Abstract
IGF and EGF regulate various physiological and pathological processes. IGF binding protein (IGFBP)-3 regulates cell proliferation in IGF-dependent and -independent fashions. Recently, we identified IGFBP-3 as a novel EGF receptor (EGFR) downstream target molecule in primary and immortalized human esophageal epithelial cells, suggesting an interplay between the EGF and IGF signaling pathways. However, the regulatory mechanisms for IGFBP-3 expression and its functional role in esophageal cell proliferation remain to be elucidated. Herein, we report that IGFBP-3 mRNA and protein were induced upon growth factor deprivation in primary and immortalized human esophageal cells through mechanisms requiring p53-independent de novo mRNA transcription and protein synthesis. This occurred in the face of the activated phosphatidylinositol 3-OH-kinase (PI3K)/mammalian target of rapamycin (mTOR) pathway. Secreted IGFBP-3 neutralized IGFs and prevented IGF-I receptor (IGF-IR) activation. In contrast, EGF suppressed IGFBP-3 mRNA and protein expression through activation of MAPK in an EGFR-tyrosine kinase-dependent manner to restore the cellular response to IGF-I. When stably overexpressed, wild-type IGFBP-3 but not I56G/L80G/L81G (GGG) mutant IGFBP-3, which has a reduced affinity to IGFs, prevented IGF-I from activating IGF-IR and Akt as well as stimulating cell proliferation. However, unlike other cell types where IGFBP-3 exerts antiproliferative effects, neither wild-type nor GGG mutant IGFBP-3 alone affected cell proliferation or EGFR activity. These results indicate that IGF signaling is subject to negative regulation through IGFBP-3 and positive regulation by EGF, the latter of which suppresses IGFBP-3. This provides a platform for understanding the novel cross talk between EGF- and IGF-mediated pathways.
Keywords: insulin-like growth factor binding protein-3, epidermal growth factor receptor, mammalian target of rapamycin, esophageal epithelial cells
Homeostasis of the stratified squamous epithelium of the esophagus is maintained by epithelial renewal involving dynamic biological processes including cell proliferation, migration, differentiation, and apoptosis, which are thought subject to regulation by various polypeptide growth factors such as EGF and IGF-I. They play a critical role in pathological processes such as carcinogenesis and wound healing. In addition to individual or differential actions, EGF and IGF-I synergistically act toward esophageal epithelial cells and other cell types (42, 49). In mouse embryonic fibroblasts, a functional IGF-I receptor (IGF-IR) is required for EGF receptor (EGFR) to exert its mitogenic and transforming activities (12). However, the molecular mechanisms underlying the cross talk between their signal transduction pathways remain to be elucidated.
EGFR tyrosine kinase is a critical oncogene implicated in the early stage of esophageal carcinogenesis. EGFR overexpression is observed frequently in esophageal cancer (31, 62). In primary and immortalized esophageal epithelial cells, EGFR overexpression leads to unique biological phenotypes, including basal cell hyperplasia in organotypic culture and corroborated in transgenic mice (1). We have recently identified IGF binding protein (IGFBP)-3 as a gene that is most highly upregulated in EGFR-overexpressing esophageal epithelial cells (53).
IGFBP-3 is a glycoprotein found in the circulation as a component of the 150-kDa ternary complex, comprising an 85-kDa acid labile glycoprotein subunit and IGF-I or IGF-II, and serves as a major carrier protein for IGFs (17, 30). IGFBP-3 is secreted by many cell types including fibroblasts, endothelial cells, and epithelial cells (19, 23, 59). While the full-length form of IGFBP-3 (43–45 kDa) has an affinity to IGFs as high as IGF-IR, it undergoes cleavage mediated by proteases such as matrix metalloproteinases, cathepsin L, and plasmin, resulting in a decrease in the affinity for IGFs and enhancement of the availability of IGFs to the cells (17). The bioactivity of IGFBP-3 may also be regulated through other posttranslational modifications, such as glycosylation and phosphorylation (17). IGFBP-3 expression is regulated by many agents and factors, including peptide growth factors, cytokines, hormones, vitamins, minerals, and chemotherapeutic agents (17, 25). IGFBP-3 is also found to be induced upon cellular senescence or mitotic quiescence (19, 23, 45). However, the mechanisms involved in the regulation of IGFBP-3 mRNA and protein expression are unknown. The antiproliferative or proapoptotic effects of IGFBP-3 have been implicated in cell growth inhibition induced by several antiproliferative agents through both IGF-dependent and -independent mechanisms in vitro. In contrast, IGFBP-3 may stimulate cell proliferation under certain circumstances (17). However, it remains to be determined how IGFBP-3 affects esophageal cell proliferation.
To gain insights into the role of IGFBP-3 in the regulation of human esophageal epithelial cell proliferation, we determined how IGFBP-3 is regulated in primary and immortalized human esophageal cells. We found that IGFBP-3 is subject to negative regulation by EGF. Whereas IGFBP-3 overexpression facilitated cell growth inhibition upon growth factor deprivation by reducing IGF-IR sensitivity to IGF, EGF restored it by reducing the IGFBP-3 level.
MATERIALS AND METHODS
Growth factors and pharmacological inhibitors
All chemicals and inhibitors were purchased from Sigma Chemical (St. Louis, MO) unless otherwise noted. Actinomycin D (Act D) was dissolved in methanol. Cycloheximide (CHX), recombinant human EGF, recombinant human insulin, and recombinant human IGF-I (Invitrogen; Carlsbad, CA) were dissolved in double-distilled water. Recombinant human Des(1–3)-IGF-I (Peninsula Laboratories; San Carlos, CA), recombinant human long R3-IGF-I (Upstate Biotechnology; Lake Placid, NY), and recombinant human IGFBP-3 (Upstate Biotechnology) were reconstituted in 10 mM acetic acid. AG-1478 (Calbiochem; San Diego, CA), PD-98059 (Cell Signaling Technology; Beverly, MA), U-0126 (Cell Signaling Technology), LY-294002 (Calbiochem), and rapamycin (Calbiochem) were dissolved in DMSO.
Culture of primary and immortalized human esophageal cells
Primary human esophageal keratinocytes EPC1 and EPC2 as well as immortalized EPC2 derivertives (EPC2-hTERT, EPC2-hTERT-puro, and EPC2-hTERT-p53R175H) have been described previously (1, 28, 54). Cells were counted using a Coulter Z1 Counter (Beckman) and grown at 37°C under 5% CO2 in keratinocyte serum-free medium (KSFM) containing 1 ng/ml EGF, 5 µg/ml insulin, 150 µg/ml bovine pituitary extract (BPE), 6.7 ng/ml triiodithyronine, and 74 ng/ml hydrocortisone (Invitrogen). For growth factor deprivation, cells were rinsed twice with Dulbecco’s PBS (DPBS) without calcium chloride and magnesium chloride and exposed to keratinocyte basal medium (KBM; Bio Whittaker; Walkersville, MD), which is devoid of the growth factors and hormones contained in KSFM. Conditioned medium (CM) was harvested by incubating subconfluent cells for 24 h with KSFM or KBM. To concentrate the CM, Centricon YM-10 Centrifugal Filter Units (10,000 nominal mol. wt. limit, Millipore; Billerica, MA) were used according to the manufacturer’s instructions.
Retrovirus-mediated transduction of mutant Ras and wild-type and mutant IGFBP-3
Stable transduction of esophageal cells with retroviral vectors has been described previously (54). cDNAs encoding dominant negative Ha-RasN17 (dnRasN17) and wild-type and mutant IGFBP-3 were isolated by PCR using Ha-RasN17 plasmids (a gift of Dr. Timothy C. Wang), pcDNA3-hIGFBP-3 (a gift of Dr. Adda Grimberg), and pCMV-hIGFBP-3-GGG (a gift of Dr. Liam J. Murphy) as templates and subcloned into the pBabe-puro retroviral vector (39) at its BamHI and EcoRI sites, resulting in the creation of pBABE-puro-HaRasN17, pBabe-puro-hIGFBP-3, and pBabe-puro-hIGFBP-3-GGG. The inserted region of the constructs was verified by DNA sequencing. Oncogenic Ras (Ha-RasV12) (46) and Ras effector loop mutants (Ha-RasV12/S35, Ha-RasV12/G37, and Ha-RasV12/C40) (32, 58) expressed using the pBabe-Puro vector are gifts of Dr. Scott W. Lowe. They were transfected into a Phoenix-Ampho packaging cell line (a gift of Dr. Garry Nolan) with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Culture supernatants from individual Phoenix-Ampho cells were used to infect EPC1, EPC2, and EPC2-hTERT cells. Cells were passaged 48 h after infection and selected with 0.5 µg/ml puromycin (Invitrogen) for 5 days. Selection was not carried out for cells transduced with mutant Ras because they were subjected to Western blot analysis within 48 h after retrovirus infection.
Real-time RT-PCR
IGFBP-3 mRNA was determined by real-time RT-PCR as described previously (53). Briefly, total RNA was isolated from cells with the RNeasy Mini Kit (Qiagen; Valencia, CA), and cDNA was synthesized with the Superscript First Strand Synthesis System (Invitrogen). Real-time PCR was performed using the SYBR green reagent (PE Applied Biosystems; Foster City, CA) and the ABI PRISM 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer’s instructions. GAPDH was used as an internal control. All PCRs were performed in triplicate. The relative expression level of IGFBP-3 mRNA was calculated by normalizing it to the GAPDH mRNA expression level. Data were analyzed using ABI PRISM 7000 sequence detection system software (Applied Biosystems).
Western blot analysis
Western blot analysis was carried out as described previously (1, 53, 54). In brief, cell lysates were denatured and fractionated on a NuPAGE Bis-Tris 4–12% gel using the Nu-PAGE System (Invitrogen) and electrotransferred to a polyvinylidene difluoride membrane (Immobilon-P, Millipore). The membrane was incubated with primary antibody [affinity-purified goat anti-human IGFBP-3 (DSL-R00536, Diagnostic Systems Laboratories; Webster, TX), anti-β-actin (Sigma Chemical), anti-human p53 (DO-1) mouse monoclonal antibody (Oncogene Research Products; San Diego, CA), anti-phospho-IGF-IR (Tyr1135/1136) rabbit monoclonal antibody (19H7, no. 3024, Cell Signaling Technology), affinity-purified rabbit anti-human IGF-IRβ (C-20, sc-713, Santa Cruz Biotechnology; Santa Cruz, CA), affinity-purified rabbit anti-phospho-Akt (Ser473) antibody (no. 9271, Cell Signaling Technology), affinity-purified rabbit anti-Akt antibody (no. 9272, Cell Signaling Technology), anti-phospho-EGFR mouse monoclonal antibody (Tyr1068, 1H12, no. 2236, Cell Signaling Technology), anti-EGFR Ab-12 (cocktail R19/48) mouse monoclonal antibody (NeoMarkers; Union City, CA), anti-Ras mouse monoclonal antibody (RAS10, Upstate Biotechnology), anti-pRB mouse monoclonal antibody (G3–245, BD Pharmingen; San Diego, CA), affinity-purified rabbit anti-phospho-checkpoint kinase 1 (Chk1; Ser317) antibody (no. 2344, Cell Signaling Technology), affinity-purified rabbit anti-phospho-Chk1 (Ser345) antibody (no. 2341, Cell Signaling Technology), or affinity-purified rabbit anti-phospho-Chk2 (Thr68) antibody (no. 2661, Cell Signaling Technology)]. The signal was detected by horseradish peroxidase-conjugated secondary antibody [donkey anti-goat IgG (sc-2020, Santa Cruz Biotechnology), donkey anti-rabbit IgG (Amersham Biosciences), or sheep anti-mouse IgG (Amersham Biosciences)], visualized by an enhanced chemiluminescence solution (ECL Plus; Amersham Biosciences Pharmacia Biotech), and exposed to X-Omat LS film (Eastman Kodak; New York, NY).
Western ligand blot analysis
Western ligand blot analysis was done as described previously (53). In brief, 10 µl of 10 times concentrated CM were electrophoresed on a NuPAGE Bis-Tris 4–12% gel under nonreduced conditions and transferred to an Immobilon-P membrane. After being blocked, the membrane was incubated with 0.02 µCi of 3-[125I]iodotyrosyl recombinant human IGF-I or IGF-II (Amersham Biosciences) per centimeter squared of membrane for 16 h at 4°C. The membrane was subjected to autoradiography using Kodak X-OMAT AR film with an intensifying screen at −80°C.
ELISA
ELISA was performed using the Human IGFBP-3 DuoSet ELISA Development System (R&D Systems; Minneapolis, MN) as described previously (53).
[3H]thymidine incorporation assay
To assess cell proliferation, DNA synthesis was measured by a [3H]thymidine incorporation assay as described previously (53, 54). In brief, cells were incubated with 1 µCi of [methyl-3H]thymidine (1 µCi•ml−1•well−1, Perkin Elmer Life Sciences; Boston, MA) for 4 h in 12-well tissue culture dishes before cells were harvested. Cell lysates were filtrated with glass microfiber filters (Whatman; Kent, ME), and the radioactivity was measured with a LS 6500 Multi-Purpose Scintillation Counter (Beckman Coulter).
Statistical analyses
Student’s t-test was used to compare data between two groups. Data represent means ± SE. P < 0.05 was considered to be statistically significant.
RESULTS
IGFBP-3 is induced upon growth factor deprivation in primary and immortalized human esophageal cells
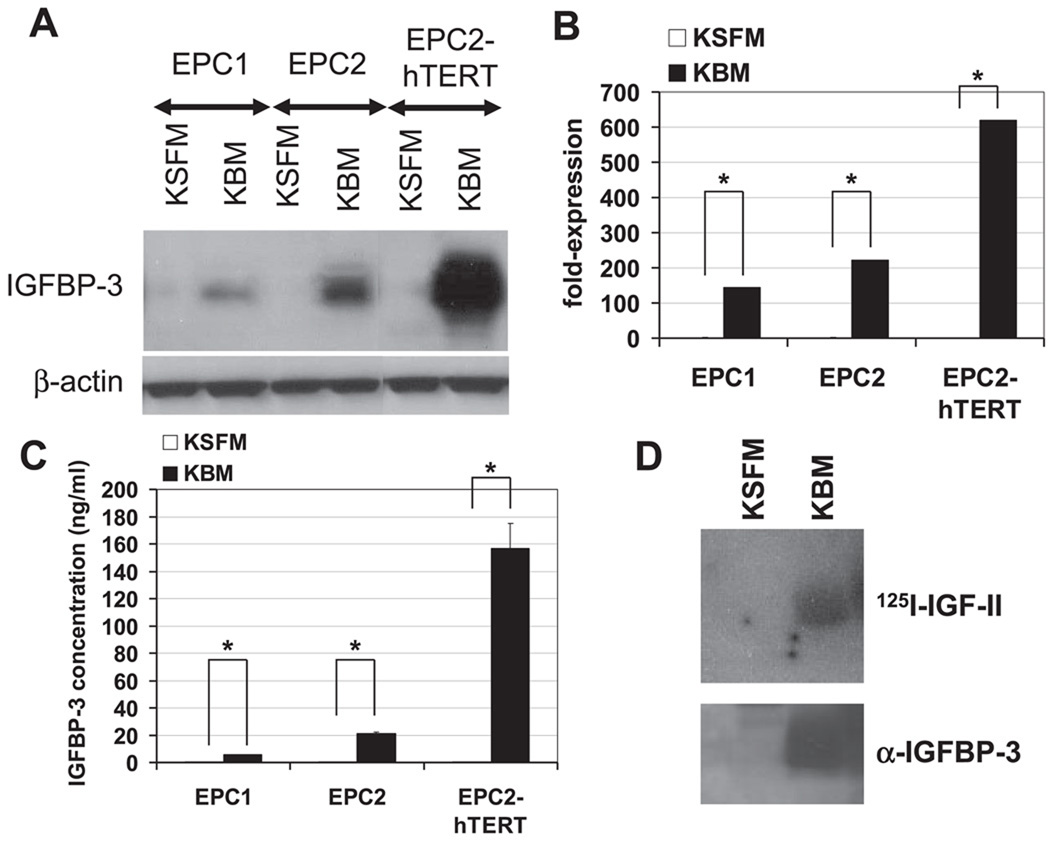
To study the role of IGFBP-3 in the regulation of esophageal cell proliferation, we first determined the expression of IGFBP-3 in standard culture as well as growth factor-deprived conditions. Cellular IGFBP-3 was barely detectable by Western blot analysis in EPC1, EPC2, and EPC2-hTERT cells when they were grown in KSFM containing growth factors (Fig. 1A). ELISA detected only a basal level of IGFBP-3 (1.5–2.4 ng/ml) in the CM (Fig. 1C and Table 1). In contrast, a marked induction of IGFBP-3 mRNA and protein was observed by real-time RT-PCR, Western blot analysis, and ELISA in these cells and their CM within 24 h after growth factor deprivation with KBM, which is devoid of growth factors (Fig. 1). Interestingly, the most potent induction of IGFBP-3 was found in EPC2-hTERT cells, an immortalized cell line. Western ligand blot analysis detected a single band with an apparent molecular mass of 45 kDa as the major IGF binding activity expressed in the CM, which was also confirmed as IGFBP-3 by Western blot analysis (Fig. 1D).
Fig. 1.
IGF binding protein (IGFBP)-3 is induced upon growth factor (GF) deprivation in primary and immortalized human esophageal cells. Subconfluent cells were incubated with either keratinocyte serum-free medium (KSFM) or keratinocyte basal medium (KBM) for 24 h before cells and condition media (CM) were harvested to determine the IGFBP-3 level. A: IGFBP-3 in cell lysates was detected by Western blot analysis. β-Actin was used as a loading control. B: IGFBP-3 mRNA expression was determined by real-time RT-PCR. The normalized IGFBP-3 mRNA level determined in cells in KSFM was set to 1. Means ± SE (n = 3 independent experiments) in a representative experiment are shown. C: IGFBP-3 concentration in CM was measured by ELISA. Protein yields in cell lysates prepared simultaneously were used to adjust the differences in cell number that may affect IGFBP-3 concentration in CM. Means ± SE (n = 3 independent experiments) in a representative experiment are shown. D: IGF binding activity of IGFBP-3 secreted in CM of EPC2-hTERT cells was assessed by Western ligand blot analysis. The blot was probed with 125I-labeled IGF-II (125I-IGF-II). The blot was reprobed with anti-IGFBP-3 to identify the IGF binding activity detected by the radiolabeled ligand.
Table 1.
IGFBP-3 levels in CM
| Cells | IGFBP-3, ng/ml | |
|---|---|---|
| EPC1 | ||
| Parental | 1.6±0.0 | |
| Puro | 1.7±0.0 | |
| IGFBP-3 (WT) | 1,562.9±5.5 | |
| EPC2 | ||
| Parental | 1.5±0.1 | |
| Puro | 1.3±0.2 | |
| IGFBP-3 (WT) | 1,606.1±21.0 | |
| hTERT | ||
| Parental | 2.4±0.0 | |
| Puro | 9.4±0.2 | |
| IGFBP-3 (WT) | 1,951.2±78.6 | |
| IGFBP-3-GGG (MT) | 2,194.8±95.9 |
Means ± SE (n = 3) in a representative experiment are shown. The IGF binding protein (IGFBP)-3 concentration in, conditioned media (CM) from EPC1, EPC2, and EPC2-hTERT cells transduced with empty vector (puro), wild-type IGFBP-3 [IGFBP-3 (WT)], and GGG mutant IGFBP-3 [IGFBP-3-GGG (MT)] was measured by ELISA. Protein yields in cell lysates prepared simulataneously were used to adjust the differences in cell number that may affect IGFBP-3 concentration in CM.
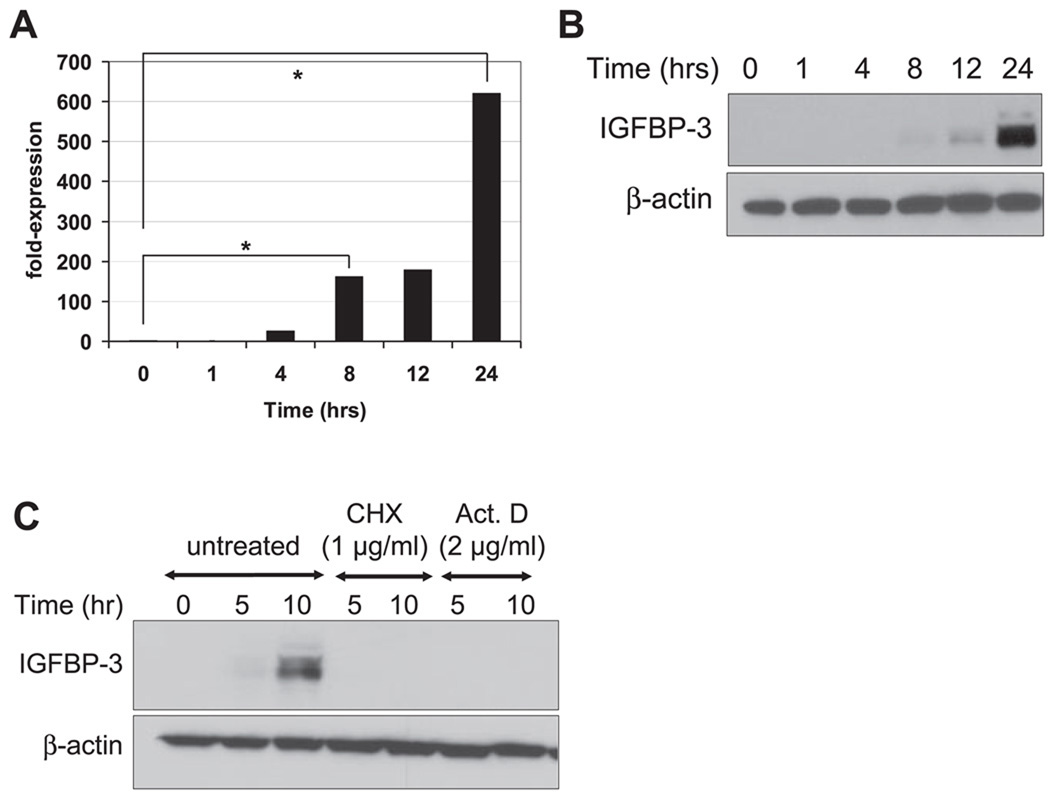
After growth factor deprivation, IGFBP-3 expression was detectable by 8 h and peaked at 24 h in EPC2-hTERT cells (Fig. 2, A and B). The induction of IGFBP-3 mRNA (Fig. 2A) was found to precede the induction of IGFBP-3 protein (Fig. 2B). The induction of IGFBP-3 appeared to require de novo transcription of IGFBP-3 mRNA and protein synthesis because it was completely blocked in EPC2-hTERT cells treated with Act D, a RNA transcription inhibitor, and CHX, a protein translation inhibitor, respectively (Fig. 2C).
Fig. 2.
IGFBP-3 induction upon GF deprivation requires de novo mRNA transcription and protein synthesis. EPC2-hTERT cells were subjected to GF deprivation with KBM for indicated the time periods before cell harvest. A: IGFBP-3 mRNA level was determined by real-time RT-PCR. The normalized IGFBP-3 mRNA level determined at time 0 was set to 1. Means ± SE (n = 3 independent experiments) in a representative experiment are shown. B and C: IGFBP-3 in cell lysates was detected by Western blot analysis. β-Actin was used as a loading control. For C, GF deprivation was carried out in the presence of cycloheximide (CHX) or actinomycin D (Act D).
Growth factor withdrawal resulted in a redistribution of the cell cycle in EPC2-hTERT cells, with an increase in the G0/G1 fraction by 9–25% and a decrease in the S phase fraction by 10–18%, and DNA synthesis was reduced by 70–80% within 24 h (Table 2 and data not shown). Apoptosis did not occur for at least 24 h in the growth factor-deprived condition because cleavage of caspase 3, a sub-G1 fraction, and nuclear chromatin condensation were not observed in EPC1, EPC2, and EPC2-hTERT cells exposed to KBM (data not shown).
Table 2.
Cell cycle analysis
| Phase |
|||
|---|---|---|---|
| G0/G1, % | G2,% | S, % | |
| Parental | |||
| KSFM | 78.7±0.2 | 4.3±0.1 | 17.0±0.1 |
| KBM | 88.6±0.1* | 4.6±0.1 | 6.8±0.0* |
| KBM + IGF-I | 73.3±0.2† | 4.8±0.2 | 22.0±0.0† |
| Puro | |||
| KSFM | 77.4±0.2 | 4.2±0.0 | 18.3±0.2 |
| KBM | 86.4±0.1* | 4.0±0.1 | 9.7±0.0* |
| KBM + IGF-I | 77.4±0.3† | 5.1±0.2 | 17.4±0.2† |
| IGFBP-3 (WT) | |||
| KSFM | 80.9±0.2 | 4.9±0.1 | 14.2±0.1 |
| KBM | 93.2±0.2* | 3.7±0.1 | 3.1 ±0.0* |
| KBM + IGF-I | 93.6±0.1 | 3.9±0.1 | 2.5±0.0 |
| IGFBP-3-GGG (MT) | |||
| KSFM | 78.0±0.1 | 5.7±0.1 | 16.3±0.1 |
| KBM | 88.7±0.1* | 4.7±0.1 | 6.6±0.0* |
| KBM + IGF-I | 76.0±0.1† | 5.4±0.1 | 18.6±0.1† |
Means ± SD (n = 3) in a representative experiment are shown. The EPC2-hTERT parental cell line and its derivatives stably transduced with empty vector, IGFBP-3 (WT), and IGFBP-3GGG (MT) were grown in keratinocyte serum-free medium (KSFM) or subjected to growth factor deprivation with keratinocyte basal medium (KBM) in the absence or presence of 100 ng/ml IGF-I for 24 h before cell harvest.
Comparison between KSFM and KBM (P < 0.0001);
comparison between KBM and KBM + IGF-I (P < 0.0001).
These results suggest that growth factors in standard media suppress IGFBP-3 expression in esophageal epithelial cells, whereas IGFBP-3 may negatively modulate their mitogenic effects.
IGFBP-3 induction upon growth factor deprivation does not depend on p53-mediated transcriptional activity in human esophageal cells
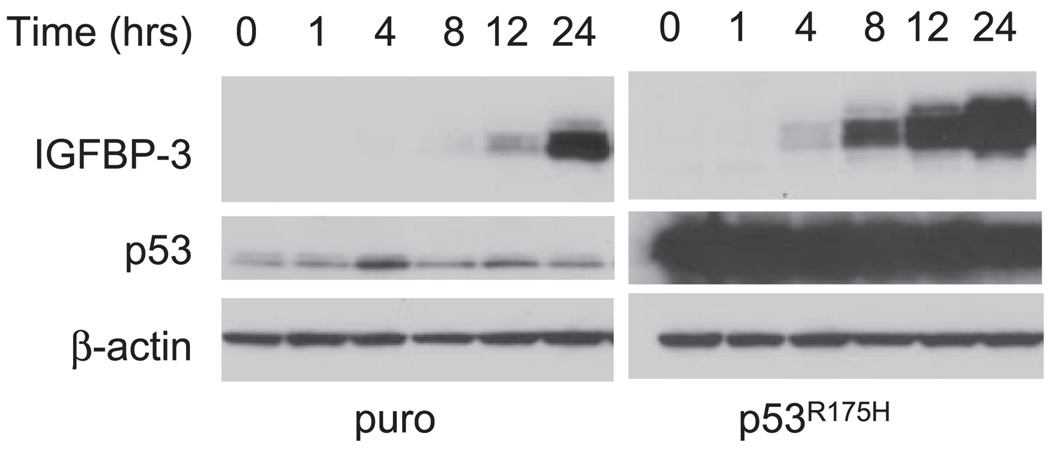
p53 tumor suppressor protein is known to transactivate IGFBP-3 gene expression through its cis-regulatory elements, which are identified in the proximal promoter region as well as in an intron of the IGFBP-3 gene (6, 8). It is possible that growth factor deprivation leads to p53 activation and induces IGFBP-3 and cell growth inhibition. We have previously documented that the p53 gene is wild type and activated by genotoxic stresses in EPC2 and EPC2-hTERT cells (28). We have further established that their derivatives stably express dominant negative mutant p53R175H. Mutant p53 proteins at this amino acid residue lack the ability to transactivate the IGFBP-3-derived promoter (44). As shown in Fig. 3, mutant p53 is highly expressed in EPC2-hTERT-p53R175H cells compared with the basal level of wild-type p53 expression observed in the control cell line (EPC2-hTERT-puro). The induction of p53 target genes such as p21WAF1/CIP1 and Bax is functionally abrogated in EPC2-hTERT-p53R175H cells, as described previously (54). We examined whether the p53 status affects IGFBP-3 induction in these cell lines. Growth factor deprivation resulted in the induction of IGFBP-3 regardless of the p53 status in these cell lines, although a mild elevation of p53 was transiently observed in the control cells at 4 h after growth factor deprivation (Fig. 3). These results suggest that IGFBP-3 can be induced in a wild-type p53-independent fashion upon growth factor deprivation in human esophageal cells but does not preclude the established role of wild-type p53 in the induction of IGFBP-3 in normal serum. Interestingly, IGFBP-3 was induced at an earlier time point in EPC2-hTERT-p53R175H cells than control cells (Fig. 3), suggesting the possibility that the mutant p53 protein exerted not only a “dominant negative” effect upon wild-type p53 but also a “gain of function” effect to enhance IGFBP-3 expression.
Fig. 3.
p53 status does not affect IGFBP-3 induction by GF deprivation. Subconfluent EPC2-hTERT cells expressing the dominant negative form of p53 (p53R175H) or empty vector-transduced control (puro) cells were subjected to GF deprivation with KBM for the indicated time periods. Expression of IGFBP-3 and p53 in cell lysates was determined by Western blot analysis. β-Actin was used as a loading control. This result was reproducible in at least 2 independent experiments.
The MAPK and PI3K–mTOR pathways regulate IGFBP-3 expression in human esophageal cells
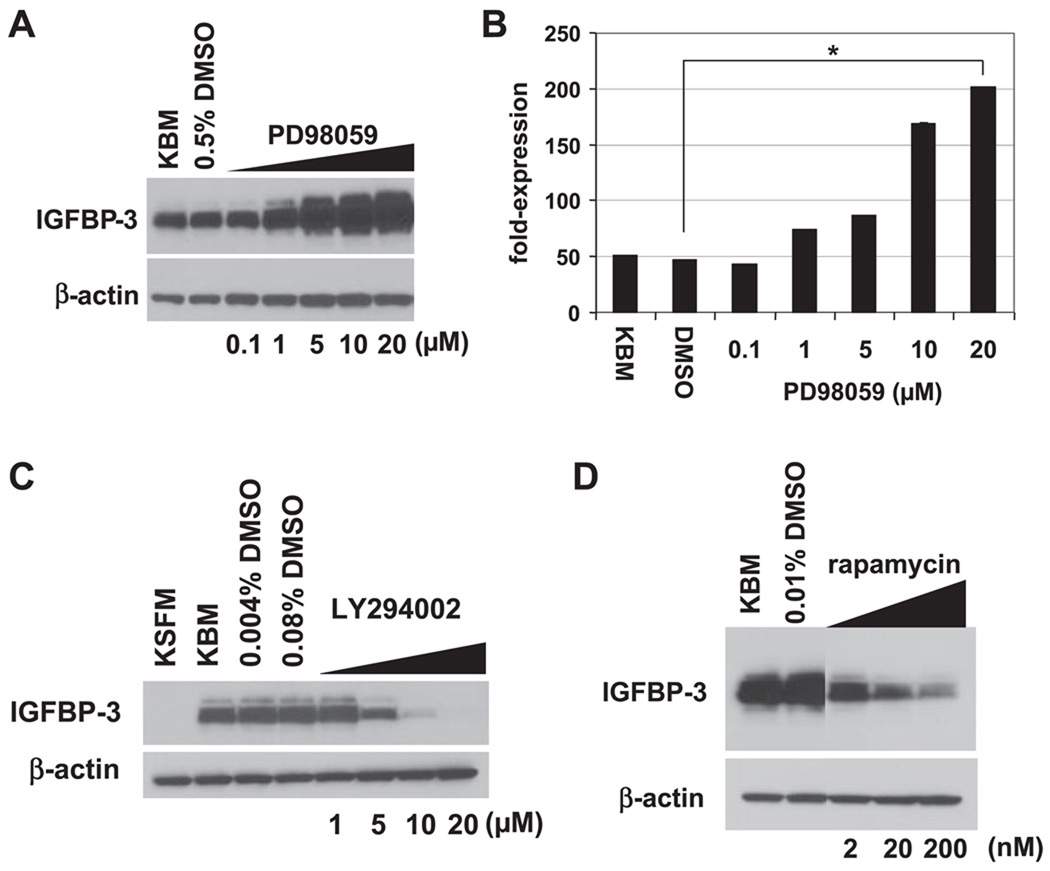
To identify the key regulatory pathways contributing to IGFBP-3 induction upon growth factor deprivation, we employed pharmacological inhibitors. First, we treated EPC2-hTERT cells with PD-98059, a MEK1 inhibitor, because a basal level of MAPK activity was detectable for at least 24 h during growth factor deprivation (data not shown). In these cells, PD-98059 dose dependently enhanced the induction of IGFBP-3 mRNA and protein in KBM (Fig. 4, A and B). Moreover, U-0126, a MEK1/2 inhibitor, also greatly enhanced IGFBP-3 protein induction in KBM (see Fig. 7C). Thus MAPK activity may inhibit IGFBP-3.
Fig. 4.
IGFBP-3 induction is regulated through the MAPK and phosphatidylinositol 3-OH-kinase (PI3K)/ mammalian target of rapamycin (mTOR) pathways. Subconfluent EPC2-hTERT cells were incubated with KBM for 24 h in the absence or presence of indicated concentrations of PD-98059 (A and B), LY-294002 (C), or rapamycin (D). DMSO was used as a vehicle for these compounds at the indicated concentrations. Cell lysates were subjected to Western blot analysis to assess the expression of IGFBP-3 (A C, and D). β-Actin was used as a loading control. In B, IGFBP-3 mRNA expression was determined by real-time RT-PCR. The normalized IGFBP-3 mRNA level determined in cells treated with KSFM was set to 1. Means ± SE (n = 3 independent experiments) in a representative experiment are shown.
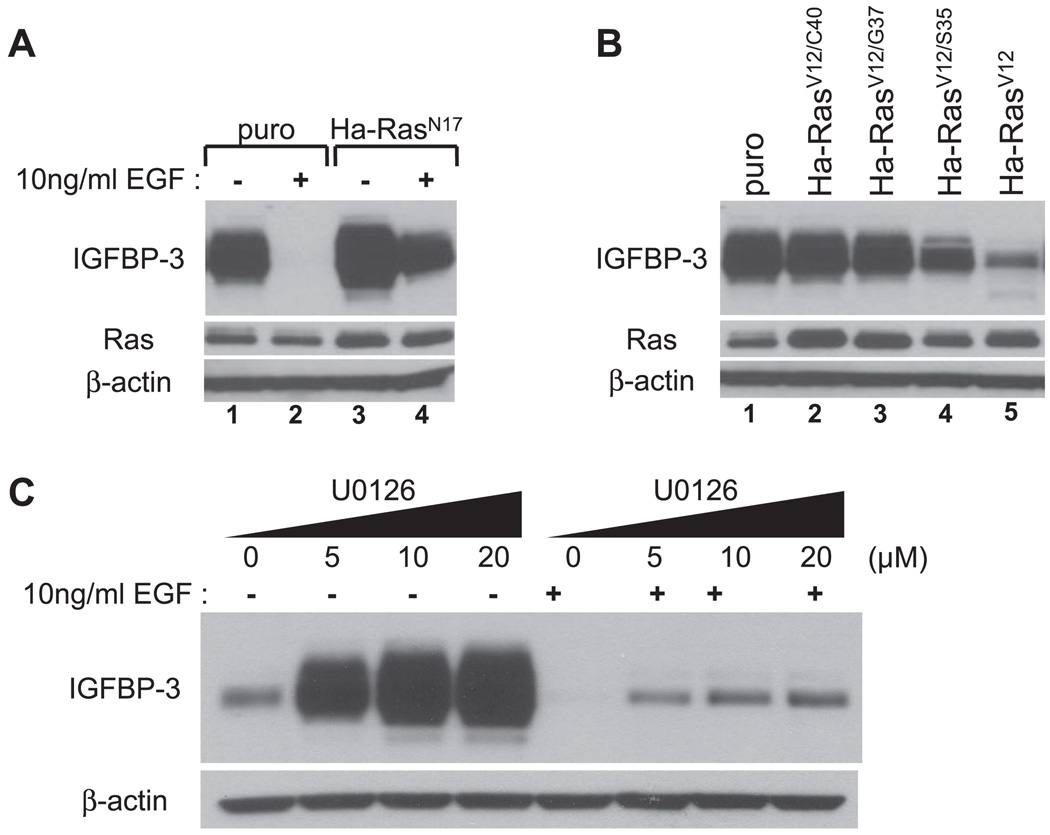
Fig. 7.
The Ras-MAPK pathway negatively regulates IGFBP-3. Subconfluent EPC2-hTERT cells were transduced with retrovirus expressing dominant negative Ha-RasN17 (A), constitutively active Ha-RasV12 and its effector loop mutants (Ha-RasV12/C40, Ha-RasV12/G37, or Ha-RasV12/S35) (B), or an empty vector (puro; A and B). Twenty-four hours after retrovirus infection, cells were subjected to GF deprivation with KBM for 24 h. In A, cells were incubated along with (+) or without (−) 10 ng/ml EGF. C: subconfluent EPC2-hTERT cells were incubated for 24 h with KBM in the absence or presence of indicated concentrations of U-0126 along with or without 10 ng/ml EGF. DMSO was used as a vehicle in the absence of U-0126. Cell lysates were subjected to Western blot analysis to assess the expression of IGFBP-3. β-Actin was used as a loading control. In A and B, total Ras was determined.
Because de novo protein synthesis is needed for IGFBP-3 induction (Fig. 2C), cells were treated with LY-294002, a PI3K inhibitor, and rapamycin, a mTOR inhibitor. Both agents inhibited the induction of IGFBP-3 protein upon growth factor deprivation in a dose-dependent fashion (Fig. 4, C and D), indicating that the PI3K/mTOR pathway may play a role in the translational regulation of IGFBP-3.
EGF suppresses IGFBP-3 and restores cellular response to IGF-I in growth factor-deprived human esophageal cells
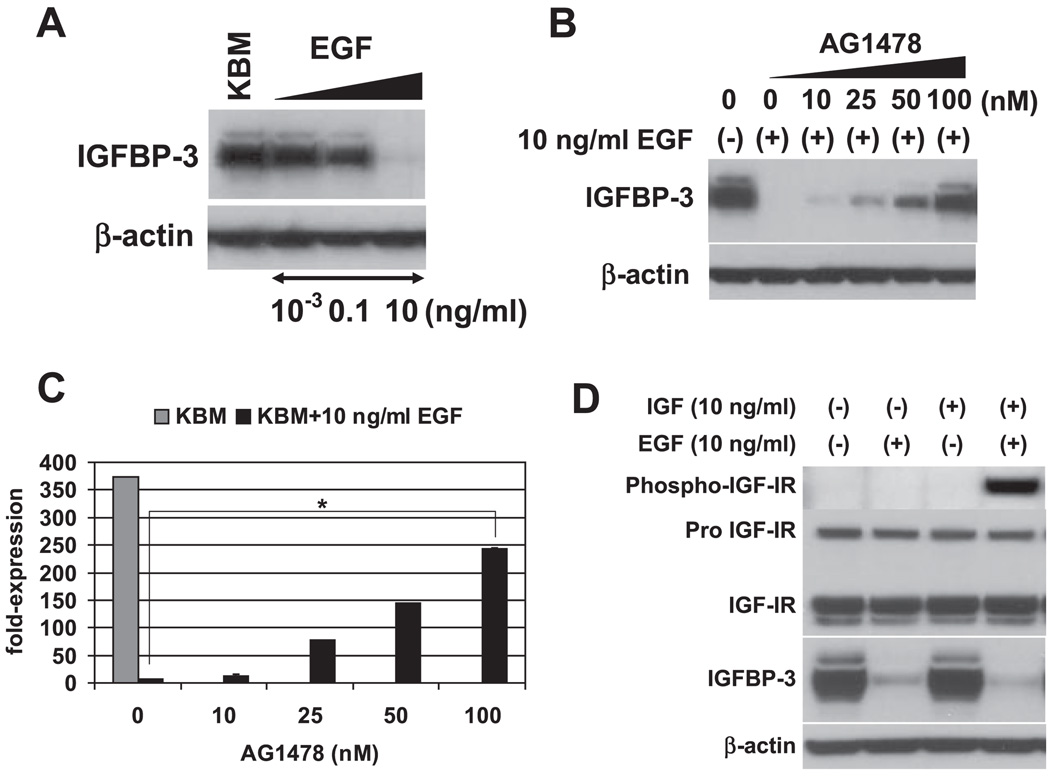
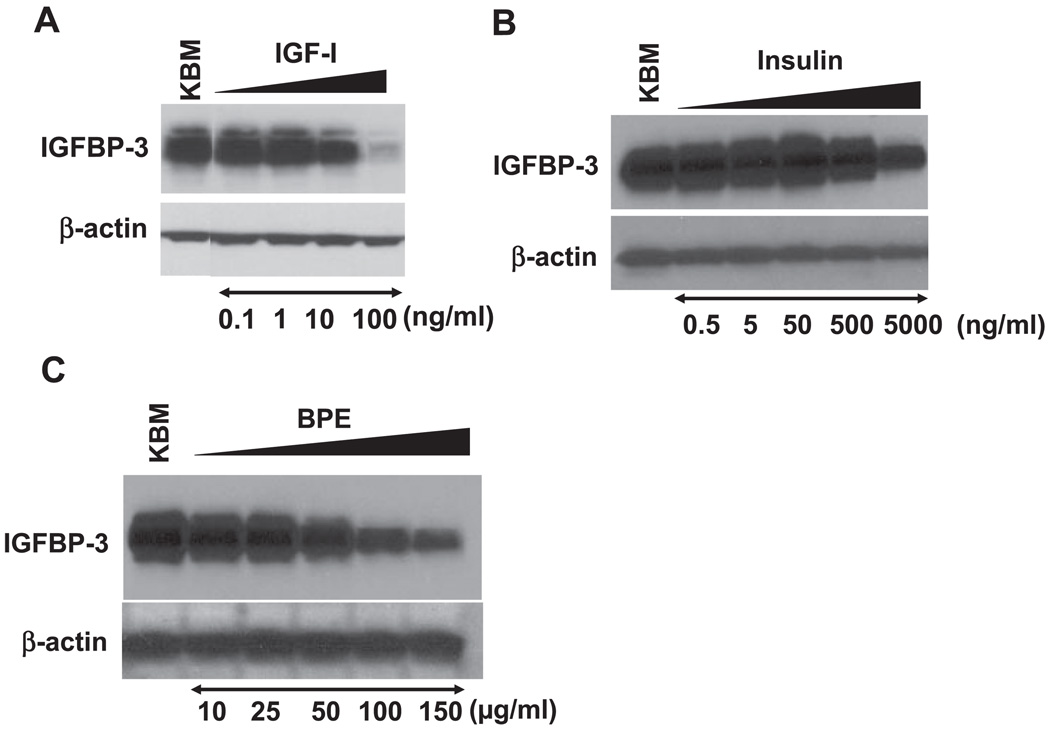
We next sought to determine the growth factor(s) that negatively regulates IGFBP-3 expression. Full KSFM contains 5 µg/ml insulin, 1 ng/ml EGF, and 150 µg/ml BPE. As shown in Figs. 5 and 6, all growth supplements added in KSFM dose dependently inhibited IGFBP-3 induction when individually added into KBM (starving medium). IGF-I also appeared to suppress IGFBP-3 in a dose-dependent fashion (Fig. 6A). These results show that multiple growth factors can inhibit IGFBP-3. It should be noted, however, that 10 ng/ml EGF, a physiologically relevant concentration, fully suppressed both IGFBP-3 mRNA and protein expression (Fig. 5, A–C), whereas high levels of insulin (5,000 ng/ml) failed to inhibit IGFBP-3 to a similar extent (Fig. 6B). IGF-I and BPE also appeared to be less potent than EGF in suppressing IGFBP-3 (Fig. 6, A and C), suggesting that EGF may be physiologically the more essential growth factor in negatively regulating IGFBP-3. To confirm the requirement of the receptor tyrosine kinase activity of EGFR for EGF-mediated inhibition of IGFBP-3 induction upon growth factor deprivation, EPC2-hTERT cells were treated with EGF in the presence of AG-1478, an EGFR-specific tyrosine kinase inhibitor. Whereas 10–100 ng/ml EGF fully suppressed IGFBP-3 induction at both mRNA and protein levels, AG-1478 antagonized such an effect in a dose-dependent fashion (Fig. 5, B and C, and data not shown). Thus EGFR activation contributes to inhibition of IGFBP-3 expression in growth factor-deprived conditions.
Fig. 5.
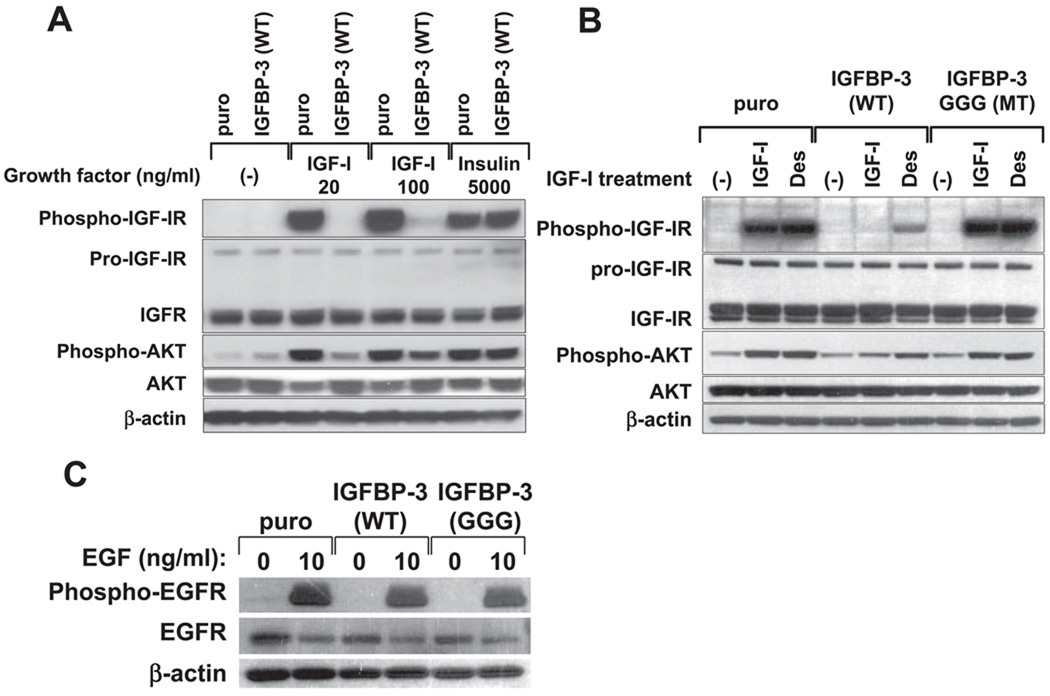
EGF inhibits IGFBP-3 induction through activation of EGF receptor (EGFR) tyrosine kinase and restores IGF-I receptor (IGF-IR) activation by IGF-I in GF-deprived cells. Subconfluent EPC2-hTERT cells were incubated with KBM for 24 h in the absence or presence of indicated concentrations of EGF before cell harvest. In B and C, cells were incubated along with the indicated concentrations of AG-1478, for which 0.001% DMSO was used as a vehicle. In D, cells were further stimulated with 10 ng/ml IGF-I for 30 min after the incubation period with KBM containing 0 or 10 ng/ml EGF. Cell lysates were subjected to Western blot analysis to assess the expression of IGFBP-3 (A B, and D) and activation of IGF-IR (Tyr1131). β-Actin and total IGF-IR were used as loading controls. In C, IGFBP-3 mRNA expression was determined by real-time RT-PCR. The normalized IGFBP-3 mRNA level determined in cells treated with 10 ng/ml EGF alone in KBM was set to 1. Means ± SE (n = 3 independent experiments) in a representative experiment are shown.
Fig. 6.
Multiple GFs suppress IGFBP-3 expression. Subconfluent EPC2-hTERT cells were subjected to GF deprivation in the absence (KBM) or presence of IGF-I (A), insulin (B), or bovine pituitary extract (BPE; C) at the indicated concentrations for 24 h before cell harvest. IGFBP-3 in cell lysates was detected by Western blot analysis. β-Actin was used as a loading control.
Because Ras is one of the prominent EGFR downstream effector molecules, dnRas (Ha-RasN17) was transduced in EPC2-hTERT cells to determine its role in the regulation of IGFBP-3. As shown in Fig. 7A, EGF-mediated IGFBP-3 inhibition (lane 2) was antagonized by Ha-RasN17 (lane 4), suggesting that EGF-induced Ras activity contributes to IGFBP-3 suppression. Interestingly, dnRas alone enhanced IGFBP-3 expression in growth factor-deprived conditions (Fig. 7A, lane 3), consistent with the above-described notion that MAPK may inhibit IGFBP-3 (Figs. 4, A and B, and 7C). To further determine how Ras affects IGFBP-3 expression, we also transduced the constitutively active form of oncogenic ras (Ha-RasV12) and its effector loop mutants in EPC2-hTERT cells. Ha-RasV12/S35, Ha-RasV12/G37, and Ha-RasV12/C40 can interact preferentially with Raf-1, Ral.GDS, and PI3K, respectively, thus allowing activation of the preferred Ras downstream pathways (32, 34, 58). When overexpressed, Ha-RasV12 suppressed IGFBP-3 induction in KBM growth factor deprivation medium (Fig. 7B, lane 5) compared with empty vector-transduced cells (Fig. 7B, lane 1). Among the effector loop mutants, Ha-RasV12/S35 but not Ha-RasV12/G37 or Ha-RasV12/C40 inhibited IGFBP-3 (Fig. 7B), suggesting that the Raf-1-mediated but not Ral.GDS-mediated or PI3K–mediated pathway may play a major role in IGFBP-3 suppression. Western blot analysis also demonstrated an elevated level of Ras expression in retrovirally transduced cells (Fig. 7B), indicating that the lack of IGFBP-3 suppression by Ha-RasV12/G37 and Ha-RasV12/C40 is not attributable to insufficient expression of these mutants. Furthermore, U-0126 partially but dose dependently prevented EGF from inhibiting IGFBP-3 (Fig. 7C). Such partial antagonism suggests the possibility that EGF may suppress IGFBP-3 through Ras-dependent as well as -independent pathways, which is in agreement with the incomplete antagonism by dnRas upon EGF suppression of IGFBP-3. Interestingly, PD-98059 (a MEK1 inhibitor) failed to demonstrate such antagonism (data not shown), suggesting that both MEK1 and MEK2 may contribute to IGFBP-3 inhibition. In addition, LY-294002 also failed to antagonize EGF inhibition of IGFBP-3 despite activation of Akt (data not shown), indicating the opposing roles in IGFBP-3 regulation between the PI3K–Akt and Ras-MAPK pathways.
We further determined how EGF-mediated suppression of IGFBP-3 influences the cellular response to IGF-I. As shown in Fig. 5D, IGF-I failed to activate IGF-IR in EPC2-hTERT cells preincubated with KBM without EGF, implying the neutralization of IGF-I by IGFBP-3 secreted in the CM upon growth factor deprivation. In contrast, IGF-I fully induced phosphorylation of IGF-IR in cells pretreated with KBM containing 10 ng/ml EGF, indicating that EGF restores the cellular response to IGF-I. Therefore, EGF may positively regulate the IGF signaling pathway through inhibition of IGFBP-3.
IGFBP-3 prevents IGF-IR activation by IGF-I in esophageal epithelial cells
To delineate the functional consequences of IGFBP-3 induction in human esophageal cells, we stably transduced wild-type and mutant IGFBP-3 in primary and immortalized human esophageal cells by retroviral vectors. Gly56/Gly80/Gly81 (GGG) mutant IGFBP-3 has three amino acids within the NH2-terminal region replaced with glycine residues, culminating in marked reduction of its affinity to IGFs, and thus provides an ideal approach to determine the IGF-independent biological functions of IGFBP-3 (9, 35, 47).
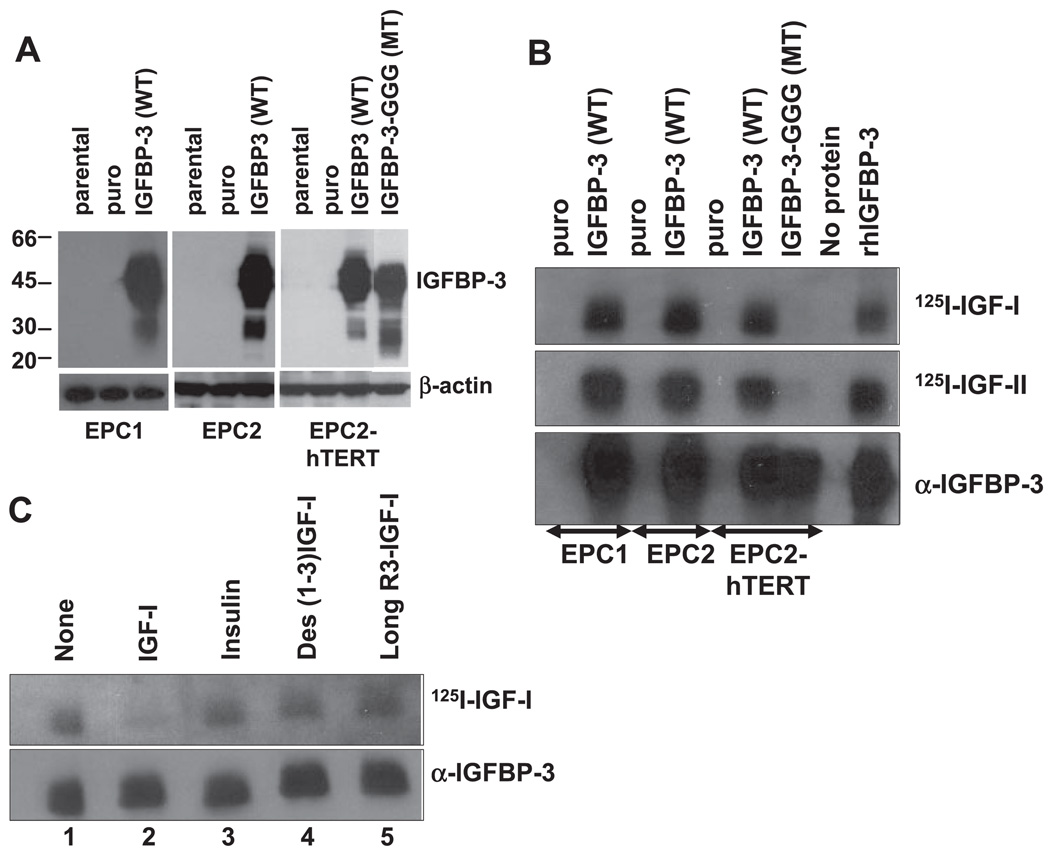
Western blot analysis confirmed IGFBP-3 overexpression in cell lysates as well as CM, whereas ELISA revealed elevated levels of IGFBP-3 secretion in CM of IGFBP-3 overexpressing EPC1, EPC2, and EPC2-hTERT cells compared with parental or empty vector-transduced cells (Fig. 8 and Table 1). Western ligand blot analysis further confirmed the IGF binding capacity for both IGF-I and IGF-II of wild-type IGFBP-3 (Fig. 8B). GGG mutant IGFBP-3 demonstrated barely detectable IGF binding activities, although protein expression was confirmed by Western blot analysis in the CM (Fig. 8B). The binding specificity of wild-type IGFBP-3 to IGF-I was confirmed by Western ligand blot analysis using radioactively unlabeled ligands as a cold competitor. As shown in Fig. 8C, a 100-fold molar excess of unlabeled IGF-I antagonized 125I-labeled IGF-I to bind to IGFBP-3 expressed in the CM of EPC2-hTERT cells overexpressing wild-type IGFBP-3. In contrast, the binding of 125I-labeled IGF-I to IGFBP-3 was observed in the presence of excessive amount of insulin, Des(1–3)-IGF-I, or long 3R–IGF-I, all of which have greatly reduced affinity, if any, to full-length IGFBP-3 but intact binding capacity to IGF-IR (2, 61, 63). These results indicate that the overexpressed wild-type IGFBP-3 protein, but not mutant IGFBP-3 protein, possesses IGF-binding capacity.
Fig. 8.
IGFBP-3 is overexpressed in primary and immortalized human esophageal cells. EPC1, EPC2, and EPC2-hTERT cells were stably transduced with retroviruses expressing wild-type IGFBP-3 (WT), GGG mutant IGFBP-3 (MT), or empty vector (puro). A: IGFBP-3 expression in cell lysates was determined by Western blot analysis. β-Actin was used as a loading control. B: IGF binding activity of IGFBP-3 secreted in CM was assessed by Western ligand blot analysis. The blot was probed with 125I-labeled IGF-I (125I-IGF-I) or 125I-IGF-II. The blot was reprobed with anti-IGFBP-3 to identify IGF binding activity detected by the radiolabeled ligands. rhIGFBP-3, recombinant human IGFBP-3. C: ligand-specific binding activity of IGFBP-3 expressed in EPC2-hTERT cells was determined by incubating the strips of membrane with 125I-IGF-I in the absence (lane 1) or presence of 100× molar excess of cold competitors [lane 2, IGF-I; lane 3, insulin; lane 4, Des(1–3)-IGF-I (Des); lane 5, long R3-IGF-I]. Blots were reprobed with anti-IGFBP-3 to confirm the equal loading of concentrated CM.
We examined further whether IGFBP-3 overexpression affects IGF receptor activation by its ligands. Western blot analysis demonstrated that IGF-IR phosphorylation was rapidly induced by IGF-I treatment in parental cells, empty vector-transduced cells, and GGG mutant IGFBP-3-expressing cells but not in wild-type IGFBP-3-transduced cells (Fig. 9, A and B), suggesting that wild-type IGFBP-3 secreted into CM prevented IGF-I from binding to IGF-IR. In contrast, a high insulin concentration induced a comparable level of IGF-IR phosphorylation in both wild-type IGFBP-3-overexpressing cells and control cells, albeit to lesser extent than was induced by IGF-I (Fig. 9A). Both Des(1–3)-IGF-I and long 3R–IGF-I appeared to be capable of activating IGF-IR in wild-type IGFBP-3-expressing cells as well as control cells and GGG mutant IGFBP-3-expressing cells, although the IGF-IR phosphorylation level was lower in wild-type IGFBP-3-expressing cells, indicating that their affinity to IGFBP-3 is weak but not absent and that overexpressed IGFBP-3 suppressed complete activation of IGF-IR by these mutant IGF-I analogs (Fig. 9B and data not shown).
Fig. 9.
IGFBP-3 suppresses the activation of IGFR signaling by IGF-I but does not affect EGFR activation by EGF. EPC2-hTERT cells transduced with wild-type IGFBP-3, GGG mutant IGFBP-3, or empty vector were stimulated with GFs in fresh KBM for 15 min after overnight GF deprivation (16 h) with KBM. Cell lysates were subjected to Western blot analysis to assess ligand-induced phosphorylation of IGFI-R (Tyr1131), Akt (Ser473), and EGFR (Tyr1068). Total IGF-IR, Akt, and EGFR were determined. β-Actin was used as a loading control. A: cells were treated with the indicated concentrations of IGF-I or insulin. B: cells were stimulated with 100 ng/ml IGF-I or Des. C: cells were stimulated with 10 ng/ml EGF.
We next assessed whether Akt, one of the pivotal downstream target molecules of the IGF-IR signaling pathway, was activated. As shown in Fig. 9, A and B, IGF-I did not induce as robust Akt phosphorylation in wild-type IGFBP-3-overex-pressing cells, although both Des(1–3)-IGF-I and long 3R–IGF-I appeared to activate Akt to some extent. In contrast, Akt phosphorylation was fully induced in parental, empty vector-transduced, and GGG mutant IGFBP-3-overexpressing cells (Fig. 9, A and B). Among the other signaling molecules examined, p44/p42 MAPK remained phosphorylated during the growth factor deprivation period for 24 h, and its level was not affected by IGF-I stimulation regardless of IGFBP-3 status (data not shown). These results indicate IGF-dependent function in IGFBP-3-overexpressing human esophageal cells and that there is an accompanying suppression of Akt activation.
IGFBP-3 overexpression inhibits the mitogenic effect of IGF-I
Both growth stimulatory and inhibitory functions of IGFBP-3 have been described in an IGF-dependent or -independent manner (17). IGFBP-3 has been implicated in cellular senescence (19, 22). IGFBP-3 was induced in primary and immortalized human esophageal cells within 24 h upon growth factor deprivation while a concomitant reduction in cell proliferation was observed (Table 2 and data not shown). EPC1 and EPC2 cells underwent replicative senescence as described previously (28). We also observed upregulation of IGFBP-3 mRNA when EPC2 cells undergo replicative senescence (data not shown). Thus we asked whether IGFBP-3 affects cell proliferation and replicative lifespan in esophageal epithelial cells.
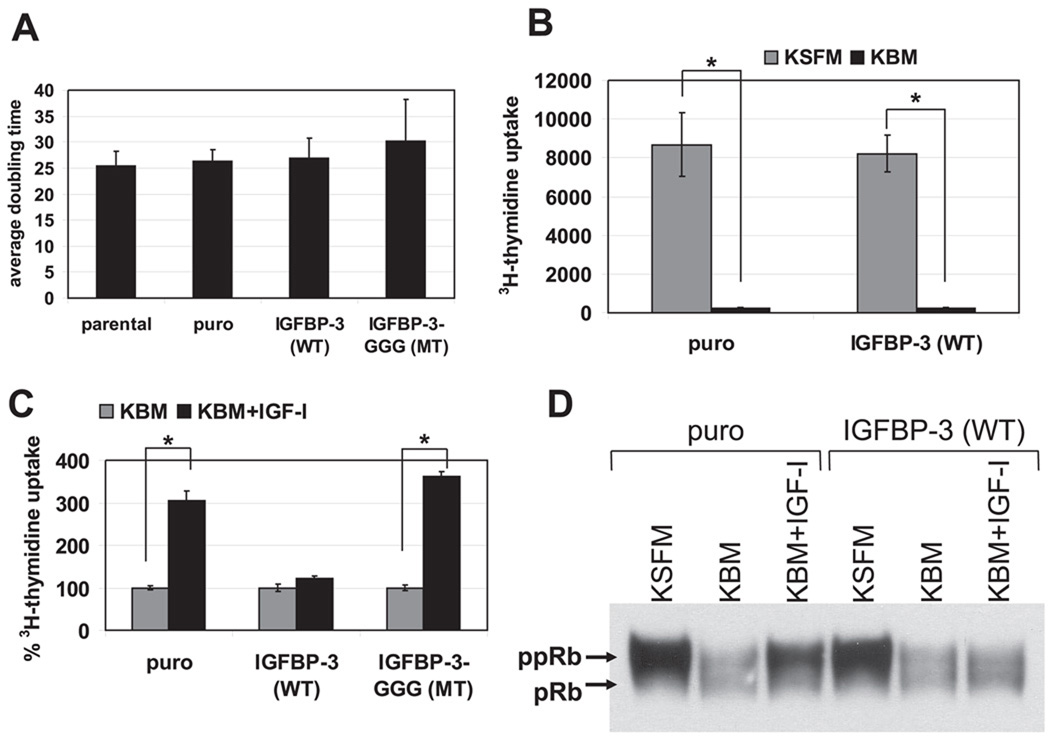
In standard cell culture conditions, neither wild-type nor mutant IGFBP-3 affected the population doubling time of EPC2-hTERT cells (Fig. 10A). In fact, wild-type IGFBP-3 overexpression did not affect cell proliferation or cell cycle distribution when cells were grown in KSFM (Fig. 10B and Table 2). In EPC1 and EPC2 cells assayed over 20 population doubling times, there was also no statistically significant difference observed in average population doubling times (65.2 ± 10.8 and 77.5 ± 22.8 h for wild-type IGFBP-3-transduced cells, respectively, compared with 70.9 ± 16.2 and 73.8 ± 17.5 h for empty vector-transduced control cells, respectively). Thus IGFBP-3 overexpression did not appear to affect the replicative lifespan of primary esophageal epithelial cells.
Fig. 10.
IGFBP-3 overexpression alone does not affect esophageal cell proliferation in regular culture conditions, whereas it suppresses the mitogenic effect of IGF-I and activates the G1 checkpoint of the cell cycle upon GF deprivation. A: average doubling time was determined in EPC2-hTERT parental cells and their derivatives over a time period of 12–13 population doublings (14 days). Values are means ± SE (n = 4) and represent 1 of at least 3 independently performed experiments with similar results. B and C: cell proliferation was assessed by [3H]thymidine uptake by cells during a 4-h incubation period before cell harvest. Values are means ± SE (n = 4) and represent 1 of at least 3 independently performed experiments with similar results for both B and C. In B, subconfluent EPC2-hTERT cells transduced with wild-type IGFBP-3 or empty vector were subjected to GF deprivation with KBM or left in the regular culture medium (KSFM). In C, EPC2-hTERT cells transduced with wild-type IGFBP-3, GGG mutant IGFBP-3, or empty vector were subjected to GF deprivation for 24 h in the presence (KBM+IGF-I) or absence (KBM) of 100 ng/ml IGF-I. [3H]thymidine uptake of the control cells (puro) was set as 100%. D: subconfluent EPC2-hTERT cells transduced with wild-type IGFBP-3 or empty vector were subjected to overnight GF deprivation (16 h) with KBM in the presence (KBM+IGF-I) or absence (KBM) of 100 ng/ml IGF-I or left in the regular culture medium (KSFM). Cell lysates were subjected to Western blot analysis to assess the phosphorylation status of pRB. ppRB, phosphorylated pRB.
Next, we examined whether IGFBP-3 overexpression influenced the cellular response to growth factor deprivation and mitogenic stimulation by IGF-I. As shown in Fig. 10B, both wild-type IGFBP-3-overexpressing cells and control cells underwent growth inhibition within 24 h after growth factor deprivation in KBM. This was also reproducibly confirmed by cell cycle analysis (Table 2). Western blot analysis showed a reduction in the phosphorylation level of pRB upon growth factor deprivation in both IGFBP-3-overexpressing cells and control cells (Fig. 10D), corroborating the G1 arrest by growth factor withdrawal. In contrast, neither Chk1 nor Chk2 was activated regardless of the IGFBP-3 expression level or IGF-I treatment (data not shown), consistent with the lack of a significant increase in the G2/M cell fraction upon growth factor deprivation (Table 2). Such cell growth inhibition was antagonized by 100 ng/ml IGF-I in parental and empty vector-transduced cells (Table 2 and Fig. 10C), which was corroborated by the pRB phosphorylation stimulated by IGF-I in control cells (Fig. 10D). In contrast, cells overexpressing wild-type IGFBP-3 but not GGG mutant IGFBP-3 failed to respond to IGF-I, without evidence of increased cell proliferation as assessed by [3H]thymidine incorporation (Fig. 10C). In wild-type IGFBP-3-overexpressing cells, IGF-I also failed to induce pRB phosphorylation (Fig. 10D). Therefore, overexpression of IGFBP-3 affected the mitogenic effect of IGF-I upon esophageal cell proliferation, implying an IGF-dependent growth inhibitory function.
IGFBP-3 may induce apoptosis under certain circumstances (17, 24). Growth factor deprivation suppressed DNA synthesis in both IGFBP-3-overexpressing cells and control cells (Fig. 10B and data not shown). However, no significant apoptosis was observed upon IGFBP-3 overexpression in EPC1, EPC2, or EPC2-hTERT cells regardless of the presence or absence of growth factors (data not shown). Finally, we examined whether IGFBP-3 overexpression induces EGFR in our esophageal epithelial cells. However, IGFBP-3 overexpression appeared to increase neither the EGFR level nor its sensitivity to EGF in EPC2-hTERT cells (Fig. 9C).
In aggregate, the above results demonstrate that EGF and other growth factors inhibit IGFBP-3 expression while IGFBP-3 negatively regulates cell proliferation mainly by restricting the G1/S cell cycle progression in an IGF-dependent manner. Although growth factor deprivation leads to the induction of IGFBP-3 and cell growth inhibition, IGFBP-3 alone does not affect cell proliferation in primary and immortalized human esophageal epithelial cells.
DISCUSSION
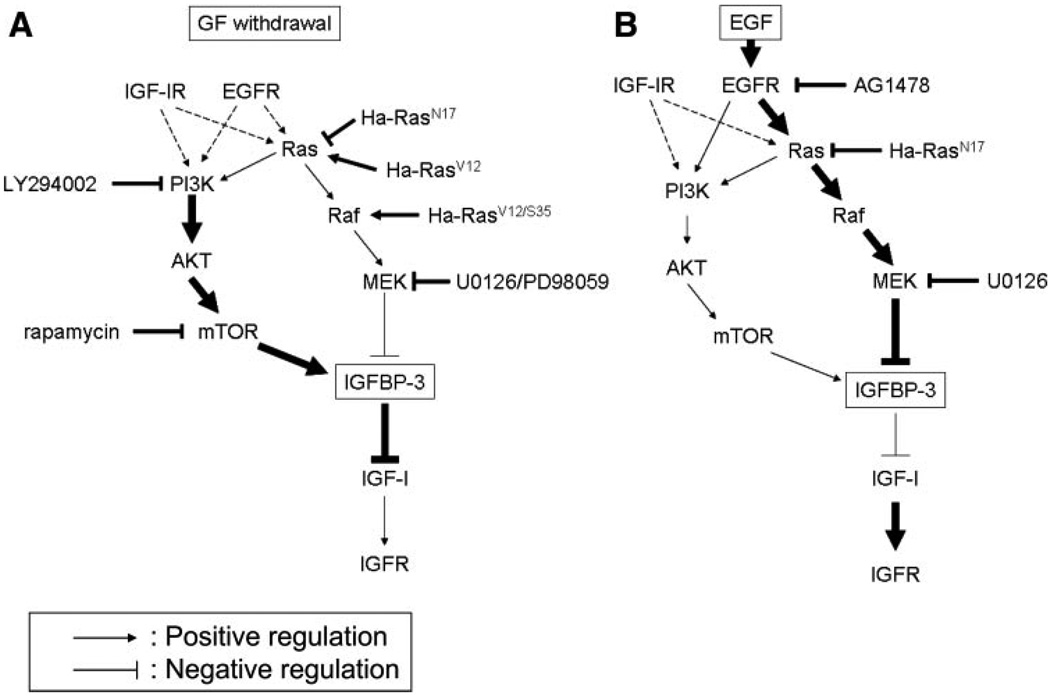
We have demonstrated that IGFBP-3 is induced by growth factor deprivation with concomitant cell growth inhibition. The IGFBP-3 induction appeared to be p53 independent and requires de novo mRNA and protein synthesis that is regulated by the MAPK and PI3K/mTOR signaling pathways (Fig. 11A). Whereas induced IGFBP-3 prevented IGF-I from activating IGF-IR, EGF greatly suppressed IGFBP-3 (Fig. 11B) and restored cellular sensitivity to IGF-I. When overexpressed, IGFBP-3 activated the G1 checkpoint of the cell cycle and inhibited cell proliferation in an IGF-dependent fashion. However, IGFBP-3 alone did not appear to be growth inhibitory or stimulatory in primary and immortalized human esophageal epithelial cells.
Fig. 11.
Proposed model of GF-mediated regulation of IGFBP-3. A: GF deprivation results in a reduction of external mitogenic signals; however, basal activity of the PI3K/ AKT/mTOR pathway permits de novo IGFBP-3 transcription and translation. mTOR may contribute to IGFBP-3 protein induction through a posttranscriptional mechanism. IGFBP-3 is secreted out and prevents exogenous IGF from stimulating the IGF receptor (IGFR). B: EGF potently suppresses IGFBP-3 transcription and translation through excessive activation of the Ras/Raf/MAPK pathway. Although EGF can activate the PI3K pathway (1), IGF-IR may play a predominant role in its activation in primary and immortalized human esophageal cells. The thick arrows and lines indicate the predominant signaling pathways for IGFBP-3 regulation upon GF deprivation (A) or EGF treatment (B). Dashed arrows indicate residual signals from IGFR or EGFR during GF deprivation. The pharmacological inhibitors and genetic constructs used in this study are noted.
Mechanisms of regulation of IGFBP-3 expression
IGFBP-3 mRNA induction by growth factor deprivation and inhibition by EGF suggests that IGFBP-3 is probably transcriptionally repressed by EGF and potentially other growth factors in the tissue culture medium. Serum deprivation induces IGFBP-3 in a p53-dependent fashion, as reported in some other systems (20, 21, 27). However, IGFBP-3 was induced in the cells with impaired p53 function upon growth factor deprivation (Fig. 3). In addition, IGFBP-3 is overexpressed in esophageal cancer cell lines with documented p53 mutation and dysfunction (53). Grimberg et al. (26) have demonstrated that p53-mediated induction of IGFBP-3 depends on the tissue type in γ-irradiated mice and thus wild-type p53 may not play an essential role in IGFBP-3 expression in esophageal epithelial cells. Recently, Barbieri et al. (3) demonstrated that ΔNp63α, a homolog of p53, directly regulates IGFBP-3 transcription in the squamous epithelium through interaction with p53-specific cis-regulatory elements in the IGFBP-3 gene. Suliman et al. (52) described that both full-length and amino-truncated forms of p63 are upregulated in the oral esophageal epithelia of p53-null mice, and the induction of p21WAF1/CIP1 may potentially be preserved through the increase of p63. Because IGFBP-3 induction was more pronounced in EPC2-hTERT cells expressing p53R175H than in control cells (Fig. 3), it is tempting to speculate about the role of p63 in the IGFBP-3 induction in esophageal cells, which may be accounted for by the gain of function effect of p53R175H (13), inhibiting transcriptional activity of p53 family member p63 by a physical interaction with the p53R175H mutant protein (18).
Inhibition of histone deacetylase leads to activation of the IGFBP-3 promoter, which is mediated by the interaction of Sp1 and Sp3 with a proximal GC-rich cis-regulatory element of the IGFBP-3 promoter (11, 55–57). Sp1 mediates the response of the IGFBP-3 promoter to the growth-regulating stimulus induced by EGF (37). Both p44/p42 MAPK and the PI3K signaling pathway regulate vascular endothelial growth factor gene expression through Sp1, independent of hypoxia (38, 40). Basal MAPK activity was present and PI3K activity was required in the induction of IGFBP-3 during growth factor in EPC2-hTERT cells subjected to growth factor deprivation (Fig. 4). Thus Sp1 may be involved in the observed IGFBP-3 transcriptional regulation. In contrast, MAPK activity appears to regulate negatively IGFBP-3 expression (Figs. 4, A and B, and 7C), consistent with the inhibition of IGFBP-3 mRNA by EGF, because MAPK is activated by EGF in EPC2 cells (1). Recently, both MAPK and PI3K pathways were implicated in the induction of IGFBP-3 mRNA in MAC-T immortalized bovine epithelial cells, which is in agreement with our observations. However, IGF-I and transforming growth factor (TGF)-α stimulated IGFBP-3 expression in this cell line (50). In contrast, suppression of IGFBP-3 mRNA expression by EGF has been observed in ectocervical epithelial cells immortalized by human papilloma virus (29), spontaneously immortalized HaCaT cells (60), and primary epidermal keratinocytes (14). We have recently demonstrated that EGF at 1 ng/ml concentration suppresses IGFBP-3 expression in TE2 and TE7 esophageal cancer cells, whereas EGFR activation resulted in a potent induction of IGFBP-3 in the TE11 esophageal cancer cell line and A431 cells (53). Such discrepancies in EGF-mediated regulation of IGFBP-3 expression may be attributable to the EGFR level or EGFR activity that may be modulated by cell-cell contact mediated by E-cadherin (41). Cell density may also affect IGFBP-3 expression (5). Other growth factors produced by cells in an autocrine fashion may also interfere with exogenous EGF. Furthermore, IGFBP-3 is up-regulated in primary and immortalized human esophageal cells that were retrovirally transduced to overexpress EGFR (53). Although this reveals a regulatory role of EGFR in IGF signaling by modulating IGFBP-3, the negative regulation of IGFBP-3 by EGF observed in the present study suggests that complex regulatory mechanisms regulate IGFBP-3 expression. We have recently observed in EPC2-hTERT cells that EGF fully activates Ras in KBM growth factor deprivation medium, whereas it paradoxically suppresses the Ras activity in KSFM despite full activation of EGFR, implying a negative feedback mechanism through EGFR to prevent excessive Ras activation in the presence of a physiologically exceeding level of insulin. Moreover, such inhibition of Ras activity by EGF was augmented in EGFR-overexpressing cells compared with control cells (M. Takaoka and H. Nakagawa, unpublished observations). Given the inhibitory effect of Ras-MAPK signaling upon IGFBP-3 expression, it is tempting to speculate that our previous observation of IGFBP-3 upregulation in EGFR-overexpressing cells may be accounted for by the suppression of Ras by EGF in KSFM. Thus the dichotomous effects observed with IGFBP-3 induction or suppression are likely dependent on the concentration of EGF and the status of the EGFR level and activation, particularly in terms of the cellular context and physiological versus stress-related conditions, and whether cells are normal or transformed. In particular, growth factor deprivation can be viewed as analogous to cellular stress in which IGFBP-3’s level and role may be quite different than under normal conditions. Certainly, divergent levels and roles are not without precedence, as observed with oncogenic Ki-ras promoting cellular proliferation or cellular senescence or normal levels versus overexpression of other oncogenes (cyclin D1 and Akt) inducing dichotomous effects.
Biological consequences of IGFBP-3 induction or overexpression
IGFBP-3 has been implicated in cell growth inhibition or apoptosis induced by serum starvation (20, 21, 27) and various antiproliferative agents such as TNF-α, TGF-β1, and vitamin D (17). In primary and immortalized esophageal cells, IGFBP-3 induced by growth factor deprivation or overexpressed by retrovirus prevented ligand binding and activation of IGF-IR (Figs. 5D, 8, and 9). In a transgenic mouse model where IGFBP-3 was ubiquitously overexpressed by the cytomegalovirus promoter, reduced proliferation of epidermal keratinocytes was observed, although there was no gross morphological change in the skin (15). In esophageal cells, however, IGFBP-3 overexpression alone did not greatly affect cell proliferation or differentiation in monolayer culture or reconstituted stratified epithelium in organotypic culture (Fig. 10 and data not shown). IGFBP-3 is frequently upregulated in esophageal cancer with EGFR overexpression (53). When stably transfected in T47D human breast cancer cells, IGFBP-3 initially exhibited the growth inhibitory effect yet eventually led to aggressive tumor growth with upregulation and activation of EGFR (10). IGFBP-3 enhances EGF-mediated activation of MAPK in MCF-10A mammary epithelial cells (36) and stimulates PI3K in MCF-7 breast carcinoma cells (43). However, in IGFBP-3-overexpressing esophageal cells, the EGFR level was not altered, and EGFR was activated to an equal extent compared with control cells (Fig. 9C). IGFBP-3 secreted in CM inhibited Akt activation by IGF-I (Fig. 9, A and B). Furthermore, intracellular IGFBP-3 did not influence MAPK or Akt activation induced by EGF or IGF-I (data not shown).
IGFBP-3 also plays a role in retinoic acid signaling through an interaction with the retinoic acid X receptor (4), TGF-β-mediated signaling by activating Smad2 (4), and cell cycle regulation through p21WAF1/CIP1 (7) and signal transducers and activators of transcription-1 (51) in an IGF-independent manner. Because IGFBP-3 is induced by hypoxia (16, 26), it may be involved in angiogenesis (33) and glucose metabolism (48). These possibilities warrant further investigation in esophageal epithelial cells.
In conclusion, our data demonstrate that IGFBP-3 is induced upon growth factor deprivation and inhibits the mitogenic effect of IGF-I in primary and immortalized human esophageal cells. EGFR positively regulates the IGF signaling pathway by negatively modulating IGFBP-3 levels, providing a novel mechanistic link between EGF and IGF signaling pathways.
ACKNOWLEDGMENTS
We thank Dr. Gary Swain and Kelly Dempsey (Morphology Core Facility at the University of Pennsylvania School of Medicine) for immunocytochemistry and morphological assessment of apoptosis and Drs. Therese B. Deramaudt, Cameron N. Johnstone, Kenji Oyama, Jess Porter, Mark J. Bowser, and Ben Rhoades (Rustgi Laboratory) for helpful discussions. Finally, we are grateful to Drs. Adda Grimberg, Meenhard M. Herlyn, and Anil K. Rustgi for the support and advice and critical review of the manuscript by Dr. Rustgi.
GRANTS
This study was supported in part by National Institutes of Health (NIH) Grant R21-DK-64249 and K01-DK-066205 (to H. Nakagawa), the American Gastroenterological Association/Foundation for Digestive Health and Nutrition Research Scholar Award (to H. Nakagawa), NIH Training Grant R25-DK-066028 (to C. E. Smith and M. K. Mashiba), NIH Grant F32-CA-108657 (to C. D. Andl), NIH Grant P01-CA-098101 (to H. Nakagawa, C. D. Andl, C. E. Smith, M. K. Mashiba, M. Takaoka, T. Okawa, and W. S. El-Deiry) and its core facilities, and the NIH Center for Molecular Studies in Digestive and Liver Diseases, its Morphology Core, Molecular Biology, and Cell Culture Core facilities, and pilot grant program (P30-DK-50306).
REFERENCES
- 1.Andl CD, Mizushima T, Nakagawa H, Oyama K, Harada H, Chruma K, Herlyn M, Rustgi AK. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278:1824–1830. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- 2.Ballard FJ, Wallace JC, Francis GL, Read LC, Tomas FM. Des(1–3)IGF-I: a truncated form of insulin-like growth factor-I. Int J Biochem Cell Biol. 1996;28:1085–1087. doi: 10.1016/1357-2725(96)00056-8. [DOI] [PubMed] [Google Scholar]
- 3.Barbieri CE, Perez CA, Johnson KN, Ely KA, Billheimer D, Pietenpol JA. IGFBP-3 is a direct target of transcriptional regulation by DeltaNp63alpha in squamous epithelium. Cancer Res. 2005;65:2314–2320. doi: 10.1158/0008-5472.CAN-04-3449. [DOI] [PubMed] [Google Scholar]
- 4.Baxter RC. Signalling pathways involved in antiproliferative effects of IGFBP-3: a review. Mol Pathol. 2001;54:145–148. doi: 10.1136/mp.54.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blat C, Villaudy J, Harel L. Density-dependent inhibition of mouse embryo fibroblast growth: involvement of IGFBP-3. Exp Cell Res. 1994;215:114–118. doi: 10.1006/excr.1994.1322. [DOI] [PubMed] [Google Scholar]
- 6.Bourdon JC, Deguin-Chambon V, Lelong JC, Dessen P, May P, Debuire B, May E. Further characterisation of the p53 responsive element—identification of new candidate genes for trans-activation by p53. Oncogene. 1997;14:85–94. doi: 10.1038/sj.onc.1200804. [DOI] [PubMed] [Google Scholar]
- 7.Boyle BJ, Zhao XY, Cohen P, Feldman D. Insulin-like growth factor binding protein-3 mediates 1α,25-dihydroxyvitamin D3 growth inhibition in the LNCaP prostate cancer cell line through p21/WAF1. J Urol. 2001;165:1319–1324. [PubMed] [Google Scholar]
- 8.Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 9.Buckway CK, Wilson EM, Ahlsen M, Bang P, Oh Y, Rosenfeld RG. Mutation of three critical amino acids of the N-terminal domain of IGF-binding protein-3 essential for high affinity IGF binding. J Clin Endocrinol Metab. 2001;86:4943–4950. doi: 10.1210/jcem.86.10.7936. [DOI] [PubMed] [Google Scholar]
- 10.Butt AJ, Martin JL, Dickson KA, McDougall F, Firth SM, Baxter RC. Insulin-like growth factor binding protein-3 expression is associated with growth stimulation of T47D human breast cancer cells: the role of altered epidermal growth factor signaling. J Clin Endocrinol Metab. 2004;89:1950–1956. doi: 10.1210/jc.2003-030914. [DOI] [PubMed] [Google Scholar]
- 11.Choi HS, Lee JH, Park JG, Lee YI. Trichostatin A, a histone deacetylase inhibitor, activates the IGFBP-3 promoter by upregulating Sp1 activity in hepatoma cells: alteration of the Sp1/Sp3/HDAC1 multiprotein complex. Biochem Biophys Res Commun. 2002;296:1005–1012. doi: 10.1016/s0006-291x(02)02001-6. [DOI] [PubMed] [Google Scholar]
- 12.Coppola D, Ferber A, Miura M, Sell C, D’Ambrosio C, Rubin R, Baserga R. A functional insulin-like growth factor I receptor is required for the mitogenic and transforming activities of the epidermal growth factor receptor. Mol Cell Biol. 1994;14:4588–4595. doi: 10.1128/mcb.14.7.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, Finlay C, Levine AJ. Gain of function mutations in p53. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 14.Edmondson SR, Murashita MM, Russo VC, Wraight CJ, Werther GA. Expression of insulin-like growth factor binding protein-3 (IGFBP-3) in human keratinocytes is regulated by EGF and TGFbeta1. J Cell Physiol. 1999;179:201–207. doi: 10.1002/(SICI)1097-4652(199905)179:2<201::AID-JCP10>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Edmondson SR, Thumiger SP, Kaur P, Loh B, Koelmeyer R, Li A, Silha JV, Murphy LJ, Wraight CJ, Werther GA. Insulin-like growth factor binding protein-3 (IGFBP-3) localizes to and modulates proliferative epidermal keratinocytes in vivo. Br J Dermatol. 2005;152:225–230. doi: 10.1111/j.1365-2133.2004.06350.x. [DOI] [PubMed] [Google Scholar]
- 16.Feldser D, Agani F, Iyer NV, Pak B, Ferreira G, Semenza GL. Reciprocal positive regulation of hypoxia-inducible factor 1alpha and insulin-like growth factor 2. Cancer Res. 1999;59:3915–3918. [PubMed] [Google Scholar]
- 17.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 18.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein S, Moerman EJ, Jones RA, Baxter RC. Insulin-like growth factor binding protein 3 accumulates to high levels in culture medium of senescent and quiescent human fibroblasts. Proc Natl Acad Sci USA. 1991;88:9680–9684. doi: 10.1073/pnas.88.21.9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granata R, De Petrini M, Trovato L, Ponti R, Pons N, Ghe C, Graziani A, Ferry RJ, Jr, Muccioli G, Ghigo E. Insulin-like growth factor binding protein-3 mediates serum starvation- and doxorubicin-induced apoptosis in H9c2 cardiac cells. J Endocrinol Invest. 2003;26:1231–1241. doi: 10.1007/BF03349163. [DOI] [PubMed] [Google Scholar]
- 21.Granata R, Trovato L, Garbarino G, Taliano M, Ponti R, Sala G, Ghidoni R, Ghigo E. Dual effects of IGFBP-3 on endothelial cell apoptosis and survival: involvement of the sphingolipid signaling pathways. FASEB J. 2004;18:1456–1458. doi: 10.1096/fj.04-1618fje. [DOI] [PubMed] [Google Scholar]
- 22.Grigoriev VG, Moerman EJ, Goldstein S. Overexpression of insulin-like growth factor binding protein-3 by senescent human fibroblasts: attenuation of the mitogenic response to IGF-I. Exp Cell Res. 1995;219:315–321. doi: 10.1006/excr.1995.1234. [DOI] [PubMed] [Google Scholar]
- 23.Grillari J, Hohenwarter O, Grabherr RM, Katinger H. Subtractive hybridization of mRNA from early passage and senescent endothelial cells. Exp Gerontol. 2000;35:187–197. doi: 10.1016/s0531-5565(00)00080-2. [DOI] [PubMed] [Google Scholar]
- 24.Grimberg A. P53 and IGFBP-3: apoptosis and cancer protection. Mol Genet Metab. 2000;70:85–98. doi: 10.1006/mgme.2000.3008. [DOI] [PubMed] [Google Scholar]
- 25.Grimberg A, Cohen P. Role of insulin-like growth factors and their binding proteins in growth control and carcinogenesis. J Cell Physiol. 2000;183:1–9. doi: 10.1002/(SICI)1097-4652(200004)183:1<1::AID-JCP1>3.0.CO;2-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimberg A, Coleman CM, Burns TF, Himelstein BP, Koch CJ, Cohen P, El-Deiry WS. p53-dependent and p53-independent induction of IGFBP-3 by DNA damage and hypoxia. J Clin Endocrinol Metab. 2005;90:3568–3574. doi: 10.1210/jc.2004-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimberg A, Liu B, Bannerman P, El-Deiry WS, Cohen P. IGFBP-3 mediates p53-induced apoptosis during serum starvation. Int J Oncol. 2002;21:327–335. [PMC free article] [PubMed] [Google Scholar]
- 28.Harada H, Nakagawa H, Oyama K, Takaoka M, Andl CD, Jacobmeier B, von Werder A, Enders GH, Opitz OG, Rustgi AK. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1:729–738. [PubMed] [Google Scholar]
- 29.Hembree JR, Agarwal C, Eckert RL. Epidermal growth factor suppresses insulin-like growth factor binding protein 3 levels in human papillomavirus type 16-immortalized cervical epithelial cells and thereby potentiates the effects of insulin-like growth factor 1. Cancer Res. 1994;54:3160–3166. [PubMed] [Google Scholar]
- 30.Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999;20:761–787. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- 31.Itakura Y, Sasano H, Shiga C, Furukawa Y, Shiga K, Mori S, Nagura H. Epidermal growth factor receptor overexpression in esopha Epidermal growth factor receptor overexpression in esophageal carcinoma. An immunohistochemical study correlated with clinicopathologic findings and DNA amplification. Cancer. 1994;74:795–804. doi: 10.1002/1097-0142(19940801)74:3<795::aid-cncr2820740303>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 32.Joneson T, White MA, Wigler MH, Bar-Sagi D. Stimulation of membrane ruffling and MAP kinase activation by distinct effectors of Ras. Science. 1996;271:810–812. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- 33.Lee YM, Bae MH, Lee OH, Moon EJ, Moon CK, Kim WH, Kim KW. Synergistic induction of in vivo angiogenesis by the combination of insulin-like growth factor-II and epidermal growth factor. Oncol Rep. 2004;12:843–848. [PubMed] [Google Scholar]
- 34.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longobardi L, Torello M, Buckway C, O’Rear L, Horton WA, Hwa V, Roberts CT, Jr, Chiarelli F, Rosenfeld RG, Spagnoli A. A novel insulin-like growth factor (IGF)-independent role for IGF binding protein-3 in mesenchymal chondroprogenitor cell apoptosis. Endocrinology. 2003;144:1695–1702. doi: 10.1210/en.2002-220959. [DOI] [PubMed] [Google Scholar]
- 36.Martin JL, Weenink SM, Baxter RC. Insulin-like growth factorbinding protein-3 potentiates epidermal growth factor action in MCF-10A mammary epithelial cells. Involvement of p44/42 and p38 mitogen-activated protein kinases. J Biol Chem. 2003;278:2969–2976. doi: 10.1074/jbc.M210739200. [DOI] [PubMed] [Google Scholar]
- 37.Merchant JL, Du M, Todisco A. Sp1 phosphorylation by Erk 2 stimulates DNA binding. Biochem Biophys Res Commun. 1999;254:454–461. doi: 10.1006/bbrc.1998.9964. [DOI] [PubMed] [Google Scholar]
- 38.Milanini J, Vinals F, Pouyssegur J, Pages G. p42/p44 MAP kinase module plays a key role in the transcriptional regulation of the vascular endothelial growth factor gene in fibroblasts. J Biol Chem. 1998;273:18165–18172. doi: 10.1074/jbc.273.29.18165. [DOI] [PubMed] [Google Scholar]
- 39.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pore N, Liu S, Shu HK, Li B, Haas-Kogan D, Stokoe D, Milanini-Mongiat J, Pages G, O’Rourke DM, Bernhard E, Maity A. Sp1 is involved in Akt-mediated induction of VEGF expression through an HIF-1-independent mechanism. Mol Biol Cell. 2004;15:4841–4853. doi: 10.1091/mbc.E04-05-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 2004;23:1739–1784. doi: 10.1038/sj.emboj.7600136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qureshi FG, Tchorzewski MT, Duncan MD, Harmon JW. EGF and IGF-I synergistically stimulate proliferation of human esophageal epithelial cells. J Surg Res. 1997;69:354–358. doi: 10.1006/jsre.1997.5080. [DOI] [PubMed] [Google Scholar]
- 43.Ricort JM, Binoux M. Insulin-like growth factor binding protein-3 stimulates phosphatidylinositol 3-kinase in MCF-7 breast carcinoma cells. Biochem Biophys Res Commun. 2004;314:1044–1049. doi: 10.1016/j.bbrc.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Ryan KM, Vousden KH. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol Cell Biol. 1998;18:3692–3698. doi: 10.1128/mcb.18.7.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarze SR, DePrimo SE, Grabert LM, Fu VX, Brooks JD, Jarrard DF. Novel pathways associated with bypassing cellular senescence in human prostate epithelial cells. J Biol Chem. 2002;277:14877–14883. doi: 10.1074/jbc.M200373200. [DOI] [PubMed] [Google Scholar]
- 46.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 47.Silha JV, Gui Y, Mishra S, Leckstrom A, Cohen P, Murphy LJ. Overexpression of gly56/gly80/gly81-mutant insulin-like growth factor-binding protein-3 in transgenic mice. Endocrinology. 2005;146:1523–1531. doi: 10.1210/en.2004-0905. [DOI] [PubMed] [Google Scholar]
- 48.Silha JV, Gui Y, Murphy LJ. Impaired glucose homeostasis in insulin-like growth factor-binding protein-3-transgenic mice. Am J Physiol Endocrinol Metab. 2002;283:E937–E945. doi: 10.1152/ajpendo.00014.2002. [DOI] [PubMed] [Google Scholar]
- 49.Simmons JG, Hoyt EC, Westwick JK, Brenner DA, Pucilowska JB, Lund PK. Insulin-like growth factor-I and epidermal growth factor interact to regulate growth and gene expression in IEC-6 intestinal epithelial cells. Mol Endocrinol. 1995;9:1157–1165. doi: 10.1210/mend.9.9.7491108. [DOI] [PubMed] [Google Scholar]
- 50.Sivaprasad U, Fleming J, Verma PS, Hogan KA, Desury G, Cohick WS. Stimulation of insulin-like growth factor (IGF) binding protein-3 synthesis by IGF-I and transforming growth factor-alpha is mediated by both phosphatidylinositol-3 kinase and mitogen-activated protein kinase pathways in mammary epithelial cells. Endocrinology. 2004;145:4213–4221. doi: 10.1210/en.2003-1377. [DOI] [PubMed] [Google Scholar]
- 51.Spagnoli A, Torello M, Nagalla SR, Horton WA, Pattee P, Hwa V, Chiarelli F, Roberts CT, Jr, Rosenfeld RG. Identification of STAT-1 as a molecular target of IGFBP-3 in the process of chondrogenesis. J Biol Chem. 2002;277:18860–18867. doi: 10.1074/jbc.M200218200. [DOI] [PubMed] [Google Scholar]
- 52.Suliman Y, Opitz OG, Avadhani A, Burns TC, El-Deiry W, Wong DT, Rustgi AK. p63 expression is associated with p53 loss in oral-esophageal epithelia of p53-deficient mice. Cancer Res. 2001;61:6467–6473. [PubMed] [Google Scholar]
- 53.Takaoka M, Harada H, Andl CD, Oyama K, Naomoto Y, Dempsey KL, Klein-Szanto AJ, El-Deiry WS, Grimberg A, Nakagawa H. Epidermal growth factor receptor regulates aberrant expression of insulinlike growth factor-binding protein 3. Cancer Res. 2004;64:7711–7723. doi: 10.1158/0008-5472.CAN-04-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takaoka M, Harada H, Deramaudt TB, Oyama K, Andl CD, Johnstone CN, Rhoades B, Enders GH, Opitz OG, Nakagawa H. Ha-Ras(G12V) induces senescence in primary and immortalized human esophageal keratinocytes with p53 dysfunction. Oncogene. 2004;23:6760–6768. doi: 10.1038/sj.onc.1207923. [DOI] [PubMed] [Google Scholar]
- 55.Tsubaki J, Choi WK, Ingermann AR, Twigg SM, Kim HS, Rosenfeld RG, Oh Y. Effects of sodium butyrate on expression of members of the IGF-binding protein superfamily in human mammary epithelial cells. J Endocrinol. 2001;169:97–110. doi: 10.1677/joe.0.1690097. [DOI] [PubMed] [Google Scholar]
- 56.Tsubaki J, Hwa V, Twigg SM, Rosenfeld RG. Differential activation of the IGF binding protein-3 promoter by butyrate in prostate cancer cells. Endocrinology. 2002;143:1778–1788. doi: 10.1210/endo.143.5.8766. [DOI] [PubMed] [Google Scholar]
- 57.Walker GE, Wilson EM, Powell D, Oh Y. Butyrate, a histone deacetylase inhibitor, activates the human IGF binding protein-3 promoter in breast cancer cells: molecular mechanism involves an Sp1/Sp3 multiprotein complex. Endocrinology. 2001;142:3817–3827. doi: 10.1210/endo.142.9.8380. [DOI] [PubMed] [Google Scholar]
- 58.White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler MH. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 59.Wraight CJ, Murashita MM, Russo VC, Werther GA. A keratinocyte cell line synthesizes a predominant insulin-like growth factor-binding protein (IGFBP-3) that modulates insulin-like growth factor-I action. J Invest Dermatol. 1994;103:627–631. doi: 10.1111/1523-1747.ep12397667. [DOI] [PubMed] [Google Scholar]
- 60.Wraight CJ, Werther GA. Insulin-like growth factor-I and epidermal growth factor regulate insulin-like growth factor binding protein-3 (IGFBP-3) in the human keratinocyte cell line HaCaT. J Invest Dermatol. 1995;105:602–607. doi: 10.1111/1523-1747.ep12323716. [DOI] [PubMed] [Google Scholar]
- 61.Xi G, Kamanga-Sollo E, Pampusch MS, White ME, Hathaway MR, Dayton WR. Effect of recombinant porcine IGFBP-3 on IGF-I and long-R3-IGF-I-stimulated proliferation and differentiation of L6 myogenic cells. J Cell Physiol. 2004;200:387–394. doi: 10.1002/jcp.20068. [DOI] [PubMed] [Google Scholar]
- 62.Yacoub L, Goldman H, Odze RD. Transforming growth factor-alpha, epidermal growth factor receptor, and MiB-1 expression in Barrett’s-associated neoplasia: correlation with prognosis. Mod Pathol. 1997;10:105–112. [PubMed] [Google Scholar]
- 63.Yamanaka Y, Wilson EM, Rosenfeld RG, Oh Y. Inhibition of insulin receptor activation by insulin-like growth factor binding proteins. J Biol Chem. 1997;272:30729–30734. doi: 10.1074/jbc.272.49.30729. [DOI] [PubMed] [Google Scholar]