Abstract
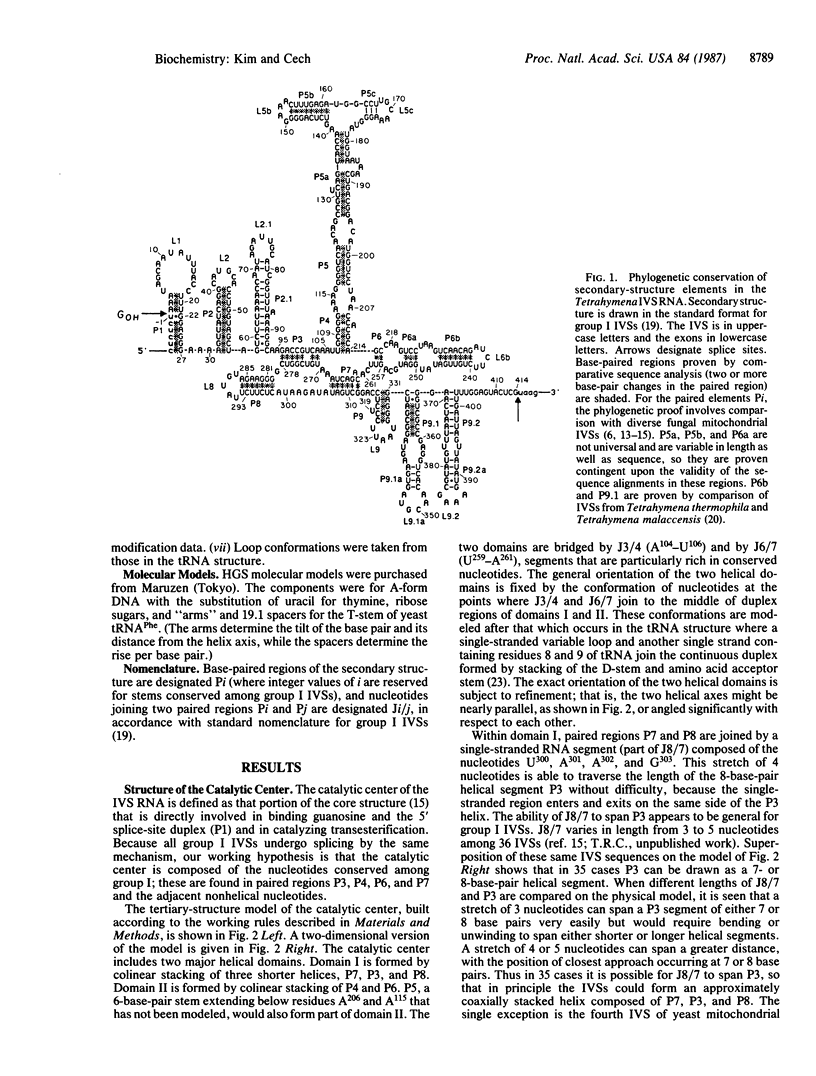
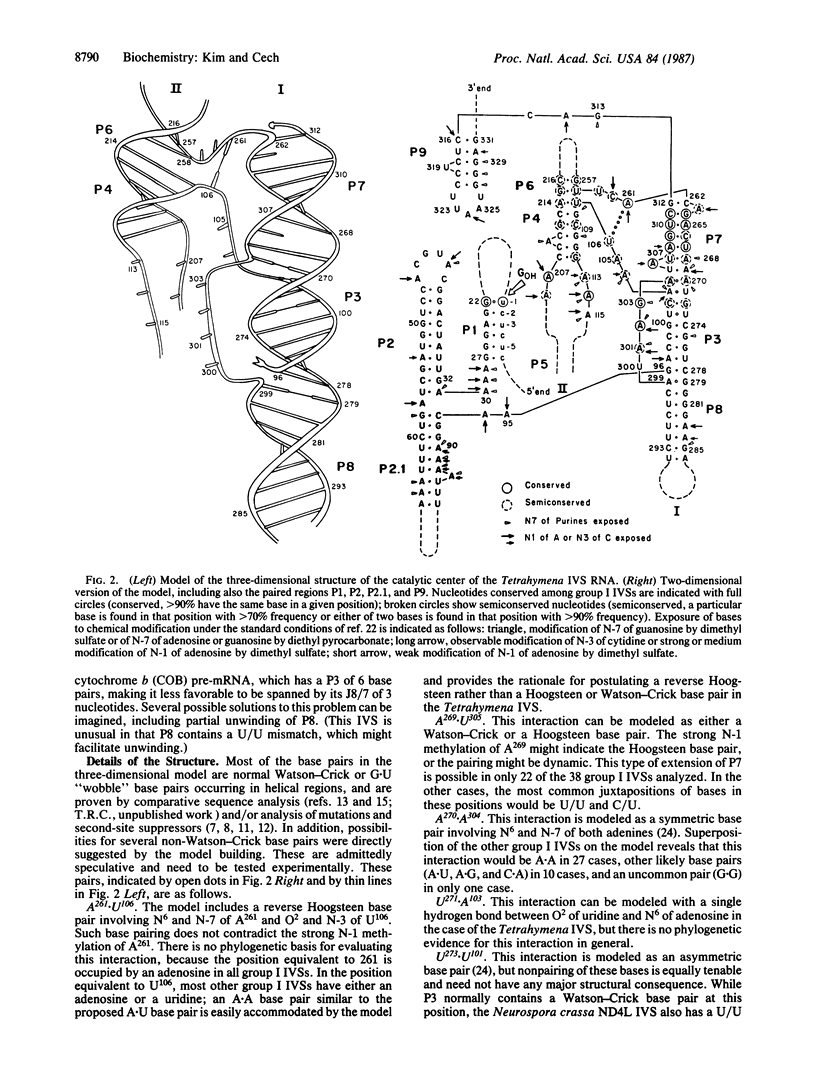
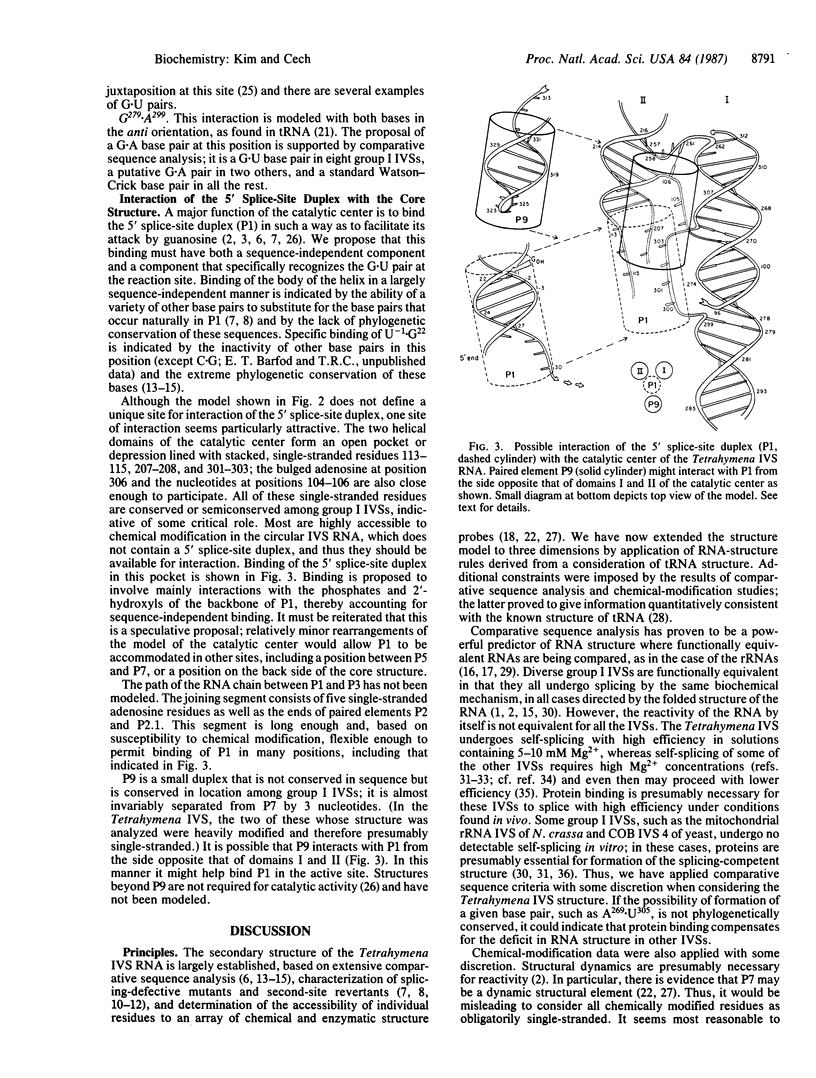
The rRNA intervening sequence of Tetrahymena is a catalytic RNA molecule, or "ribozyme." A tertiary-structure model of the active site of this ribozyme has been constructed based on comparative sequence analysis of related group I intervening sequences, data on the accessibility of each nucleotide to chemical and enzymatic probes, and principles of RNA folding derived from a consideration of the structure of tRNA determined by x-ray crystallography. In the model, the catalytic center has a two-helix structural framework composed of the base-paired segments of the group I conserved sequence elements. The structural framework supports and orients the conserved nucleotides that are adjacent to the base-paired sequence elements; these conserved nucleotides are proposed to form the active site and to bind the 5' splice-site duplex and the guanine nucleotide substrate. Tests of the model are proposed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass B. L., Cech T. R. Ribozyme inhibitors: deoxyguanosine and dideoxyguanosine are competitive inhibitors of self-splicing of the Tetrahymena ribosomal ribonucleic acid precursor. Biochemistry. 1986 Aug 12;25(16):4473–4477. doi: 10.1021/bi00364a001. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986 Oct 24;47(2):207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- Burger G., Werner S. The mitochondrial URF1 gene in Neurospora crassa has an intron that contains a novel type of URF. J Mol Biol. 1985 Nov 20;186(2):231–242. doi: 10.1016/0022-2836(85)90100-7. [DOI] [PubMed] [Google Scholar]
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Breitenberger C., Heckman J. E., Dujon B., RajBhandary U. L. Cytochrome b gene of Neurospora crassa mitochondria. Partial sequence and location of introns at sites different from those in Saccharomyces cerevisiae and Aspergillus nidulans. J Biol Chem. 1984 Jan 10;259(1):504–511. [PubMed] [Google Scholar]
- Burke J. M., Irvine K. D., Kaneko K. J., Kerker B. J., Oettgen A. B., Tierney W. M., Williamson C. L., Zaug A. J., Cech T. R. Role of conserved sequence elements 9L and 2 in self-splicing of the Tetrahymena ribosomal RNA precursor. Cell. 1986 Apr 25;45(2):167–176. doi: 10.1016/0092-8674(86)90380-6. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987 Jun 19;236(4808):1532–1539. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]
- Chu F. K., Maley G. F., West D. K., Belfort M., Maley F. Characterization of the intron in the phage T4 thymidylate synthase gene and evidence for its self-excision from the primary transcript. Cell. 1986 Apr 25;45(2):157–166. doi: 10.1016/0092-8674(86)90379-x. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- De La Salle H., Jacq C., Slonimski P. P. Critical sequences within mitochondrial introns: pleiotropic mRNA maturase and cis-dominant signals of the box intron controlling reductase and oxidase. Cell. 1982 Apr;28(4):721–732. doi: 10.1016/0092-8674(82)90051-4. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Gampel A., Tzagoloff A. In vitro splicing of the terminal intervening sequence of Saccharomyces cerevisiae cytochrome b pre-mRNA. Mol Cell Biol. 1987 Jul;7(7):2545–2551. doi: 10.1128/mcb.7.7.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M., Inoue T., Cech T. R. Mechanism of recognition of the 5' splice site in self-splicing group I introns. Nature. 1986 Jul 3;322(6074):86–89. doi: 10.1038/322086a0. [DOI] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M. Protein-dependent splicing of a group I intron in ribonucleoprotein particles and soluble fractions. Cell. 1986 Aug 29;46(5):669–680. doi: 10.1016/0092-8674(86)90342-9. [DOI] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M. RNA splicing in neurospora mitochondria: self-splicing of a mitochondrial intron in vitro. Cell. 1984 Dec;39(3 Pt 2):631–641. doi: 10.1016/0092-8674(84)90470-7. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Kim S. H. Correlation between chemical modification and surface accessibility in yeast phenylalanine transfer RNA. Biopolymers. 1983 Apr;22(4):1145–1166. doi: 10.1002/bip.360220410. [DOI] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Michel F., Cummings D. J. Analysis of class I introns in a mitochondrial plasmid associated with senescence of Podospora anserina reveals extraordinary resemblance to the Tetrahymena ribosomal intron. Curr Genet. 1985;10(1):69–79. doi: 10.1007/BF00418495. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. A., Macino G. Three class I introns in the ND4L/ND5 transcriptional unit of Neurospora crassa mitochondria. Mol Gen Genet. 1987 Feb;206(2):318–325. doi: 10.1007/BF00333590. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Engberg J. Sequence comparison of the rDNA introns from six different species of Tetrahymena. Nucleic Acids Res. 1985 Oct 25;13(20):7445–7455. doi: 10.1093/nar/13.20.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Price J. V., Kieft G. L., Kent J. R., Sievers E. L., Cech T. R. Sequence requirements for self-splicing of the Tetrahymena thermophila pre-ribosomal RNA. Nucleic Acids Res. 1985 Mar 25;13(6):1871–1889. doi: 10.1093/nar/13.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W. Enzymatic activity of the conserved core of a group I self-splicing intron. Nature. 1986 Jul 3;322(6074):83–86. doi: 10.1038/322083a0. [DOI] [PubMed] [Google Scholar]
- Tanner N. K., Cech T. R. Self-catalyzed cyclization of the intervening sequence RNA of Tetrahymena: inhibition by methidiumpropyl.EDTA and localization of the major dye binding sites. Nucleic Acids Res. 1985 Nov 11;13(21):7759–7779. doi: 10.1093/nar/13.21.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W. Assessment of a model for intron RNA secondary structure relevant to RNA self-splicing--a review. Gene. 1984 Jun;28(3):277–291. doi: 10.1016/0378-1119(84)90145-8. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Ray J. A., Edwards S. W., Scazzocchio C., Davies R. W. The Tetrahymena rRNA intron self-splices in E. coli: in vivo evidence for the importance of key base-paired regions of RNA for RNA enzyme function. Cell. 1985 Feb;40(2):371–380. doi: 10.1016/0092-8674(85)90151-5. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Scazzocchio C., Brown T. A., Davies R. W. Close relationship between certain nuclear and mitochondrial introns. Implications for the mechanism of RNA splicing. J Mol Biol. 1983 Jul 5;167(3):595–605. doi: 10.1016/s0022-2836(83)80100-4. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986 Jan 31;231(4737):470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- van der Horst G., Tabak H. F. Self-splicing of yeast mitochondrial ribosomal and messenger RNA precursors. Cell. 1985 Apr;40(4):759–766. doi: 10.1016/0092-8674(85)90335-6. [DOI] [PubMed] [Google Scholar]



