd, 0.23 ± 0.02; n = 20), compared with YFP (Faxon/Fsd, 0.30 ± 0.05; n = 12) (Fig. 8A,B,D). Kv3.1aHA highly colocalized with KIF5B-YFP in clusters along axons (Fig. 8B). Overexpressed KIF5B-YFP may increase the KIF5 motors that are available for transporting Kv3.1a channels to penetrate the AIS and enter distal axons. Next, we found that even the tailless KIF5B1–760-YFP, missing the Kv3.1 T1-binding site, significantly enhanced the axonal level of Kv3.1aHA (Faxon/Fsd, 0.58 ± 0.04; n = 20) (Fig. 8C,D). Kv3.1aHA not only entered proximal axons but also partially colocalized with KIF5B1–760-YFP in clusters in axonal endings (Fig. 8C). The tailless KIF5B1–760-YFP may dimerize with the endogenous KIF5B to form a dimeric motor with two motor domains but only one tail domain, which apparently was sufficient to transport Kv3.1. Furthermore, we examined the effect of coexpressed KIF5B-YFP on the axonal level of Kv1.2HA. KIF5B-YFP (Faxon/Fsd, 0.79 ± 0.06; n = 18) did not increase the axonal level of Kv1.2HA compared with YFP (Faxon/Fsd, 0.75 ± 0.07; n = 17), suggesting that its effect on Kv3.1aHA was specific.
Figure 8.
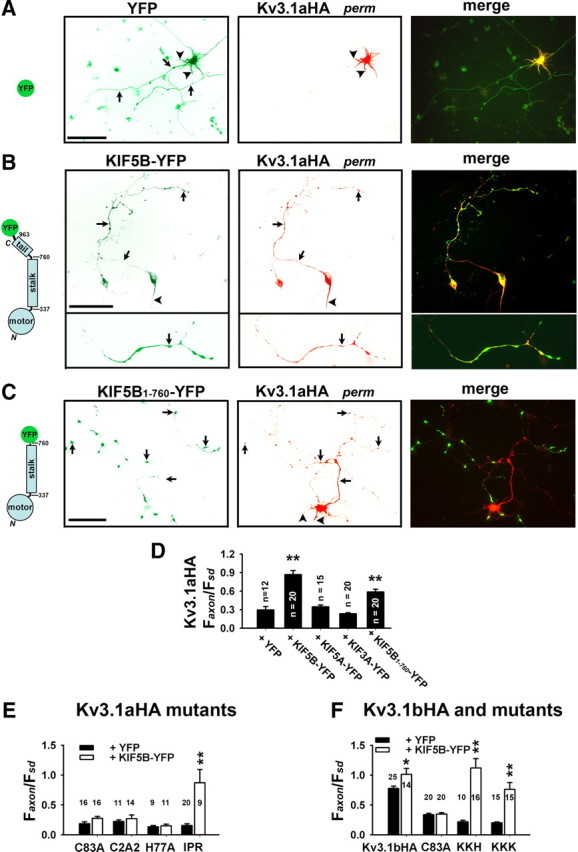
Overexpression of KIF5B constructs assists dendritic Kv3.1a to penetrate the AIS and enter distal axons. Kv3.1aHA and kinesin fusion proteins were cotransfected into cultured hippocampal neurons at 5 DIV. Two days later, transfected neurons were stained with anti-HA and anti-MAP2 antibodies under permeabilized conditions. A, Kv3.1aHA (red) mainly localized in somatodendritic regions in the presence of YFP (green). B, Kv3.1aHA (red) was targeted into axons and colocalized with KIF5B-YFP (green) in clusters along axons (top). An axonal segment is shown with three times higher magnification (bottom). C, The effect of coexpressed KIF5B1–760-YFP on Kv3.1aHA targeting. Black arrows, Axons; black arrowheads, dendrites. D, Summary of the effects of kinesin constructs on Kv3.1aHA targeting. E, Summary of the effects of KIF5B-YFP coexpression on the relative axonal level of Kv3.1aHA mutants. In the presence of YFP or KIF5B-YFP, the Faxon/Fsd ratios are 0.19 ± 0.03, 0.27 ± 0.03 for Kv3.1aHAC83A; 0.22 ± 0.03, 0.27 ± 0.06 for Kv3.1aHAC2A2; 0.13 ± 0.02, 0.14 ± 0.03 for Kv3.1aHAH77A; 0.16 ± 0.03, 0.87 ± 0.22 for Kv3.1aHAIPR, respectively. F, Summary of the effects of KIF5B-YFP on axonal levels of Kv3.1bHA and its mutants. In the presence of YFP or KIF5B-YFP, the Faxon/Fsd ratios are 0.78 ± 0.04, 1.01 ± 0.10 for Kv3.1bHA; 0.34 ± 0.02, 0.35 ± 0.02 for Kv3.1bHAC83A; 0.22 ± 0.03, 1.12 ± 0.15 for Kv3.1bHAKKH; 0.20 ± 0.01, 0.76 ± 0.11 for Kv3.1bHAKKK, respectively. Scale bars, 100 μm. *p < 0.05; **p < 0.001, one-way ANOVA followed by Dunnett's test for p values. Error bars indicate SEM.
Our data showed that overexpressed KIF5B-YFP transported dendritic Kv3.1aHA into axons. We wondered what is the determinant in this process. In our previous study, we generated Kv3.1b mutants with altered ATM in the C-terminal region. These mutants are restricted in somatodendritic regions (Xu et al., 2007). In the present study, we made Kv3.1 mutants with disrupted Zn2+-binding site in T1 and these mutants also failed to enter axons (Fig. 6). These mutants were cotransfected with KIF5B-YFP into neurons. Remarkably, overexpressed KIF5B-YFP brought all the Kv3.1 C-terminal mutants with an impaired ATM to penetrate the AIS and enter distal axons, but failed to affect any of the T1 mutants (Fig. 8E,F). Expression of KIF5B-YFP further increased Kv3.1bHA axonal levels (Fig. 8F). Therefore, overexpressed KIF5B-YFP assisted dendritic Kv3.1aHA and even Kv3.1 mutants with a faulty ATM to penetrate the AIS and enter the axon. Interestingly, this effect apparently requires properly assembled T1 domains, consistent with the hypothesis that T1 tetramerization is essential for binding to KIF5 and hence Kv3.1 targeting in axons (Figs. 5, 6).
Colocalization and comovement of fluorescently tagged Kv3.1 and KIF5B
Since our results suggest that KIF5 motors transport Kv3.1 channels, KIF5 and Kv3.1 may not only colocalize but also comove along axons. To visualize the potential comovement using two-color time-lapse imaging, we first fused fluorescent molecules (CFP and YFP) to Kv3.1a, Kv3.1b, and KIF5B. When expressed in hippocampal neurons, YFP-Kv3.1a (a YFP fused to the N terminus of Kv3.1a) and YFP-Kv3.1b (a YFP fused to the N terminus of Kv3.1b) were concentrated in somatodendritic regions. The axonal level of YFP-Kv3.1b was only moderately higher than that of YFP-Kv3.1a (Fig. 9A,B, left). It is possible that the sizable YFP molecule (or CFP, ∼220 aa) reduces the difference of the N/C-terminal interaction for two splice variants, but the Kv3.1–KIF5B interaction was apparently unaffected. When coexpressed with KIF5B-YFP, axonal levels of both CFP-Kv3.1a and CFP-Kv3.1b markedly increased, and both highly colocalized with KIF5B-YFP in punctuate clusters along axons (Fig. 9A,B, right), consistent with the results obtained with HA-tagged Kv3.1 (Fig. 8).
Figure 9.
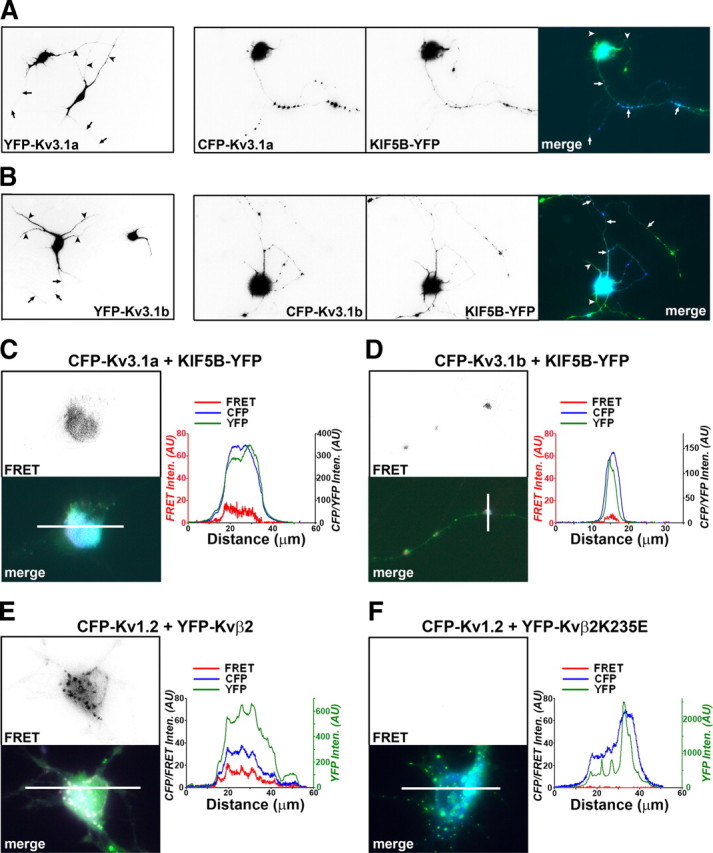
KIF5B-YFP assists CFP-Kv3.1a and CFP-Kv3.1b to penetrate the AIS and enter distal axons via direct binding revealed by the FRET imaging. Hippocampal neurons from the E18 culture were transfected with fluorescently tagged Kv3.1 and KIF5 constructs at 5 DIV, and imaged 2–3 d later. CFP- or YFP-tagged Kv3.1a (A, left) and Kv3.1b (B, left) were mainly concentrated in somatodendritic regions. The level of YFP-Kv3.1b in axons was only moderately higher than that of YFP-Kv3.1a. It is possible that after fusing with CFP or YFP (∼220 aa) at the N termini, the difference of axonal levels between Kv3.1a and Kv3.1b decreased. The axonal level of CFP-Kv3.1a (blue in merged) markedly increased in the presence of KIF5B-YFP (green in merged) (A, right), consistent with the result using Kv3.1aHA in Figure 8. CFP-Kv3.1a and KIF5B-YFP highly colocalized in clusters along axons (A, right). Similarly, the axonal level of CFP-Kv3.1b (blue in merged) markedly increased in the presence of KIF5B-YFP (green in merged) (B, right). CFP-Kv3.1b and KIF5B-YFP highly colocalized in clusters along axons (B, right). In nonmerged images, signals are inverted. Arrows, Axons; arrowheads, dendrites. Scale bars: A, B, 100 μm. C, FRET signals were detected in neuronal soma expressing CFP-Kv3.1a and KIF5B-YFP. The fluorescence intensity profiles along the white line within the merged image are shown on the right. D, FRET signals were detected in axons expressing CFP-Kv3.1b and KIF5B-YFP. In C and D, the scale for FRETcorrected (FRET in red) is on the left, and the scale for CFP (blue) and YFP (dark green) is on the right. E, Robust FRET signals were detected in neuronal soma expressing CFP-Kv1.2 and YFP-Kvβ2. F, No FRET signal was detected in neuronal soma expressing CFP-Kv1.2 and YFP-Kvβ2K235E. In E and F, the scale for CFP (blue) and FRETcorrected (FRET in red) is on the left, and the scale for YFP (green) is on the right. AU, Arbitrary unit.
We further examined whether CFP-Kv3.1a and CFP-Kv3.1b bind to KIF5B-YFP in axons of live neurons by using the FRET imaging. Indeed, FRET signals were observed from both CFP-Kv3.1a/KIF5B-YFP (11 of 14 neurons) and CFP-Kv3.1b/KIF5B-YFP (12 of 17 neurons), and in both soma and axons (Fig. 9C,D). Although significantly higher than background, the FRET signals were not robust, and they were weaker than those generated by a positive control pair, CFP-Kv1.2 and YFP-Kvβ2 (14 of 14 neuronal soma had significant FRET signals) (Fig. 9E). The FRET imaging of CFP-Kv1.2 and YFP-Kvβ2 was shown in our previous study (Gu et al., 2006). We also showed that a single point mutation in Kvβ2, located at the T1–Kvβ2 interface, eliminated the Kv1.2 and Kvβ2 binding (Gu et al., 2006). In the present study, we found that no FRET signal was detected between CFP-Kv1.2 and YFP-Kvβ2K235E (0 of 15 neuronal soma had clear FRET signals) (Fig. 9F), consistent with our previous study. Thus, we had both good positive and negative controls for the present study. FRET signals between CFP-Kv3.1 and KIF5B-YFP were not robust, which may be attributable to the following potential reasons: (1) the distance between CFP located at the Kv3.1 N terminus and YFP located in the KIF5B C terminus may not be very close; (2) the CFP and YFP may be not in the orientation optimal for FRET; (3) the interaction between Kv3.1 and KIF5 may be transient, which is actually consistent with the role of KIF5 in transporting Kv3.1 channel proteins.
To determine whether Kv3.1 and KIF5 can comove, we performed the two-color time-lapse imaging. Although most puncta of YFP-Kv3.1a, YFP-Kv3.1b, or KIF5B-YFP, were stationary, some moved in either anterograde or retrograde direction (Fig. 10A–F; supplemental Movies 1, 2, available at www.jneurosci.org as supplemental material). There was no major difference among their anterograde (and retrograde) transport velocities (Fig. 10E). Interestingly, average anterograde velocities of YFP-Kv3.1a and YFP-Kv3.1b (even KIF5B-YFP) puncta were much slower than that of APP-YFP (average anterograde velocity of ∼1.5 μm/s measured with our own setup). More than one-half of the moving puncta contained both fluorescently tagged Kv3.1 and KIF5B, in both anterograde (Fig. 10C; supplemental Movie 3, available at www.jneurosci.org as supplemental material) and retrograde (Fig. 10D; supplemental Movie 4, available at www.jneurosci.org as supplemental material) directions. Because of its direct binding to KIF5, Kv3.1 channels might act as both cargos and adaptors for the motor. There was no major difference in travel velocity and distance of Kv3.1 axonal transport in the absence or presence of KIF5B-YFP (Fig. 10E,F). Moreover, the cotransporting events were observed in both proximal and distal axons. Together, our results suggest that KIF5 is the major kinesin motor that transports Kv3.1 channels along axons.
Figure 10.
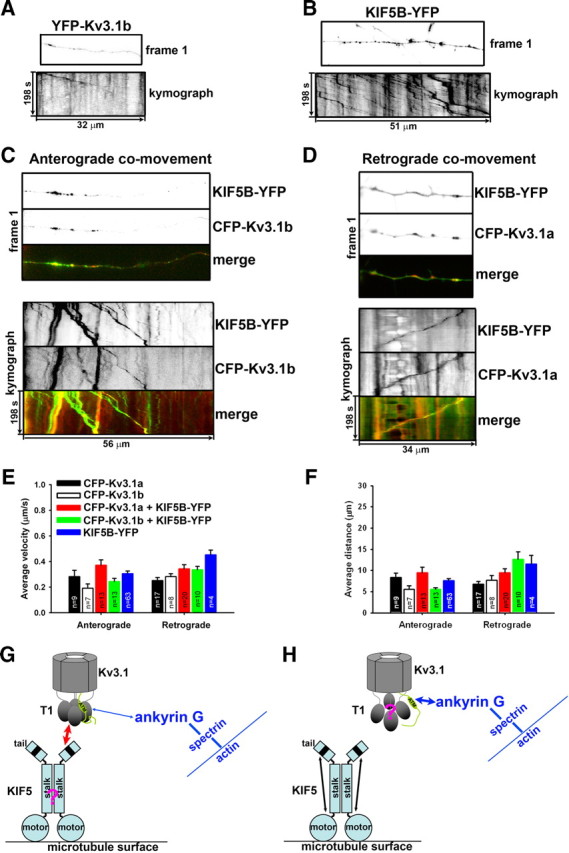
Axonal cotransport of Kv3.1 and KIF5. A, Anterograde transport of YFP-Kv3.1b. The hippocampal axon that expressed YFP-Kv3.1b alone was visualized with time-lapse imaging. The first frame (top) and kymograph (bottom) are shown with distance (in micrometers) and time (in seconds) labeled. Left side, Toward soma; right side, toward axonal growth cone. Signals are inverted. B, Anterograde transport of KIF5B-YFP. C, Anterograde cotransport of KIF5B-YFP and CFP-Kv3.1b. The first frame and kymograph of KIF5B-YFP (green in merged image) and CFP-Kv3.1b (red in merge image) are provided. Time (in seconds) and distance (in micrometers) are labeled on merged kymograph. D, Retrograde cotransport of KIF5B-YFP (green in the merged) and CFP-Kv3.1a (red in the merged). E, Summary of average velocities for axonal transport and cotransport of indicated fusion proteins. Anterograde and retrograde velocities were as follows (in μm/s): 0.28 ± 0.05 and 0.25 ± 0.02 for C(Y)FP-Kv3.1a; 0.20 ± 0.02 and 0.32 ± 0.03 for C(Y)FP-Kv3.1b; 0.37 ± 0.04 and 0.34 ± 0.03 for CFP-Kv3.1a and KIF5B-YFP comovement; 0.24 ± 0.02 and 0.34 ± 0.03 for CFP-Kv3.1b and KIF5B-YFP; 0.31 ± 0.02 and 0.45 ± 0.04 for KIF5B-YFP alone. The “n” for each bar indicates the number of particles. F, Summary of the average travel distances. Anterograde and retrograde travel distances were as follows (in μm): 8.33 ± 1.05 and 6.76 ± 0.65 for C(Y)FP-Kv3.1a; 5.82 ± 0.68 and 8.92 ± 0.80 for C(Y)FP-Kv3.1b; 9.51 ± 1.28 and 9.49 ± 0.90 for CFP-Kv3.1a and KIF5B-YFP comovement; 5.60 ± 0.35 and 12.62 ± 1.75 for CFP-Kv3.1b and KIF5B-YFP comovement; 7.56 ± 0.54 and 11.47 ± 2.09 for KIF5B-YFP. Error bars indicate SEM. G, H, A proposed model for the potential interplay of KIF5 and ankyrin G during transportation of Kv3.1 channels through the AIS. G, A KIF5 motor directly binds to the Kv3.1 T1 tetramer via its C-terminal tail and transports the channel tetramer. Kv3.1 channel may or may not also bind to ankyrin G through its C terminus assembled together with the T1 tetramer. For clarification, only one C-terminal tail is shown here. H, Disassembly or conformational change of the T1 tetramer dissociates the channel from KIF5 and allows a stronger binding of ankyrin G to the C terminus. The process may involve more intermediate states in addition to the two shown here.
A recent study identified an ankyrin G- and F-actin-dependent cytoplasmic filter at the AIS, which emerges within 2 d after axon/dendrite differentiation, allowing axonal entry of small molecules (<10 kDa), but not large ones (>70 kDa) (Song et al., 2009). A Kv3.1b channel tetramer (with posttranslational modifications) is ∼360 kDa. Therefore, an axonal Kv3.1 channel must be actively transported through the AIS, consistent with the conclusion in the present study. Based on our data, we propose the following hypothesis. Post-Golgi vesicles containing Kv3.1a and/or Kv3.1b are carried by KIF5 in the soma, since Kv3.1a and Kv3.1b have the exact same T1 domain, the KIF5-binding domain. When they reach the AIS, Kv3.1 channels may undergo multiple rounds of association and dissociation with ankyrin G and KIF5 (Fig. 10G,H). The splice domain in Kv3.1b allows the channel to bind strongly to ankyrin G (Xu et al., 2007) and thus to stay in the AIS for enough time, allowing it to be reloaded to KIF5 motors moving toward distal axons. This elegant balance may be altered by the overexpression of KIF5B-YFP, which carried even dendritic Kv3.1a channels through the AIS into distal axons (Figs. 8, 9).
Discussion
In this study, we have identified a novel interaction between Kv3.1 channel and KIF5/kinesin I motor. This interaction is mediated by a direct binding between the Kv3.1 N-terminal T1 domain and a conserved domain in KIF5 tail. Importantly, our data suggest that the binding requires proper T1 tetramerization. We have also provided several lines of evidence showing KIF5 is the major motor for axonal transport of Kv3.1 channels.
The voltage-sensing and pore-forming subunit of Kv3.1 directly binds to the kinesin heavy chain
The dendritic transport of NMDA and AMPA receptors, by KIF17 and KIF5, respectively, requires two different sets of adaptor proteins (Setou et al., 2000, 2002; Hirokawa and Takemura, 2005). A recent study shows that delivery of GABAA receptors to synapses is performed by KIF5 also via an adaptor protein, Huntingtin-associated protein 1 (Twelvetrees et al., 2010).
In this study, we have identified KIF5 as a binding protein of the conserved T1 domain of Kv3.1 channels and subsequently confirmed the Kv3.1b–KIF5 binding by coimmunoprecipitation from solubilized brain crude membranes (Fig. 1B). Using pull-down assays, we have mapped the Kv3.1-binding site to a 70 residue region in KIF5 tail (Fig. 2). This region pulled down Kv3.1 T1 but not Kv1.2 T1 or Kv4.2 T1 (Fig. 5A–C), suggesting that the direct binding between Kv3 and KIF5 is most likely specific within the Kv channel family. Furthermore, the purified KIF5B tail domain efficiently precipitated Kv3.1 T1 (Fig. 5D–F), confirming the binding is direct. Importantly, the binding was significantly weakened in the presence of EDTA (Fig. 5D,E), which disassembles T1 tetramers into monomers by chelating Zn2+ (Bixby et al., 1999; Nanao et al., 2003), and was completely abolished by mutating the Zn2+-binding site in T1 (Fig. 6A,B). Mutating the Zn2+-binding site disrupts Kv3.1 T1 tetramerization (Bixby et al., 1999; Nanao et al., 2003). These results suggest that T1 tetramerization plays a crucial role in the effective Kv3.1–KIF5 binding. The fact that the EDTA incubation did not completely abolish the GST-Tail865–934–His-31T1 binding (Fig. 5F) may result from the incomplete disassociation of Kv3.1 T1 tetramers.
Our data suggest that the KIF5 tail domain preferentially binds to T1 tetramers but not monomers. How may T1 tetramerization affect the T1-tail binding? The possibilities, including (1) the formation of a proper binding surface on the tetramer, (2) the cooperativity between adjacent monomers in the tetrameric form in their binding to dimerized tail domains, and (3) allosteric effect induced by tetramerization, remain to be elucidated in future studies. How many KIF5 tail domains can a Kv3 T1 tetramer bind to? A tailless KIF5B construct, KIF5B1–760-YFP, when expressed into neurons, likely dimerizes with endogenous KIF5B and generates some KIF5 motors with only one tail. The result that KIF5B1–760-YFP brought dendritic Kv3.1a to cross the AIS (Fig. 8C,D), suggests a Kv3.1 T1 tetramer can bind to at least one KIF5 tail domain. Furthermore, why do so many proteins interact with KIF5 tail but do not end up in the same location? As a major motor protein, KIF5 transports many different cargos and is highly regulated at multiple layers. It will be interesting to determine how these proteins may compete or collaborate with Kv3.1 for binding to this region in future studies.
Interestingly, in addition to the Kv3 C-terminal ATM-ankyrin G binding (Xu et al., 2007), the direct binding between the Kv3 T1 domain and the 70 residue region in KIF5 tail is also likely evolutionarily conserved. Kv3 T1 domains are highly conserved from fruit fly to human. The binding site, the 70 residue region in KIF5 tail, also shares >75% identity from fruit fly to human (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). Therefore, our studies have uncovered a conserved core mechanism governing intracellular transport of Kv3 channels.
Distinct mechanisms govern axonal transport of Kv1 and Kv3 channels
Kv1 and Kv3 are two major types of Kv channels operating in axons. The exact mechanisms underlying their axonal transport are likely very different (Gu et al., 2006; Xu et al., 2007). The notion has been further reinforced by the present study. First, purified GST-31N but not GST-12N pulled down KIF5B-YFP expressed in HEK293 cells (Fig. 1C). Second, the purified KIF5B tail domain (GST-Tail) precipitated the His-tagged Kv3.1 T1 but not Kv1.2 T1 (Fig. 5A,B). Third, coexpression of KIF5B-YFP significantly increased the axonal levels of both Kv3.1aHA and Kv3.1bHA, but not that of Kv1.2HA (Fig. 8D). Finally, the T1 domains of Kv1.2 and Kv3.1, when fused to the N terminus of TfR-GFP, exhibited different effects on polarized targeting of the reporter protein. Kv3.1T1-TfR-GFP was restricted in clusters in soma and proximal dendrites (Xu et al., 2007), whereas Kv1.2T1-TfR-GFP entered distal axons (Gu et al., 2003), suggesting that the T1 domains within Kv3.1T1-TfR-GFP might not tetramerize properly. As a result, the fusion protein cannot be recognized and transported by the KIF5 motor into axons. In contrast, the Kv1.2 T1 domain appeared more flexible in adopting the context of reporter proteins (Gu et al., 2003).
KIF5B was reported to play an important role in axonal transport of Kv1.3 channels (Rivera et al., 2007) and in forward trafficking of cardiac Kv1.5 channels (Zadeh et al., 2009). How these channels are loaded onto the KIF5B motor remains unclear. The association and transport of different cargos of KIF5 motors are differentially regulated (Hirokawa and Takemura, 2005; Hirokawa et al., 2009). Our results show that, unlike Kv3.1 T1, Kv1.2 T1 does not directly bind to KIF5B tail (Fig. 5A,B), and therefore suggest the potential KIF5B-mediated transport of Kv1 may be very different from that of Kv3.1. Moreover, electron microscopy showed multiple tiny bridges linking an intracellular vesicle to microtubules, suggesting that a vesicle cargo can be transported by more than one type of kinesin motor (Hirokawa and Takemura, 2005). Therefore, it is possible that a Kv channel complex can be transported by different kinesin motors simultaneously, or by different types of motors in different types of cells, developmental stages, and activity states. These are important questions for future investigation.
Microtubule-based trafficking of Kv channels in axons
Despite their differences, Kv1 and Kv3 channels do share a common strategy. In this study, we show T1 tetramerization is important for the Kv3.1–KIF5 binding (Figs. 5, 6), which is critical for KIF5-based axonal targeting of Kv3.1 channels (Figs. 3, 4, 6, 8). Our previous study showed that residues on the interface (and the T1–β interface) but not on the outer surface of the Kv1.2 T1 tetramer are critical for axonal targeting of CD4-Kv1.2T1 (Gu et al., 2003), suggesting proper tetramerization is required for Kv1 axonal transport likely through the Kv1–Kvβ binding. Furthermore, these two major types of axonal Kv channels are linked to kinesin motor proteins via their T1 domains for long-distance and microtubule-based trafficking. The important role of the N-terminal T1 domains of Kv channels in long-distance trafficking is consistent with the notion that Kv channel T1 domains can form tetramers right after emerging from the ribosomal exiting tunnel once synthesis of the T1–S1 linker is completed (Kosolapov et al., 2004).
Their C-terminal domains are linked to local retention or trafficking, either through ankyrin G and the actin cytoskeleton (Xu et al., 2007), or through a tyrosine-based endocytic motif and a PDZ (postsynaptic density-95/Discs large/zona occludens-1) domain ligand (Lai and Jan, 2006). In contrast, dendritic ion channels are linked to kinesin motors via their C termini (Setou et al., 2000, 2002; Chu et al., 2006).
Binding to Kv3.1/ankyrin G may affect the activity of KIF5 during axonal entry
By interacting with both ankyrin G at the AIS (Xu et al., 2007) and the KIF5 motor, Kv3.1 channels are in a unique position to shed light on the interaction between the AIS filter and kinesin motors. The transport efficacy of the motor-cargo complex, but not the cargo or the motor alone, was suggested to determine the axonal entry (Song et al., 2009). The cargo binding to the motor may change the conformation of the motor and affect the recognition of microtubules and the direction of transport (Setou et al., 2002; Hirokawa and Takemura, 2005). Therefore, the binding between the Kv3.1 C terminus and ankyrin G may build enough tension to change the conformation and hence the activity of KIF5 motors. The following two interesting questions remain to be determined in future investigation. How does Kv3.1 shift between actin- and microtubule-based trafficking? How might the N/C-terminal interaction of Kv3.1 be involved in this process?
Furthermore, the transcripts of Kv3.1a and Kv3.1b, often coexisting in the same neuron, are downregulated and upregulated in the perinatal stage, respectively (Perney et al., 1992). As a result, there are likely more Kv3.1b subunits than Kv3.1a subunits in a mature neuron. Since Kv3.1a and Kv3.1b subunits can coassemble into heterotetramers, neuronal maturation may shift Kv3.1 channels from dendrites to axons via alternative splicing. Moreover, expression of Kv3 channels, KIF5, and ankyrin G varies in different neurons, and other players may be involved in fine-tuning the mechanism by regulating the Kv3 binding to ankyrin G and/or KIF5. These are also interesting topics for future investigation.
Footnotes
This work was supported by a Career Transition Fellowship Award from the National Multiple Sclerosis Society (Grant TA3012A1) and National Institute of Neurological Disorders and Stroke–National Institutes of Health (NIH) Research Grant R01 NS062720 (C.G.). We thank Drs. R. Tsien and R. Vale for sharing reagents, The Ohio State University Mass Spectrometry and Proteomics Facility for mass spectrometry analysis, University of California, Davis–NIH NeuroMab facility (a nonprofit supplier) for high-quality monoclonal antibodies used in this study. All animal experiments have been conducted in accordance with the NIH Animal Use Guidelines.
References
- Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V, Chen L. Ankyrins and cellular targeting of diverse membrane proteins to physiological sites. Curr Opin Cell Biol. 2001;13:61–67. doi: 10.1016/s0955-0674(00)00175-7. [DOI] [PubMed] [Google Scholar]
- Bixby KA, Nanao MH, Shen NV, Kreusch A, Bellamy H, Pfaffinger PJ, Choe S. Zn2+-binding and molecular determinants of tetramerization in voltage-gated K+ channels. Nat Struct Biol. 1999;6:38–43. doi: 10.1038/4911. [DOI] [PubMed] [Google Scholar]
- Cai Q, Gerwin C, Sheng ZH. Syntabulin-mediated anterograde transport of mitochondria along neuronal processes. J Cell Biol. 2005;170:959–969. doi: 10.1083/jcb.200506042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Liang CW, Muralidharan S, Kao JP, Tang CM, Thompson SM. Unique roles of SK and Kv4.2 potassium channels in dendritic integration. Neuron. 2004;44:351–364. doi: 10.1016/j.neuron.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Choe S. Potassium channel structures. Nat Rev Neurosci. 2002;3:115–121. doi: 10.1038/nrn727. [DOI] [PubMed] [Google Scholar]
- Chu PJ, Rivera JF, Arnold DB. A role for Kif17 in transport of Kv4.2. J Biol Chem. 2006;281:365–373. doi: 10.1074/jbc.M508897200. [DOI] [PubMed] [Google Scholar]
- Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- Cushman SJ, Nanao MH, Jahng AW, DeRubeis D, Choe S, Pfaffinger PJ. Voltage dependent activation of potassium channels is coupled to T1 domain structure. Nat Struct Biol. 2000;7:403–407. doi: 10.1038/75185. [DOI] [PubMed] [Google Scholar]
- Diefenbach RJ, Mackay JP, Armati PJ, Cunningham AL. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry. 1998;37:16663–16670. doi: 10.1021/bi981163r. [DOI] [PubMed] [Google Scholar]
- Diefenbach RJ, Diefenbach E, Douglas MW, Cunningham AL. The heavy chain of conventional kinesin interacts with the SNARE proteins SNAP25 and SNAP23. Biochemistry. 2002;41:14906–14915. doi: 10.1021/bi026417u. [DOI] [PubMed] [Google Scholar]
- Du J, Zhang L, Weiser M, Rudy B, McBain CJ. Developmental expression and functional characterization of the potassium-channel subunit Kv3.1b in parvalbumin-containing interneurons of the rat hippocampus. J Neurosci. 1996;16:506–518. doi: 10.1523/JNEUROSCI.16-02-00506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson MG, Alseikhan BA, Peterson BZ, Yue DT. Preassociation of calmodulin with voltage-gated Ca2+ channels revealed by FRET in single living cells. Neuron. 2001;31:973–985. doi: 10.1016/s0896-6273(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindhart JG, Chen J, Faulkner M, Gandhi R, Doerner K, Wisniewski T, Nandlestadt A. The kinesin-associated protein UNC-76 is required for axonal transport in the Drosophila nervous system. Mol Biol Cell. 2003;14:3356–3365. doi: 10.1091/mbc.E02-12-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EM, Watanabe S, Chang SY, Joho RH, Huang ZJ, Leonard CS, Rudy B. Specific functions of synaptically localized potassium channels in synaptic transmission at the neocortical GABAergic fast-spiking cell synapse. J Neurosci. 2005;25:5230–5235. doi: 10.1523/JNEUROSCI.0722-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EM, Clark BD, Zagha E, Nahmani M, Erisir A, Rudy B. K+ channels at the axon initial segment dampen near-threshold excitability of neocortical fast-spiking GABAergic interneurons. Neuron. 2008;58:387–400. doi: 10.1016/j.neuron.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS. Kinesin molecular motors: transport pathways, receptors, and human disease. Proc Natl Acad Sci U S A. 2001;98:6999–7003. doi: 10.1073/pnas.111145298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Sorkin A, Cooper DM. Persistent interactions between the two transmembrane clusters dictate the targeting and functional assembly of adenylyl cyclase. Curr Biol. 2001;11:185–190. doi: 10.1016/s0960-9822(01)00044-6. [DOI] [PubMed] [Google Scholar]
- Gu C, Jan YN, Jan LY. A conserved domain in axonal targeting of Kv1 (Shaker) voltage-gated potassium channels. Science. 2003;301:646–649. doi: 10.1126/science.1086998. [DOI] [PubMed] [Google Scholar]
- Gu C, Zhou W, Puthenveedu MA, Xu M, Jan YN, Jan LY. The microtubule plus-end tracking protein EB1 is required for Kv1 voltage-gated K+ channel axonal targeting. Neuron. 2006;52:803–816. doi: 10.1016/j.neuron.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Gyoeva FK, Sarkisov DV, Khodjakov AL, Minin AA. The tetrameric molecule of conventional kinesin contains identical light chains. Biochemistry. 2004;43:13525–13531. doi: 10.1021/bi049288l. [DOI] [PubMed] [Google Scholar]
- Hille B. Sunderland, MA: Sinauer; 2001. Ion channels of excitable membranes. [Google Scholar]
- Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Hoffman DA, Magee JC, Colbert CM, Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature. 1997;387:869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- Huang JD, Brady ST, Richards BW, Stenolen D, Resau JH, Copeland NG, Jenkins NA. Direct interaction of microtubule- and actin-based transport motors. Nature. 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- Jahng AW, Strang C, Kaiser D, Pollard T, Pfaffinger P, Choe S. Zinc mediates assembly of the T1 domain of the voltage-gated K channel 4.2. J Biol Chem. 2002;277:47885–47890. doi: 10.1074/jbc.M208416200. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Cloned potassium channels from eukaryotes and prokaryotes. Annu Rev Neurosci. 1997;20:91–123. doi: 10.1146/annurev.neuro.20.1.91. [DOI] [PubMed] [Google Scholar]
- Jenkins SM, Bennett V. Ankyrin-G coordinates assembly of the spectrin-based membrane skeleton, voltage-gated sodium channels, and L1 CAMs at Purkinje neuron initial segments. J Cell Biol. 2001;155:739–746. doi: 10.1083/jcb.200109026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Okada Y, Tanaka Y, Harada A, Terada S, Hirokawa N. KIF5C, a novel neuronal kinesin enriched in motor neurons. J Neurosci. 2000;20:6374–6384. doi: 10.1523/JNEUROSCI.20-17-06374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kole MH, Letzkus JJ, Stuart GJ. Axon initial segment Kv1 channels control axonal action potential waveform and synaptic efficacy. Neuron. 2007;55:633–647. doi: 10.1016/j.neuron.2007.07.031. [DOI] [PubMed] [Google Scholar]
- Kosolapov A, Tu L, Wang J, Deutsch C. Structure acquisition of the T1 domain of Kv1.3 during biogenesis. Neuron. 2004;44:295–307. doi: 10.1016/j.neuron.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Kunjilwar K, Strang C, DeRubeis D, Pfaffinger PJ. KChIP3 rescues the functional expression of Shal channel tetramerization mutants. J Biol Chem. 2004;279:54542–54551. doi: 10.1074/jbc.M409721200. [DOI] [PubMed] [Google Scholar]
- Lai HC, Jan LY. The distribution and targeting of neuronal voltage-gated ion channels. Nat Rev Neurosci. 2006;7:548–562. doi: 10.1038/nrn1938. [DOI] [PubMed] [Google Scholar]
- Li M, Jan YN, Jan LY. Specification of subunit assembly by the hydrophilic amino-terminal domain of the Shaker potassium channel. Science. 1992;257:1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- Lien CC, Jonas P. Kv3 potassium conductance is necessary and kinetically optimized for high-frequency action potential generation in hippocampal interneurons. J Neurosci. 2003;23:2058–2068. doi: 10.1523/JNEUROSCI.23-06-02058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Losonczy A, Makara JK, Magee JC. Compartmentalized dendritic plasticity and input feature storage in neurons. Nature. 2008;452:436–441. doi: 10.1038/nature06725. [DOI] [PubMed] [Google Scholar]
- Macioce P, Gambara G, Bernassola M, Gaddini L, Torreri P, Macchia G, Ramoni C, Ceccarini M, Petrucci TC. Beta-dystrobrevin interacts directly with kinesin heavy chain in brain. J Cell Sci. 2003;116:4847–4856. doi: 10.1242/jcs.00805. [DOI] [PubMed] [Google Scholar]
- Martina M, Yao GL, Bean BP. Properties and functional role of voltage-dependent potassium channels in dendrites of rat cerebellar Purkinje neurons. J Neurosci. 2003;23:5698–5707. doi: 10.1523/JNEUROSCI.23-13-05698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor DL, Lin YF, Mobley BC, Avelar A, Jan YN, Jan LY, Berger JM. The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel. Cell. 2000;102:657–670. doi: 10.1016/s0092-8674(00)00088-x. [DOI] [PubMed] [Google Scholar]
- Nanao MH, Zhou W, Pfaffinger PJ, Choe S. Determining the basis of channel-tetramerization specificity by x-ray crystallography and a sequence-comparison algorithm: Family Values (FamVal) Proc Natl Acad Sci U S A. 2003;100:8670–8675. doi: 10.1073/pnas.1432840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong LL, Lim AP, Er CP, Kuznetsov SA, Yu H. Kinectin-kinesin binding domains and their effects on organelle motility. J Biol Chem. 2000;275:32854–32860. doi: 10.1074/jbc.M005650200. [DOI] [PubMed] [Google Scholar]
- Perney TM, Marshall J, Martin KA, Hockfield S, Kaczmarek LK. Expression of the mRNAs for the Kv3.1 potassium channel gene in the adult and developing rat brain. J Neurophysiol. 1992;68:756–766. doi: 10.1152/jn.1992.68.3.756. [DOI] [PubMed] [Google Scholar]
- Rivera J, Chu PJ, Lewis TL, Jr, Arnold DB. The role of Kif5B in axonal localization of Kv1 K+ channels. Eur J Neurosci. 2007;25:136–146. doi: 10.1111/j.1460-9568.2006.05277.x. [DOI] [PubMed] [Google Scholar]
- Rudy B, McBain CJ. Kv3 channels: voltage-gated K+ channels designed for high-frequency repetitive firing. Trends Neurosci. 2001;24:517–526. doi: 10.1016/s0166-2236(00)01892-0. [DOI] [PubMed] [Google Scholar]
- Schulteis CT, Nagaya N, Papazian DM. Subunit folding and assembly steps are interspersed during Shaker potassium channel biogenesis. J Biol Chem. 1998;273:26210–26217. doi: 10.1074/jbc.273.40.26210. [DOI] [PubMed] [Google Scholar]
- Seeger MA, Rice SE. Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J Biol Chem. 2010;285:8155–8162. doi: 10.1074/jbc.M109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417:83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- Shen NV, Pfaffinger PJ. Molecular recognition and assembly sequences involved in the subfamily-specific assembly of voltage-gated K+ channel subunit proteins. Neuron. 1995;14:625–633. doi: 10.1016/0896-6273(95)90319-4. [DOI] [PubMed] [Google Scholar]
- Shen NV, Chen X, Boyer MM, Pfaffinger PJ. Deletion analysis of K+ channel assembly. Neuron. 1993;11:67–76. doi: 10.1016/0896-6273(93)90271-r. [DOI] [PubMed] [Google Scholar]
- Song AH, Wang D, Chen G, Li Y, Luo J, Duan S, Poo MM. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 2009;136:1148–1160. doi: 10.1016/j.cell.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- Strang C, Kunjilwar K, DeRubeis D, Peterson D, Pfaffinger PJ. The role of Zn2+ in Shal voltage-gated potassium channel formation. J Biol Chem. 2003;278:31361–31371. doi: 10.1074/jbc.M304268200. [DOI] [PubMed] [Google Scholar]
- Tu L, Santarelli V, Sheng Z, Skach W, Pain D, Deutsch C. Voltage-gated K+ channels contain multiple intersubunit association sites. J Biol Chem. 1996;271:18904–18911. doi: 10.1074/jbc.271.31.18904. [DOI] [PubMed] [Google Scholar]
- Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL, MacAskill AF, Rostaing P, Lumb MJ, Humbert S, Triller A, Saudou F, Yan Z, Kittler JT. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron. 2010;65:53–65. doi: 10.1016/j.neuron.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher H, Mohapatra DP, Trimmer JS. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev. 2008;88:1407–1447. doi: 10.1152/physrev.00002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDongen AM, Frech GC, Drewe JA, Joho RH, Brown AM. Alteration and restoration of K+ channel function by deletions at the N- and C-termini. Neuron. 1990;5:433–443. doi: 10.1016/0896-6273(90)90082-q. [DOI] [PubMed] [Google Scholar]
- Weiser M, Bueno E, Sekirnjak C, Martone ME, Baker H, Hillman D, Chen S, Thornhill W, Ellisman M, Rudy B. The potassium channel subunit KV3.1b is localized to somatic and axonal membranes of specific populations of CNS neurons. J Neurosci. 1995;15:4298–4314. doi: 10.1523/JNEUROSCI.15-06-04298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yu W, Jan YN, Jan LY, Li M. Assembly of voltage-gated potassium channels. Conserved hydrophilic motifs determine subfamily-specific interactions between the alpha-subunits. J Biol Chem. 1995;270:24761–24768. doi: 10.1074/jbc.270.42.24761. [DOI] [PubMed] [Google Scholar]
- Xu M, Cao R, Xiao R, Zhu MX, Gu C. The axon-dendrite targeting of Kv3 (Shaw) channels is determined by a targeting motif that associates with the T1 domain and ankyrin G. J Neurosci. 2007;27:14158–14170. doi: 10.1523/JNEUROSCI.3675-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadeh AD, Cheng Y, Xu H, Wong N, Wang Z, Goonasekara C, Steele DF, Fedida D. Kif5b is an essential forward trafficking motor for the Kv1.5 cardiac potassium channel. J Physiol. 2009;587:4565–4574. doi: 10.1113/jphysiol.2009.178442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N, Jan YN, Jan LY. An artificial tetramerization domain restores efficient assembly of functional Shaker channels lacking T1. Proc Natl Acad Sci U S A. 2000;97:3591–3595. doi: 10.1073/pnas.060016797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhang CL, Messing A, Chiu SY. Temperature-sensitive neuromuscular transmission in Kv1.1 null mice: role of potassium channels under the myelin sheath in young nerves. J Neurosci. 1998;18:7200–7215. doi: 10.1523/JNEUROSCI.18-18-07200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]