Abstract
The Krüppel-like transcription factor (KLF) family participates in diverse aspects of cellular growth, development, differentiation, and activation. Recently, several groups have identified new connections between the function of these factors and leukocyte responses in health and disease. Gene targeting of individual KLFs in mice has uncovered novel and unexpected physiologic roles among myeloid and lymphocyte cell lineage maturation, particularly in the bone marrow niche and blood. In addition, several KLF family members are downstream targets of stimuli and signaling pathways critical to T-cell trafficking, T regulatory cell differentiation or suppressor function, monocyte/macrophage activation or renewal, and B memory cell maturation or activation. Indeed, KLFs have been implicated in subtypes of leukemia, lymphoma, autoimmunity, and in acute and chronic inflammatory disease states, such as atherosclerosis, diabetes, and airway inflammation, raising the possibility that KLFs and their upstream signals are of therapeutic interest. This review focuses on the relevant literature of Krüppel-like factors in leukocyte biology and their implications in clinical settings.
Introduction
Leukocyte development requires the coordination of stage-specific transcription factors to help orchestrate the processes by which a progenitor cell emerges as a functional leukocyte. Indeed, aberrant expression or function of many of these transcription factors has been associated with several disease conditions, such as leukemia, lymphoma, autoimmunity, and chronic inflammation. Moreover, recent studies have indicated that Krüppel-like factors (KLFs) may be among those key trans-acting factors contributing to the orchestration of several aspects of leukocyte biology, including cell lineage commitment, differentiation, and function.
The original Krüppel factor was characterized in Drosophila melanogaster as a “gap” segmentation gene, homozygous mutation of which resulted in the absence of thorax and anterior abdomen in embryos.1–4 Thus, the German researchers named this gene Krüppel (English “cripple”). A conserved family of nuclear proteins encoded by Drosophila Krüppel were identified in 1986 and exhibited a striking structural similarity to the DNA-binding “finger motif” of transcription factor IIIA.5 The first mammalian gene with homology to Krüppel was identified in 1993, and its encoded protein was named erythroid Krüppel-like factor (EKLF) in accordance with its erythroid cell–specific expression.6 The function of EKLF was demonstrated by the fact that EKLF bound to human and murine adult β-globin CACCC elements via its DNA-binding domain, whereas the non–DNA-binding domain mediated transcriptional activation.7 The importance of EKLF in differentiation and development was later demonstrated by loss-of-function studies showing that homozygous EKLF−/− mice developed a fatal β-thalassemia during fetal liver erythropoiesis.8,9
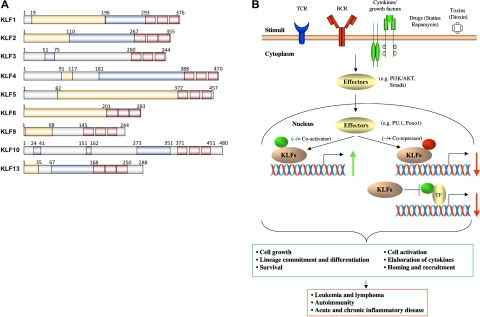
To date, members of the mammalian KLF family number 17.10 Identified by various experimental approaches, KLF1 (EKLF) through KLF17 have been termed according to their chronologic order of identification (Figure 1). Each family member is a zinc finger transcription factor. The distinguishing feature of KLFs compared with other zinc finger-containing proteins, therefore, is the presence of a highly conserved DNA-binding domain composed of 3 C2H2 zinc fingers at or near the C-terminus.11–13 As such, most KLFs are able to bind the CACCC element and GC box consensus sequences. Furthermore, the KLFs share a highly conserved 7-residue sequence, TGEKP(Y/F)X, between zinc fingers.14 The non–DNA-binding regions of each, however, are highly divergent and can function as trans-activation or trans-repression domains. Collectively, these features distinguish the KLFs from the larger family of zinc-finger transcription factors (Figure 1A).
Figure 1.
Schematic representation of trans-acting domains and molecular mechanisms of leukocyte-associated KLFs. (A) Comparison of KLF subdomains for: KLF1,89 KLF2,20,90 KLF3,91,92 KLF4,93 KLF5,94,95 KLF6,96 KLF9,97–99 KLF10,44,100 and KLF13.49,50 The “zinc fingers” are represented by red boxes. The trans-activation domains are indicated by orange boxes, whereas the trans-repression domains are indicated by blue boxes. (B) Role of KLFs in regulating signaling pathways and transcriptional targets that affect leukocytes in health and disease. On activation by stimuli, such as the TCR, BCR, cytokines/growth factors, drugs (statins, rapamycin), or environmental pollutants (dioxin), the upstream cytoplasmic (eg, AKT/PI3K) or nuclear (eg, PU.1 or FOXO1) effector proteins transduce signals that can activate or inhibit nuclear KLFs. KLFs, in turn, induce or repress target genes (alone or in association with coactivators or corepressors) that affect leukocyte cell growth and differentiation, survival, activation, or homing and recruitment, ultimately affecting various disease states.
By regulating gene transcription, KLFs are involved in many physiologic and pathologic processes, such as cell differentiation, proliferation, cell growth, and apoptosis during normal development or under different disease conditions (Figure 1B).13,15,16 This review focuses on the transcriptional control of leukocyte cell biology by several of the KLF family members. We describe the action of several KLFs in the regulation of T cells, monocyte/macrophages, granulocytes, and B cells and their involvement in homeostatic and disease states. The understanding of these processes will shed light on how leukocyte responses are transcriptionally controlled and will provide new targets for therapeutic manipulation.
KLFs in T-cell biology
T-cell development is a multistep process coordinated by a set of transcription factors activated by signaling pathways involved in hematopoietic and intrathymic differentiation and peripheral T-cell specialization.17,18 Within the thymic microenvironment, precursor cells from the bone marrow encounter signals that ultimately generate functionally distinct types of T cells, such as CD4+ T “helper” cells, including Th1, Th2, Th17, and T regulatory cells (Tregs), CD8+ cytotoxic T cells, NKT cells, αβ- or γδ-T cells, among others.17,18 Several KLF family members, including KLF2, KLF4, KLF10, KLF13, KLF5, and KLF6, have been implicated in the differentiation, activation, quiescence, or homing of various T-cell subsets (Figure 2; Table 1).
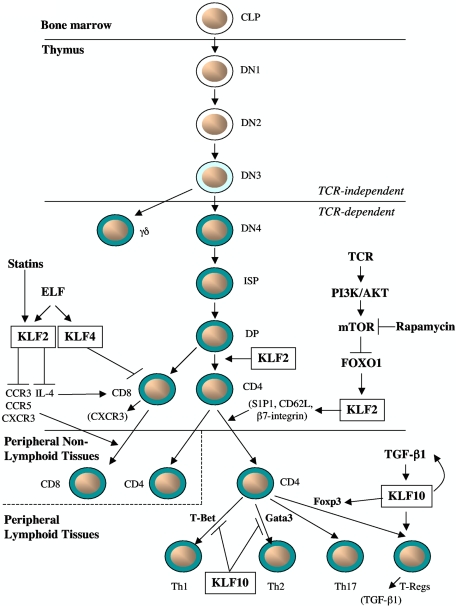
Figure 2.
Role of KLFs in T-cell development, activation, and trafficking. In response to activation of TCR signaling, KLF2 expression is reduced, whereas the immunomodulatory drugs “statins” and rapamycin increase KLF2 expression. KLF2 is required for: (1) maintaining the quiescent state of single-positive CD4+ or CD8+ T cells; (2) induction of T-cell trafficking markers S1P1, CD62L, β7-integrin on thymic CD4+ T cells to allow for egress into peripheral lymphoid tissues; and (3) expression of chemokine receptors, such as CCR3, CCR5, and CXCR3, by autonomously or nonautonomously regulating levels of IL-4 in CD8+ T cells. The Ets transcription factor ELF activates KLF4 expression to negatively regulate naive CD8+ T-cell proliferation and induces KLF2 to promote T-cell homing. In response to TGF-β1, KLF10 expression is induced and promotes Treg cell differentiation by targeting both TGF-β1 and Foxp3 as part of a positive feedback loop; in contrast, KLF10 inhibits Th1 and Th2-mediated pathways. KLF10 also promotes Treg cell suppressor function independent of Foxp3 by increasing the expression of TGF-β1 in Tregs.
Table 1.
Summary of published findings related to Kruppel-like factors in leukocyte biology and disease
| Cell type/factor | Function/observation | References |
|---|---|---|
| T cells | ||
| KLF2 | Expressed in single-positive CD4+ and CD8+ T cells, induced by statins and rapamycin, and rapidly extinguished after T-cell activation via the PI3K/AKT signaling pathway. | 24, 30, 33, 34 |
| Regulates T-cell trafficking by targeting S1P1, CD62L, β7-integrin, and chemokine receptors through both autonomous and nonautonomous mechanisms. | 27, 31, 33, 35–36 | |
| Overexpression in CD8+ effector T cells mimics statins' anti-inflammatory effects and ameliorates CD8+ T cell–mediated myocarditis. | 30 | |
| KLF4 | Expressed in CD8+ T cells where it maintains T-cell quiescence downstream of Ets transcription factor ELF. | 37 |
| Regulates expression of p21 in CD8+ T cells. | ||
| KLF4 promoter is densely methylated in PBMCs from patients with adult T-cell leukemia (ATL) compared with healthy controls. | 42 | |
| Overexpression prevents ATL cell proliferation and induces apoptosis. | ||
| KLF10 | Expressed highly in T regulatory cells than CD4+CD25− T cells. | 45, 46 |
| Induced in response to TGF-β1 and the Itch ubiquitin ligase. | ||
| Targets both TGF-β1 and Foxp3 promoters to promote T reg cell differentiation as part of a positive feedback loop. | 45 | |
| Promotes T reg cell suppressor function independent of Foxp3 by increasing the expression of TGF-β1. | ||
| Inhibits Th1 and Th2 differentiation pathways. | ||
| Adoptive transfer of KLF10−/− T cells accelerates atherosclerosis in ApoE−/− scid/scid mice. | ||
| KLF10−/− 'TGF-β1 in vitro converted T regs fail to suppress airway inflammation. | 46 | |
| Genetic variant in KLF10 3′-UTR associated with type 2 diabetes. | 47 | |
| KLF13 | Expressed in response to T-cell activation. | 48, 49 |
| KLF13−/− mice have T cells with reduced RANTES expression and enlarged thymi and spleen. | ||
| Increases T-cell survival by decreasing BCL-XL. | 50 | |
| KLF5 | Regulates lineage-specific germline TCRβ expression. | 53 |
| Binds to GC-rich area of the TCR Dβ1 promoter. | ||
| KLF6 | Regulates iNOS promoter activity in Jurkat T cells. | 55 |
| Activated by hypoxia, heat shock, or serum starvation. | ||
| Monocyte/macrophage | ||
| KLF4 | Robustly expressed in PBMCs and in a stage-specific manner during myelopoiesis. | 61 |
| Overexpression in CMPs or HSCs induces exclusive monocyte differentiation. | ||
| PU.1 binds to the KLF4 promoter. | ||
| High expression levels of KLF4 and c-Myc allows for prolonged self-renewal of differentiated macrophages deficient in MafB and c-Maf. | 66 | |
| Promotes macrophage activation by: 1) inhibiting TGF-β1 signaling via competition with Smad3 binding for p300/CBP; and 2) regulating expression of HMGB1, a non-histone DNA-binding nuclear protein that serves as a cytokine mediator. | 60, 68 | |
| KLF4−/− fetal liver transplant chimeric mice have nearly depleted circulating inflammatory macrophages in blood and tissues and reduced numbers of resident macrophages. | 62 | |
| Induced by H2O2 in PBMCs from patients with chronic myeloid leukemia. | 69 | |
| KLF2 | Reduced expression in response to LPS and in patients with coronary artery disease; expression increased by statins. | 70, 71 |
| Overexpression represses proinflammatory genes (eg, COX-2, CD40L, MCP-1). | ||
| Inhibits transcriptional activity of NF-κB and AP-1 by recruitment of P/CAF. | ||
| KLF2+/−/ApoE−/− mice develop increased atherosclerosis with increased macrophage lipid uptake and aP2 expression. | 72 | |
| KLF1 | Expressed in macrophages. | 73 |
| Regulates transcription of IL-12p40. | ||
| KLF3 | Expressed in several leukocyte subsets. | 74 |
| KLF3−/− mice develop a myeloproliferative disorder. | ||
| Granulocytes | ||
| KLF4 | Overexpression in CMPs or promyelocytic HL-60 cells inhibits granulocyte differentiation and promotes monocyte differentiation. | 61, 62 |
| KLF4−/− CMPs showed increased granulocyte differentiation and reduced monocyte differentiation in clonogenic assays | 61 | |
| B cells | ||
| KLF4 | Expressed at low levels in pro-B cells; expression increased as cells matured from pre-B cells to mature B cells. Higher expression in naïve mature B cells compared with memory B cells. | 80, 82–83 |
| Regulates cyclin D2. | 82 | |
| FOXO transcription factors target KLF4 promoter in response to BCR activation and PI3K signaling. | 83 | |
| B cell–specific KLF4−/− mice showed either: | ||
| (1) Modest decrease in pre-B cells in bone marrow and mature B cells in spleen; and (2) no differences in B cell development, function, or activation. | 82 | |
| Expressed at low levels in human B cell lymphomas | 83 | |
| Overexpression blocks transformation of pre-B cell by BCR-ABL and depletes leukemic pre-B cells in vivo. | 84 | |
| Imatinib and KLF4 additively induce apoptosis in BCR-ABL transformed pro/pre-B cells. | ||
| KLF9 | Expressed higher in naïve B cells than in memory B cells. | 80 |
| Overexpression reduces B cell proliferation in memory B cells and induces the naïve B cell phenotype. | ||
| KLF2 | Expressed strongly after pre-BCR activation and in resting pre-B cells. | |
| Overexpression increases survival of anti-IgM and anti-CD40 stimulated memory B cells. | 81 | |
| Highly expressed in blood antibody-secreting cells (ASCs) along with S1P1, which can effect IgG plasma cell homing. | 86 |
KLF2
KLF2 (lung Krüppel-like factor) is expressed in lung, endothelial cells, and lymphocytes.19–24 KLF2 deficiency is embryonic lethal because of severe hemorrhage resulting from defects in blood vessel integrity.19 In T-cell biology, KLF2 has been demonstrated to regulate T-cell properties of quiescence, survival, activation, and migration.22,25–28
KLF2 was initially found in T cells to maintain the quiescent state of single-positive CD4+ or CD8+ T cells.22,25 KLF2 also inhibits Jurkat T-cell growth by inhibiting DNA synthesis.29 Mechanistically, both activation and inhibitory domains of KLF2 are required to suppress Jurkat T-cell proliferation. In addition, KLF2 can enhance the promoter activity of the cell-cycle inhibitor p21CIP by binding to a GC-rich, Sp1-3-binding site.29 Given that KLF2 is rapidly extinguished after T-cell activation and it directly regulates IL-2 expression in the early stage of T-cell activation, KLF2 has been implicated in playing a role in T-cell activation.22,27,30 For example, HMG-CoA reductase inhibitors (statins) diminished T-cell proliferation and interferon-γ (IFN-γ) expression and are dependent, at least in part, on KLF2 expression; furthermore, forced overexpression of KLF2 in CD8+ effector T cells ameliorated a mouse model of CD8+ T cell-mediated myocarditis.31 Moreover, KLF2 may exert effects on T-cell survival. In the absence of KLF2, mature and single-positive CD4+ or CD8+ T cells have been shown to exist in an active state and are prone to apoptosis.22 In contrast to these findings, Carlson et al observed that KLF2−/− thymocytes displayed an alternative phenotype, independent of cell apoptosis, and characterized by reduced expression of critical T-cell trafficking molecules, including the sphingosine-1-phosphate receptor (S1P1), CD62L, and β7-integrin.28 KLF2 can bind and transactivate the S1P1 promoter, which is critically associated with thymocyte emigration and recirculation through peripheral organs.28 Indeed, KLF2-deficient mice phenocopy S1P1 mice and bear a profound lymphopenia in the periphery.22,28 Similar emigration defects were recently observed in KLF2-deficient γδ T cells.32 Although the emigration defects identified by Carlson et al28 may appear in conflict with the T-cell apoptosis defects identified by Kuo et al,22 variation in generation of model systems could potentially underlie differences observed: injection of KLF2−/− ES cells into Rag-2−/− blastocysts22 versus adoptive transfer of embryonic day 12.5 fetal livers from KLF2−/− donors into irradiated Rag2−/− recipients.28 Furthermore, earlier time points used for analyses may not be able to exclude long-term effects on T-cell activation or survival; thus, KLF2 may play dual roles on T-cell emigration and survival.28 Interestingly, environmental exposure to pollutants, such as dioxin, promote premature expression of KLF2 in the early stages of thymocyte cell development that precede acute cell loss and thymus atrophy, an effect that is dependent on the aryl hydrocarbon receptor.33 Dioxin exposure alters the expression of several KLF2 target genes, including S1P1, β7-integrin, and cyclin-dependent kinase inhibitor 2D, which may underlie the dioxin-associated toxicity of the immune system.33 In resting peripheral T cells, the FOXO1 transcription factor also controls T-cell homing by activating CD62L, in part, by binding to the KLF2 promoter and inducing KLF2 expression,34 effects that are potentiated by the mTOR inhibitor, rapamycin, and reversed in response to T-cell receptor (TCR) activation.35
Alternative roles for KLF2 in T-cell trafficking have been proposed. Sebzda et al demonstrated that KLF2-deficient CD4+ T cells possessed increased proinflammatory chemokine receptors, such as CXCR3, CCR3, and CCR5. Up-regulation of these receptors would be expected to allow for KLF2-deficient T cells to egress from the thymus and enter nonlymphoid sites in response to inflammatory chemokines.36 However, Weinreich et al proposed a different mechanism using conditional KLF2-deficient mice that were crossed with CD4-Cre mice. These mice demonstrated that KLF2-deficient CD4+ T cells produce higher levels of IL-4, leading to the induction of the chemokine receptor CXCR3 on neighboring cells, including the CD8+ T cells.37 These findings suggest that KLF2 may possess both autonomous and nonautonomous effects on inducing chemokine receptor expression and highlight that KLF2 may be able to control bystander cell phenotypes, such as naive T cells. Finally, the upstream Ets transcription factor ELF4 is known to regulate the proliferation and homing of CD8+ T cells, in part, by reducing expression of KLF2, CCR7, and CD62L, placing KLF2 within a transcriptional hierarchy regulating CD8+ T-cell homeostasis.38
KLF4
KLF4 (gut Krüppel-like factor) was initially identified in the epithelial lining of the gut and skin.39 Indeed, KLF4 knockout mice exhibit abnormal epidermal differentiation and, as a consequence, die within hours after birth40; in addition, KLF4 has been found to be essential for colonic goblet cell differentiation.41 KLF4 has also been implicated as both a tumor suppressor and an oncogene.42 In adult T-cell leukemia (ATL), Yasunaga et al demonstrated that the KLF4 promoter possesses dense DNA methylation, whereas in normal peripheral blood mononuclear cells (PBMCs) there was little methylation, suggesting that KLF4 methylation status may prognosticate disease severity.43 Increasing methylation of KLF4 corresponded to carrier status and ATL onset. Moreover, treatment of cells with demethylating agents was able to rescue the expression of KLF4 transcripts. Forced overexpression of KLF4 induced apoptosis of these leukemic cells,43 an effect that highlighted KLF4's role as a tumor suppressor in the context of ATL. KLF4 and p21CIP expression was also found reduced in CD8+ T cells deficient in the Ets transcription factor ELF4, an effect that promotes CD8+ T-cell hyperproliferation.38 Mechanistically, ELF4 directly targeted the KLF4 promoter, suggesting that KLF4 functions to maintain T-cell quiescence downstream of ELF4, possibly by activating p21CIP in CD8+ T cells.38
KLF10
KLF10 (transforming growth factor β [TGF-β]-inducible early gene 1 or Tieg1) was first identified by the Spelsberg laboratory in human osteoblasts and classified as a TGF-β-responsive gene.44 Indeed, gene targeting studies in mice revealed that KLF10 plays a critical role in osteoblast-mediated mineralization and osteoclast differentiation.45 Emerging evidence has further unveiled novel roles for KLF10 in T-cell biology.46,47 It is well known that Tregs play a critical role in the maintenance of self-tolerance and immune suppression. Foxp3 has been considered a definitive marker for Tregs. Recently, Venuprasad et al showed that the E3 ubiquitin ligase Itch regulates the conjugation of ubiquitin to KLF10 and cooperates with KLF10 to regulate in vitro expression of Foxp3.47 The authors also demonstrated that “TGF-β converted” Tregs from CD4+CD25− T cells of KLF10−/− mice failed to suppress airway inflammation in vivo. Recent studies by our laboratory also revealed distinct roles for KLF10 during Treg cell differentiation and function.46 Specifically, CD4+CD25− T cells overexpressing KLF10 induced both TGF-β1 and Foxp3 expression, and decreased T-bet (Th1 lineage marker) and GATA3 (Th2 lineage marker) expression. Consistent with this finding, CD4+CD25− T cells from KLF10−/− mice showed hyperactivity characterized by enhanced differentiation along Th1 and Th2 pathways, elaboration of high levels of Th1 and Th2 cytokines, and loss of suppression by wild-type (WT) Tregs. Importantly, CD4+CD25+ Tregs from KLF10−/− mice exhibited reduced suppressor function with decreased expression and secretion of TGF-β1. Notably, KLF10−/− CD4+CD25+ Tregs have impaired suppressor function independent of Foxp3 expression, an effect that was able to be completely rescued by exogenous administration of TGF-β1. Finally, adoptive transfer of KLF10−/− CD4+CD25− T cells in atherosclerotic-prone ApoE−/−/scid/scid mice accelerated atherosclerotic lesion formation, compared with transfer of WT CD4+CD25− T cells.46 Mechanistically, we demonstrated that KLF10 can transactivate both TGF-β1 and Foxp3 promoters in response to TGF-β1 exposure, ultimately promoting and maintaining Treg cell function. Indeed, KLF10−/− mice possess reduced numbers of peripheral Tregs.46,47 Taken together, we have found that KLF10 is an important regulator of CD4+CD25− T-cell activation, Treg cell differentiation, and Treg cell suppressor function46; collectively, these effects are likely to be important not only to our understanding of atherosclerotic pathogenesis, but also that of other chronic inflammatory or autoimmune diseases. Interestingly, a genetic variant of KLF10 (in the 3′UTR+ 1002 A > C) was modestly associated with type 2 diabetes patients, compared with normoglycemic, healthy controls.48
KLF13
In contrast to KLF2, KLF13 is induced in response to T-cell activation. KLF13, also known as RANTES Factor of Late Activated T Lymphocytes-1, was first identified through expression cloning as a transactivator for the chemokine RANTES, which promotes T-cell activation. It was determined that the intact CTCCC box on the RANTES promoter is necessary for RANTES Factor of Late Activated T Lymphocytes-1-mediated RANTES transcription.49,50 Recently, Zhou et al reported that KLF13 is able to increase RANTES expression and promote survival by decreasing BCL-XL expression.51 Activated T lymphocytes in KLF13−/− mice exhibited decreased RANTES expression. KLF13−/− mice also presented increased lymphoid cells that were characterized by enlarged thymi and spleens, an effect that might be the result of decreased apoptosis.51 KLF13 was found to transcriptionally regulate RANTES expression by recruiting a large trans-acting complex to the RANTES promoter composed of MAPK Nemo-like kinase, p300/CBP, PCAF, and the ATPase BRG-1.52 In contrast, KLF13 has been shown to inhibit transcription of BCL-XL by deacetylating histones mediated by corepressors.51 These findings suggest that KLF13 may play a dual role in inflammation by which it may enhance the immune response via inducement of leukocyte recruitment, including lymphocytes, monocytes, and eosinophils, and deactivating this response via apoptosis.
KLF5
KLF5 is also known as BTEB-2 or IKLF. Other than its roles in cardiovascular remodeling and angiogenesis,53 KLF5 has been shown to be involved in modulating lineage-specific germline TCR-β expression.54 At the TCR-β locus, the Eβ enhancer and the Dβ1 promoter regulate germline transcription near the TCR Dβ1 gene segment. KLF5 can bind to a GC-rich area of the Dβ1 promoter to transactivate gene expression, whereas a dominant-negative KLF5 construct inhibited Dβ1 promoter activity. Future in vivo studies are required to verify the roles of KLF5 in regulating TCR-β expression and in T-cell lineage development.
KLF6
KLF6 has been shown to regulate the expression of multiple genes and is associated with wound healing and suppression of tumorigenesis.55 KLF6 was found to induce iNOS promoter activity in Jurkat T cells, primary T lymphocytes, and COS-7 cells under a variety of pathophysiologic conditions, such as hypoxia, heat shock, and serum starvation.56 Nitric oxide (NO) production after iNOS activation is also involved in would healing and tissue repair.57 Moreover, forced expression of iNOS and NO is known to have antitumor effects.58 Taken together, it is possible that KLF6 transactivation of the iNOS promoter may explain, at least in part, the common biologic effects of KLF6 and NO. Additional studies involving KLF6-deficient mice will help validate the role of KLF6 in T cell-mediated NO production and its effects on tumorigenesis.
KLFs in monocyte/macrophage biology
Monocytes/macophages play crucial roles in both innate and adaptive immunity and are actively involved in the pathogenesis of acute (eg, sepsis) and chronic inflammatory diseases (eg, atherosclerosis, rheumatoid arthritis, and inflammatory bowel disease) and in cancer.59,60 KLF4, KLF2, KLF1, and KLF3 have been reported to be associated with monocyte/macrophage differentiation or activation (Figure 3; Table 1).
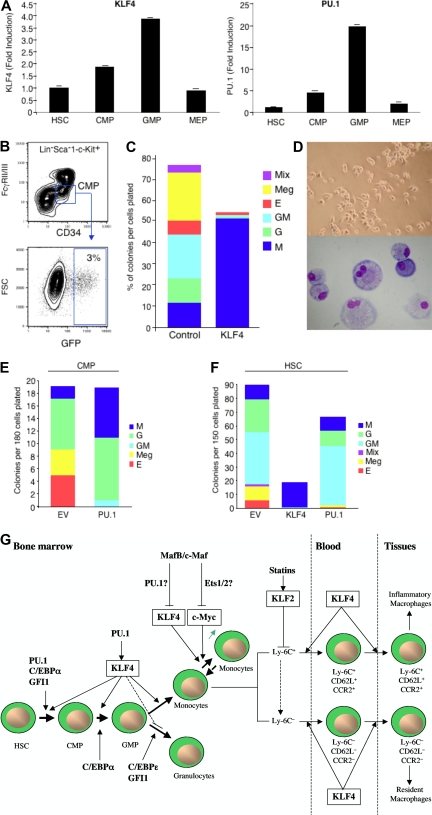
Figure 3.
Role of KLFs in myeloid development, renewal, and activation. (A) KLF4 and PU.1 are expressed in a stage-specific pattern during myelopoiesis (adapted with permission; see Feinberg et al62). (B-F) Overexpression of KLF4 in CMPs or HSCs promotes exclusive monocyte differentiation, whereas PU.1 promotes both monocytic and granulocytic differentiation (adapted with permission; see Feinberg et al62). (G) Schematic overview of KLFs in monocyte biology. KLF4 is a downstream target gene of PU.1 that promotes monocyte differentiation from hematopoietic stem cell progenitors. High expression levels of KLF4 and c-Myc enable differentiated monocytes/macrophages capable of self-renewal, an effect regulated by MafB and c-Maf and potentially mediated by their respective downstream targets, PU.1 and Ets1/2. Statins induce KLF2, which can repress genes involved in macrophage activation. KLF4 promotes the development of inflammatory monocytes (Ly-6C+, CD62L+, CCR2+) in blood and tissues and, to a lesser extent, resident macrophages in tissues (Ly6C−, CD62L−, CCR2−).
KLF4
Recent studies by our group and others have demonstrated that KLF4 is a critical regulator of monocyte commitment, differentiation, and macrophage activation.61–63 KLF4 is expressed in a monocyte-restricted and stage-specific pattern during myelopoiesis.62 Forced expression of KLF4 in promyelocytic HL-60 cells confers the phenotype of mature monocytes characterized by highly induced expression of the myeloid markers, CD11b and CD14, and accompanied by monocytic morphologic changes. Moreover, clonogenic analyses of KLF4 overexpression in primary common myeloid progenitors (CMPs) or hematopoietic stem cells (HSCs) resulted in nearly exclusive monocyte differentiation (Figure 3A-F). Conversely, KLF4-deficiency inhibited monocyte, but increased granulocyte, differentiation from these progenitor cells.62 Alder et al demonstrated that “KLF4−/− chimeras,” generated by transplantation of KLF4−/− fetal liver cells into lethally irradiated WT mice, experienced markedly impaired monocyte differentiation, as evidenced by reduced numbers of monocytic cells in bone marrow, resident monocytes (CD115+Gr1−) in spleen, and the near absence of circulating inflammatory monocytes (CD115+Gr1+) in blood.63 Mechanistic studies verified that intact KLF4 was able to markedly transactivate the monocytic-specific CD14 promoter by binding to KLF consensus sites.62 Furthermore, KLF4 was identified as a downstream target gene of PU.1,62 an important Ets transcription factor for specifying progenitor cell fate along several lineages, including macrophages, granulocytes, B and T cells, and NK cells.64,65 Collectively, these findings place KLF4 in a transcriptional hierarchy for specifying myeloid cell lineage fate along the monocyte pathway. In contrast, PU.1 may target the CCAAT enhancer-binding protein-α (C/EBPα) to regulate granulocytic pathway differentiation.66 Finally, recent work by Aziz et al demonstrated that combined deficiency of MafB and c-Maf allowed for self-renewal of differentiated functional macrophages, an effect dependent on the induction of 2 pluripotent stem cell-inducing factors, KLF4 and c–Myc.67 Indeed, combined overexpression of KLF4 and c-Myc enabled serial replating capacity to in vitro differentiated WT macrophages. MafB and c-Maf-deficient macrophages possessed higher expression levels of PU.1,67 the myeloid activator of KLF4,62 suggesting that KLF4 may participate as a downstream regulator of both macrophage differentiation and renewal.
KLF4 is not only associated with monocyte differentiation, but also macrophage activation. KLF4 can be markedly induced in macrophages by proinflammatory stimuli, including lipopolysaccharide (LPS), IFN-γ, and tumor necrosis factor-α, and can be inhibited by the anti-inflammatory stimulus TGF-β1.61 Other studies have also demonstrated that KLF4 is an IFN-γ target gene in carcinoma cell lines, an effect mediated by Stat1.68 KLF4 overexpression in J774a macrophage cell line induced the macrophage activation marker, iNOS, and inhibited TGF-β1 and Smad3 target gene plasminogen activator inhibitor-1. Conversely, KLF4 knockdown alleviated iNOS induction by IFN-γ and/or LPS, whereas it enhanced the responsiveness to TGF-β1 and Smad3 signaling.61 Mechanistically, KLF4 can transactivate the iNOS promoter via KLF binding sites and interact with the nuclear factor-κB (NF-κB) family member p65. In contrast, KLF4 has been shown to inhibit TGF-β1 and Smad3-mediated plasminogen activator inhibitor-1 induction by an antagonistic competition with Smad3 for the coactivator p300/CBP.61 In the RAW264.7 macrophage cell line, KLF4 has also been demonstrated as capable of regulating the high-mobility group box 1 nonhistone DNA-binding nuclear protein that serves as a cytokine mediator of various systemic and local inflammatory disease states.69 Finally, Alder et al demonstrated that “KLF4−/− chimeras” were nearly devoid of circulating inflammatory monocytes (CD115+Gr1+),63 which underscore a critical role for KLF4 in both monocyte differentiation and activation. Future studies that explore overexpression or deficiency of KLF4 in a macrophage-specific manner will help further define the role of this particular KLF in specific monocytic populations in acute and chronic inflammation.
Oxidative stress is hypothesized to be a contributory factor to the success of chemotherapeutic treatment for leukemias. A recent study by Li et al demonstrated that in response to hydrogen peroxide, KLF4 expression is induced in PBMCs from patients with chronic myeloid leukemia.70 Furthermore, overexpression of KLF4 in these PBMCs reduced cell growth and induced apoptosis, as evidenced by induction of bax and inhibition of bcl-2. In contrast, inhibition of KLF4 had the opposite effect, implicating KLF4 as an important potential regulator of chemotherapy-induced reactive oxygen species-mediated apoptosis.
KLF2
KLF4 and KLF2 are phylogenetically related KLF family members.11–13 KLF2 is also expressed in monocytes; however, accumulating studies have revealed an anti-inflammatory role for KLF2 in these cells.22,71 Das et al demonstrated that KLF2 expression is reduced on LPS stimulation in the THP-1 monocyte-like cell line.71 Moreover, KLF2 expression was reduced approximately 30% in PBMCs from patients with coronary artery disease, compared with age-matched healthy controls. Forced expression of KLF2 in the monocyte cell line J774a inhibited monocytic activation, characterized by attenuated proinflammatory gene expression, such as COX-2, CD40L, IL-1β, tumor necrosis factor-α, and MCP-1. In contrast, KLF2 knock-down led to enhanced gene expression of MCP-1 and COX-2. In mouse foot pads, overexpression of KLF2 inhibited carrageenan-induced inflammation. Mechanistic studies have revealed that KLF2 inhibited the transcriptional activity of both NF-κB and AP-1, in part via the recruitment of the transcriptional coactivator p300/CBP-associated factor.71 Moreover, it has been shown by Tuomisto et al that the HMG-CoA reductase inhibitor simvastatin exerts anti-inflammatory effects on macrophages specifically by increasing KLF2 expression.72 Finally, macrophages from heterozygous KLF2-deficient/ApoE−/− mice showed increased lipid uptake and expression of the lipid chaperone ap2/FABP4, effects associated with increased atherosclerosis in these mice.73 Collectively, these studies support the notion that KLF2 is also implicated in regulating macrophage activation.
KLF1 and KLF3
Until recently, KLF1 was thought to be expressed exclusively in red blood cells; however, a recent study by Luo et al has identified KLF1 expression in macrophages.74 Furthermore, KLF1 was found to differentially modulate transcription of IL-12 p40. Given that IL-12p40 is an important factor in the activation of innate immunity and development of Th1 responses, KLF1 might play a functional role in inflammation. KLF3 (basic Krüppel-like factor) was the first KLF member identified as being expressed in myeloid cells. Interestingly, KLF3-deficient mice present with a myeloproliferative disorder.75 The underlying mechanism for KLF3-mediated effects remains to be elucidated; however, reduced expression of the SHP-1 phosphatase gene in KLF3−/− mice has indicated the possible involvement of related pathway(s).75
KLFs in granulocyte biology
Granulocytes possess both overlapping and complementary functional roles with macrophages in their orchestration of the innate and adaptive immune response.59,60 Accumulating studies have demonstrated that granulocytes and macrophages originate from common early bone marrow precursors called CMPs and granulocyte/macrophage progenitors.76,77 However, further differentiation and function into mature granulocytes involve the participation of a distinct set of transcription factors. There are only a few reports detailing the roles of KLFs in granulocyte biology. Forced overexpression of KLF4 in CMPs was found to promote near-exclusive monocyte differentiation, whereas KLF4-deficient CMPs displayed reduced monocytes and increased granulocytes in clonogenic assays.62 These findings were also verified in the bipotential promyelocytic HL-60 cell line, in which forced overexpression of KLF4 blocked all-trans-retinoic acid granulocytic differentiation.63 Thus, KLF4 may promote monocyte differentiation and actively inhibit granulocytic differentiation. Further studies will be required to elucidate the molecular mechanisms by which this occurs and to validate effects on granulopoiesis in vivo. For example, it will be of interest to examine whether KLF4 inhibits CCAAT enhancer binding protein (eg, C/EBP-ϵ) expression or function important for terminal granulocyte differentiation (Figure 3; Table 1).
KLFs in B cells
Differentiation of B cells into antibody-secreting cells (ASCs) is key to humoral immune response. Exposure of naive B cells to a foreign antigen activates B-lymphocytes and generates either plasma cells or memory B cells. plasma cells can secrete large quantities of neutralizing antibody, whereas memory B cells, which can be activated rapidly and persist over long periods of time, mediate the long-term immune response.78
There are several transcription factors that are important in B-cell commitment and differentiation. For example, deletion of either Ikaros or PU.1 leads to an arrest of HSC differentiation into common lymphoid progenitors, an effect associated with the absence of B cell-specific markers.64,79 The transcription factors E box binding protein 2A, early B cell factor-1, and paired box5 are each necessary for the early commitment to the B-cell lineage.79 Paired box5, in particular, has been implicated in the maintenance of B-cell identity. In contrast, the transcription factor FOXO1 plays critical roles at later stages of B-cell development, including the early pro-B, pre-B, and transitional B-cell compartments.80 Accumulating studies have implicated KLF2, KLF4, and KLF9 in different stages of B cell development and function, as detailed below (Figure 4; Table 1).81,82
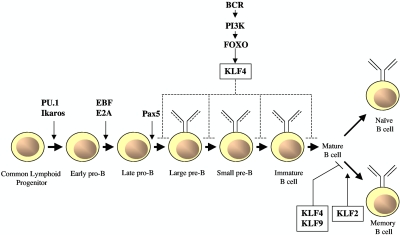
Figure 4.
Role of KLFs in B-cell differentiation and activation. In response to activation of the BCR, FOXO transcription factors target KLF4 which, on overexpression, inhibits B-cell proliferation and promotes G1 cell-growth arrest. Conversely, B cell–specific, KLF4-deficient mice have displayed modest differentiation defects of pre-B cells in bone marrow and mature B cells in spleen; however, no differences were observed in these types of B cells using a similar conditional knockout model.84 KLF4 or KLF9 overexpression reduces the number of proliferating memory B cells, and their behavior resembles that of naive B cells. In contrast, KLF2 increases the survival of anti-IgM and anti–CD40-stimulated memory B cells.
KLF4 is expressed at low levels in pro-B cells and its expression increases as the cells mature into precursor (pre)-B cells and resting mature B cells.83 Mice deficient for KLF4 in their B-cell populations experienced a defect in B-cell receptor (BCR)-mediated proliferation response because of the inability of KLF4 to bind to the cyclin D2 promoter and regulate its expression.83 In addition to the defects in proliferation, Klaewsongkram et al found that B cell-specific, KLF4-deficient mice possessed a modest decrease in the number of pre-B cells in bone marrow and mature B cells in the spleen but had no change in number of progenitor (pro)-B cells in the bone marrow.83 However, in contrast to the findings observed by Klaewsongkram et al,83 another group using a similar B cell-specific KLF4-deficient mouse model found no differences in B-cell development, function, or activation.84 The discrepancies between these 2 antithetical findings may be the result of differences between genetic backgrounds of the mice, or environmental or experimental conditions, or may indicate a redundant role for KLF4 in maintaining B-cell quiescence.84 Regardless, overexpression of KLF4 negatively regulates B-cell proliferation. Furthermore, KLF4 was identified as a FOXO1 target gene that was able to induce G1 cell-cycle arrest in proliferating B-cell blasts.84
In addition to KLF4, KLF2 plays a role in B-cell development. A genome-wide expression profiling in pro-B cells from a Rag-2 deficient mouse, where the expression of pre-BCR as well as pre-BCR-mediated clonal expansion can be controlled, identified KLF2 and its target S1P1 to be important in pre-B cell clonal expansion and migration. Both KLF2 and S1P1 were strongly expressed after pre-BCR induction, and the highest expression levels were observed in freshly isolated primary small resting pre-B cells.85
KLFs are also important in secondary antibody responses. A study by Good and Tangye81 showed that memory B cells, which are rapidly activated during secondary responses (for review, see McHeyzer-Williams and McHeyzer-Williams78), express low levels of KLF4 and KLF9 compared with naive B cells.81 Overexpression of KLF4 or KLF9 reduced the number of B cells being recruited into division and delayed their entry into division.81 In addition, overexpression of either KLF4 or KLF9 converted memory B cells into cells with a “naive” B-cell phenotype, an effect consistent with lower expression levels of these KLFs in memory B cells compared with naive B cells.81 The mechanism of the regulation of B-cell number through KLF4 and KLF9 is not clear but is thought to be independent of p21CIP in this model system. Because cyclin D1 is activated by NF-κB and KLF4 can repress cyclin D1 expression in other cell types,62 one possibility is that KLF4 and/or KLF9 may interact with NF-κB downstream of CD40 and BCR signaling to regulate B-cell proliferation.81
Another KLF family member that may participate in secondary antibody responses is KLF2. KLF2 is expressed at higher levels in memory B cells, compared with their germinal center precursors, and may be important in memory B-cell differentiation. Unlike KLF4 and KLF9, overexpression of KLF2 does not decrease the proliferative capacity of the memory B cells but rather increases the survival of anti-IgM and anti–CD40-stimulated B cells.82 In contrast, overexpression of KLF3 or KLF9 had no effect on antigen receptor and CD40-engaged B cells. Collectively, these studies implicate KLF2 as part of a transcriptional program in memory B cells that allows them to survive at higher frequencies and to respond to antigenic reexposure, effects important to the acquisition of lifelong resistance to pathogens.82 KLF2 is also expressed in high levels in blood ASCs, a finding that raises the possibility that KLF2, through S1P1, may regulate plasma cell homing and determine whether IgG ASCs leave the spleen and accumulate in the bone marrow.86
In addition to their role in the development and differentiation of normal B cells, KLFs are also implicated in B-cell malignancies. For example, Kharas et al identified KLF4 as a tumor suppressor gene in B-cell malignancies.87 It has been found to be expressed at low levels in several types of human B-cell lymphomas, B-cell lines transformed by ABL oncogenes, and leukemias; furthermore, overexpression of KLF4 can inhibit proliferation and induce apoptosis.87 It has been postulated that the mechanism of this regulation occurs through increasing the expression of p21CIP and decreasing the expression of c-Myc and cyclin D2. Furthermore, in vivo leukemogenesis studies showed that KLF4 can block transformation of pre-B cells by BCR-ABL.87 Finally, treatment with the tyrosine kinase inhibitor imatinib to KLF4 overexpressing, BCR-ABL-transformed pro/pre-B cells generated an additive effect on cell death, suggesting that imatinib- and KLF4-mediated apoptosis can occur in parallel or by nonredundant pathways.87 Future studies are required to clarify the relative transcriptional hierarchy of KLFs in relation to other critical nodal factors that regulate key aspects of normal and aberrant B-cell differentiation and function.
In conclusion, the studies discussed here reveal that KLFs can have profound effects on cell fate decisions, differentiation, and function in several leukocyte subsets in vitro and in vivo. As such, these observations bear relevancy for leukocyte development and dysfunction in both health and disease and provide the rationale for using KLFs as potential therapeutic targets. Nonetheless, a number of critical issues remain. For example, within a given leukocyte cell type, multiple KLFs may be expressed yet they may function quite differently despite binding to similar CACCC sites in the promoter region of target genes. This raises several intriguing possibilities that may dominate the KLF mechanism of action: (1) accessibility to a KLF DNA-binding site may be governed as much by the adjacent flanking sequence and neighboring DNA-binding sites of a promoter as the expression levels of the KLF within the cell; (2) the presence of KLF interacting proteins (including other KLF family members) in a cell may switch the function of a KLF from a positive to a negative regulator; (3) chromatin remodeling or post-translational modifications of leukocyte KLFs may provide another layer of gene regulation (eg, Oishi et al identified that on “deSUMOylation” of KLF5, it is converted from a repressor to a transcriptional activator of energy metabolism88); and (4) the temporal expression pattern of KLFs in response to various cytokines, growth factors, or stages of differentiation may influence the functional response in a given leukocyte subset or tissue, an effect that may either promote, inhibit, or sustain a specific stimulus or signaling pathway. In addition, correlating and contrasting KLF structure-function relationships in leukocyte biology may enable an improved understanding of their physiologic and pathologic functions. The roles of KLFs in other leukocyte subsets, such as dendritic cells, may shed light on the induction of effective antigen-specific immunity to pathogens. Further insights into the function of leukocyte KLFs and their target genes will reveal new mechanisms that will explain how these factors control leukocyte development, growth, and activation and might ultimately be manipulated to treat disorders, such as acute and chronic inflammation, autoimmunity, or leukemia.
Acknowledgments
This work was supported by the National Institutes of Health (grants HL080174 and HL091076, M.W.F.; grant F32HL086157, Z.C.; and grant F32HL088819, A.K.W.), the American Cancer Society (Research Scholar Grant RSG0719501-LIB, M.W.F.), the Boston Area Diabetes Endocrinology Research Center (Pilot Grant P30DK057521, M.W.F.), and a Cardiovascular Medicine Discovery Award (M.W.F.).
Authorship
Contribution: Z.C., X.S., B.I., A.K.W., and M.W.F. reviewed the literature and each wrote portions of the paper and figures; and M.W.F. had overall editorial responsibility for the manuscript and figures.
Conflict-of-interest disclosure: M.W.F. and the Brigham and Women's Hospital have a patent pending on the use of KLF10 as a therapeutic target and diagnostic. The remaining authors declare no competing financial interests.
Correspondence: Mark W. Feinberg, Department of Medicine, Cardiovascular Division, Brigham and Women's Hospital, Harvard Medical School, 77 Avenue Louis Pasteur, NRB-742F, Boston, MA 02115; e-mail: mfeinberg@rics.bwh.harvard.edu.
References
- 1.Wieschaus E, Nusslein-Volhard C, Kluding H. Kruppel, a gene whose activity is required early in the zygotic genome for normal embryonic segmentation. Dev Biol. 1984;104(1):172–186. doi: 10.1016/0012-1606(84)90046-0. [DOI] [PubMed] [Google Scholar]
- 2.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 3.Preiss A, Rosenberg UB, Kienlin A, Seifert E, Jackle H. Molecular genetics of Kruppel, a gene required for segmentation of the Drosophila embryo. Nature. 1985;313(5997):27–32. doi: 10.1038/313027a0. [DOI] [PubMed] [Google Scholar]
- 4.Jackle H, Rosenberg UB, Preiss A, et al. Molecular analysis of Kruppel, a segmentation gene of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1985;50:465–473. doi: 10.1101/sqb.1985.050.01.058. [DOI] [PubMed] [Google Scholar]
- 5.Schuh R, Aicher W, Gaul U, et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell. 1986;47(6):1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 6.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13(5):2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieker JJ, Southwood CM. The erythroid Kruppel-like factor transactivation domain is a critical component for cell-specific inducibility of a beta-globin promoter. Mol Cell Biol. 1995;15(2):852–860. doi: 10.1128/mcb.15.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375(6529):316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 9.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375(6529):318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 10.van Vliet J, Crofts LA, Quinlan KG, Czolij R, Perkins AC, Crossley M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87(4):474–482. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Turner J, Crossley M. Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci. 1999;24(6):236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- 12.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4(2):206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg MW, Lin Z, Fisch S, Jain MK. An emerging role for Kruppel-like factors in vascular biology. Trends Cardiovasc Med. 2004;14(6):241–246. doi: 10.1016/j.tcm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Bieker JJ. Isolation, genomic structure, and expression of human erythroid Kruppel-like factor (EKLF). DNA Cell Biol. 1996;15(5):347–352. doi: 10.1089/dna.1996.15.347. [DOI] [PubMed] [Google Scholar]
- 15.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Kruppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40(10):1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T, Aizawa K, Matsumura T, Nagai R. Vascular implications of the Kruppel-like family of transcription factors. Arterioscler Thromb Vasc Biol. 2005;25(6):1135–1141. doi: 10.1161/01.ATV.0000165656.65359.23. [DOI] [PubMed] [Google Scholar]
- 17.O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327(5969):1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taghon T, Rothenberg EV. Molecular mechanisms that control mouse and human TCR-alphabeta and TCR-gammadelta T cell development. Semin Immunopathol. 2008;30(4):383–398. doi: 10.1007/s00281-008-0134-3. [DOI] [PubMed] [Google Scholar]
- 19.Wani MA, Means RT, Jr, Lingrel JB. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 1998;7(4):229–238. doi: 10.1023/a:1008809809843. [DOI] [PubMed] [Google Scholar]
- 20.Wani MA, Wert SE, Lingrel JB. Lung Kruppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J Biol Chem. 1999;274(30):21180–21185. doi: 10.1074/jbc.274.30.21180. [DOI] [PubMed] [Google Scholar]
- 21.Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11(22):2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo CT, Veselits ML, Leiden JM. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival [see comments]. Science. 1997;277(5334):1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 23.Dekker RJ, van Soest S, Fontijn RD, et al. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2). Blood. 2002;100(5):1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 24.SenBanerjee S, Lin Z, Atkins GB, et al. KLF2 is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199(10):1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc-dependent pathway. Nat Immunol. 2001;2(8):698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- 26.Di Santo JP. Lung Krupple-like factor: a quintessential player in T cell quiescence. Nat Immunol. 2001;2(8):667–668. doi: 10.1038/90598. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Lingrel JB. Kruppel-like factor 2, a novel immediate-early transcriptional factor, regulates IL-2 expression in T lymphocyte activation. J Immunol. 2005;175(5):3060–3066. doi: 10.4049/jimmunol.175.5.3060. [DOI] [PubMed] [Google Scholar]
- 28.Carlson CM, Endrizzi BT, Wu J, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442(7100):299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 29.Wu J, Lingrel JB. KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene. 2004;23(49):8088–8096. doi: 10.1038/sj.onc.1207996. [DOI] [PubMed] [Google Scholar]
- 30.Schober SL, Kuo CT, Schluns KS, Lefrancois L, Leiden JM, Jameson SC. Expression of the transcription factor lung Kruppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. J Immunol. 1999;163(7):3662–3667. [PubMed] [Google Scholar]
- 31.Bu DX, Tarrio M, Grabie N, et al. Statin-induced Kruppel-like factor 2 expression in human and mouse T cells reduces inflammatory and pathogenic responses. J Clin Invest. 2010;120(6):1961–1970. doi: 10.1172/JCI41384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odumade OA, Weinreich MA, Jameson SC, Hogquist KA. Kruppel-like factor 2 regulates trafficking and homeostasis of gammadelta T cells. J Immunol. 2010;184(11):6060–6066. doi: 10.4049/jimmunol.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMillan BJ, McMillan SN, Glover E, Bradfield CA. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induces premature activation of the KLF2 regulon during thymocyte development. J Biol Chem. 2007;282(17):12590–12597. doi: 10.1074/jbc.M611446200. [DOI] [PubMed] [Google Scholar]
- 34.Fabre S, Carrette F, Chen J, et al. FOXO1 regulates L-Selectin and a network of human T-cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181(5):2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 35.Sinclair LV, Finlay D, Feijoo C, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9(5):513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9(3):292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 37.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31(1):122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada T, Park CS, Mamonkin M, Lacorazza HD. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat Immunol. 2009;10(6):618–626. doi: 10.1038/ni.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271(33):20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22(4):356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 41.Katz JP, Perreault N, Goldstein BG, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129(11):2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7(11):1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 43.Yasunaga J, Taniguchi Y, Nosaka K, et al. Identification of aberrantly methylated genes in association with adult T-cell leukemia. Cancer Res. 2004;64(17):6002–6009. doi: 10.1158/0008-5472.CAN-04-1422. [DOI] [PubMed] [Google Scholar]
- 44.Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23(23):4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramaniam M, Gorny G, Johnsen SA, et al. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol Cell Biol. 2005;25(3):1191–1199. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao Z, Wara AK, Icli B, et al. Kruppel-like factor KLF10 targets transforming growth factor-beta1 to regulate CD4(+)CD25(−) T cells and T regulatory cells. J Biol Chem. 2009;284(37):24914–24924. doi: 10.1074/jbc.M109.000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venuprasad K, Huang H, Harada Y, et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9(3):245–253. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutierrez-Aguilar R, Benmezroua Y, Balkau B, et al. Minor contribution of SMAD7 and KLF10 variants to genetic susceptibility of type 2 diabetes. Diabetes Metab. 2007;33(5):372–378. doi: 10.1016/j.diabet.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Song A, Chen YF, Thamatrakoln K, Storm TA, Krensky AM. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity. 1999;10(1):93–103. doi: 10.1016/s1074-7613(00)80010-2. [DOI] [PubMed] [Google Scholar]
- 50.Song A, Patel A, Thamatrakoln K, et al. Functional domains and DNA-binding sequences of RFLAT-1/KLF13, a Kruppel-like transcription factor of activated T lymphocytes. J Biol Chem. 2002;277(33):30055–30065. doi: 10.1074/jbc.M204278200. [DOI] [PubMed] [Google Scholar]
- 51.Zhou M, McPherson L, Feng D, et al. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J Immunol. 2007;178(9):5496–5504. doi: 10.4049/jimmunol.178.9.5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahn YT, Huang B, McPherson L, Clayberger C, Krensky AM. Dynamic interplay of transcriptional machinery and chromatin regulates “late” expression of the chemokine RANTES in T lymphocytes. Mol Cell Biol. 2007;27(1):253–266. doi: 10.1128/MCB.01071-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagai R, Suzuki T, Aizawa K, Shindo T, Manabe I. Significance of the transcription factor KLF5 in cardiovascular remodeling. J Thromb Haemost. 2005;3(8):1569–1576. doi: 10.1111/j.1538-7836.2005.01366.x. [DOI] [PubMed] [Google Scholar]
- 54.Yang XO, Doty RT, Hicks JS, Willerford DM. Regulation of T-cell receptor D beta 1 promoter by KLF5 through reiterated GC-rich motifs. Blood. 2003;101(11):4492–4499. doi: 10.1182/blood-2002-08-2579. [DOI] [PubMed] [Google Scholar]
- 55.Narla G, Heath KE, Reeves HL, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294(5551):2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 56.Warke VG, Nambiar MP, Krishnan S, et al. Transcriptional activation of the human inducible nitric-oxide synthase promoter by Kruppel-like factor 6. J Biol Chem. 2003;278(17):14812–14819. doi: 10.1074/jbc.M300787200. [DOI] [PubMed] [Google Scholar]
- 57.Yamasaki K, Edington HD, McClosky C, et al. Reversal of impaired wound repair in iNOS-deficient mice by topical adenoviral-mediated iNOS gene transfer. J Clin Invest. 1998;101(5):967–971. doi: 10.1172/JCI2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Juang SH, Xie K, Xu L, Wang Y, Yoneda J, Fidler IJ. Use of retroviral vectors encoding murine inducible nitric oxide synthase gene to suppress tumorigenicity and cancer metastasis of murine melanoma. Cancer Biother Radiopharm. 1997;12(3):167–175. doi: 10.1089/cbr.1997.12.167. [DOI] [PubMed] [Google Scholar]
- 59.Hume DA. The mononuclear phagocyte system. Curr Opin Immunol. 2006;18(1):49–53. doi: 10.1016/j.coi.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 60.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280(46):38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 62.Feinberg MW, Wara AK, Cao Z, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26(18):4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alder JK, Georgantas RW, 3rd, Hildreth RL, et al. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol. 2008;180(8):5645–5652. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU. 1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 65.Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, Di Santo JP. Differential requirement for the transcription factor PU. 1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97(9):2625–2632. doi: 10.1182/blood.v97.9.2625. [DOI] [PubMed] [Google Scholar]
- 66.Dahl R, Simon MC. The importance of PU. 1 concentration in hematopoietic lineage commitment and maturation. Blood Cells Mol Dis. 2003;31(2):229–233. doi: 10.1016/s1079-9796(03)00152-9. [DOI] [PubMed] [Google Scholar]
- 67.Aziz A, Soucie E, Sarrazin S, Sieweke MH. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 2009;326(5954):867–871. doi: 10.1126/science.1176056. [DOI] [PubMed] [Google Scholar]
- 68.Chen ZY, Shie JL, Tseng CC. STAT1 is required for IFN-gamma-mediated gut-enriched Kruppel-like factor expression. Exp Cell Res. 2002;281(1):19–27. doi: 10.1006/excr.2002.5633. [DOI] [PubMed] [Google Scholar]
- 69.Liu J, Liu Y, Zhang H, Chen G, Wang K, Xiao X. KLF4 promotes the expression, translocation, and release of HMGB1 in RAW264.7 macrophages in response to LPS. Shock. 2008;30(3):260–266. doi: 10.1097/shk.0b013e318162bef7. [DOI] [PubMed] [Google Scholar]
- 70.Li Z, Zhao J, Li Q, et al. KLF4 promotes hydrogen-peroxide-induced apoptosis of chronic myeloid leukemia cells involving the bcl-2/bax pathway. Cell Stress Chaperones. 2010 Apr 19; doi: 10.1007/s12192-010-0199-5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Das H, Kumar A, Lin Z, et al. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc Natl Acad Sci U S A. 2006;103(17):6653–6658. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tuomisto TT, Lumivuori H, Kansanen E, et al. Simvastatin has an anti-inflammatory effect on macrophages via upregulation of an atheroprotective transcription factor, Kruppel-like factor 2. Cardiovasc Res. 2008;78(1):175–184. doi: 10.1093/cvr/cvn007. [DOI] [PubMed] [Google Scholar]
- 73.Atkins GB, Wang Y, Mahabeleshwar GH, et al. Hemizygous deficiency of Kruppel-like factor 2 augments experimental atherosclerosis. Circ Res. 2008;103(7):690–693. doi: 10.1161/CIRCRESAHA.108.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo Q, Ma X, Wahl SM, Bieker JJ, Crossley M, Montaner LJ. Activation and repression of interleukin-12 p40 transcription by erythroid Kruppel-like factor in macrophages. J Biol Chem. 2004;279(18):18451–18456. doi: 10.1074/jbc.M400320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turner J, Crossley M. Basic Kruppel-like factor functions within a network of interacting haematopoietic transcription factors. Int J Biochem Cell Biol. 1999;31(10):1169–1174. doi: 10.1016/s1357-2725(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 76.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 77.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26(6):726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 78.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 79.Ramirez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol. 2010;22(2):177–184. doi: 10.1016/j.coi.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dengler HS, Baracho GV, Omori SA, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9(12):1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Good KL, Tangye SG. Decreased expression of Kruppel-like factors in memory B cells induces the rapid response typical of secondary antibody responses. Proc Natl Acad Sci U S A. 2007;104(33):13420–13425. doi: 10.1073/pnas.0703872104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bhattacharya D, Cheah MT, Franco CB, et al. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J Immunol. 2007;179(10):6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klaewsongkram J, Yang Y, Golech S, Katz J, Kaestner KH, Weng NP. Kruppel-like factor 4 regulates B cell number and activation-induced B cell proliferation. J Immunol. 2007;179(7):4679–4684. doi: 10.4049/jimmunol.179.7.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yusuf I, Kharas MG, Chen J, et al. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008;20(5):671–681. doi: 10.1093/intimm/dxn024. [DOI] [PubMed] [Google Scholar]
- 85.Schuh W, Meister S, Herrmann K, Bradl H, Jack HM. Transcriptome analysis in primary B lymphoid precursors following induction of the pre-B cell receptor. Mol Immunol. 2008;45(2):362–375. doi: 10.1016/j.molimm.2007.06.154. [DOI] [PubMed] [Google Scholar]
- 86.Kabashima K, Haynes NM, Xu Y, et al. Plasma cell S1P1 expression determines secondary lymphoid organ retention versus bone marrow tropism. J Exp Med. 2006;203(12):2683–2690. doi: 10.1084/jem.20061289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kharas MG, Yusuf I, Scarfone VM, et al. KLF4 suppresses transformation of pre-B cells by ABL oncogenes. Blood. 2007;109(2):747–755. doi: 10.1182/blood-2006-03-011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oishi Y, Manabe I, Tobe K, et al. SUMOylation of Kruppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-delta. Nat Med. 2008;14(6):656–666. doi: 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- 89.Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-beta signalling [published erratum appears in Nature. 1996;384(6610):648]. Nature. 1996;383(6602):691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 90.Conkright MD, Wani MA, Lingrel JB. Lung Kruppel-like factor contains an autoinhibitory domain that regulates its transcriptional activation by binding WWP1, an E3 ubiquitin ligase. J Biol Chem. 2001;276(31):29299–29306. doi: 10.1074/jbc.M103670200. [DOI] [PubMed] [Google Scholar]
- 91.Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin SH. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol Cell Biol. 1996;16(4):1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turner J, Crossley M. Cloning and characterization of mCtBP2, a corepressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17(17):5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yet SF, McA'Nulty MM, Folta SC, et al. Human EZF, a Kruppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem. 1998;273(2):1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- 94.Shi H, Zhang Z, Wang X, Liu S, Teng CT. Isolation and characterization of a gene encoding human Kruppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res. 1999;27(24):4807–4815. doi: 10.1093/nar/27.24.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Z, Teng CT. Phosphorylation of Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function. Nucleic Acids Res. 2003;31(8):2196–2208. doi: 10.1093/nar/gkg310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ratziu V, Lalazar A, Wong L, et al. Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci U S A. 1998;95(16):9500–9505. doi: 10.1073/pnas.95.16.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Imataka H, Sogawa K, Yasumoto K, et al. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11(10):3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kobayashi A, Sogawa K, Imataka H, Fujii-Kuriyama Y. Analysis of functional domains of a GC box-binding protein, BTEB. J Biochem. 1995;117(1):91–95. doi: 10.1093/oxfordjournals.jbchem.a124727. [DOI] [PubMed] [Google Scholar]
- 99.Sogawa K, Kikuchi Y, Imataka H, Fujii-Kuriyama Y. Comparison of DNA-binding properties between BTEB and Sp1. J Biochem. 1993;114(4):605–609. doi: 10.1093/oxfordjournals.jbchem.a124224. [DOI] [PubMed] [Google Scholar]
- 100.Cook T, Gebelein B, Belal M, Mesa K, Urrutia R. Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J Biol Chem. 1999;274(41):29500–29504. doi: 10.1074/jbc.274.41.29500. [DOI] [PubMed] [Google Scholar]