Abstract
Background
Citrus canker is a disease caused by Xantomonas citri subsp.citri (Xac), and has emerged as one of the major threats to the worldwide citrus crop because it affects all commercial citrus varieties, decreases the production and quality of the fruits and can spread rapidly in citrus growing areas. In this work, the first proteome of Xac was analyzed using two methodologies, two-dimensional liquid chromatography (2D LC) and tandem mass spectrometry (MS/MS).
Results
In order to gain insight into the metabolism of Xac, cells were grown on two different media (NB - Nutrient Broth and TSE - Tryptone Sucrose broth enriched with glutamic acid), and proteins were proteolyzed with trypsin and examined by 2D LC-MS/MS. Approximately 39% of all predicted proteins by annotation of Xac were identified with their component peptides unambiguously assigned to tandem mass spectra. The proteins, about 1,100, were distributed in all annotated functional categories.
Conclusions
This is the first proteomic reference map for the most aggressive strain of Xanthomonas pathogen of all orange varieties. The compilation of metabolic pathways involved with bacterial growth showed that Xac expresses a complete central and intermediary metabolism, replication, transcription and translation machineries and regulation factors, distinct membrane transporters (ABC, MFS and pumps) and receptors (MCP, TonB dependent and metabolites acquisition), two-component systems (sensor and regulatory components) and response regulators. These data corroborate the growth curve in vitro and are the first reports indicating that many of these genome annotated genes are translated into operative in Xac. This proteomic analysis also provided information regarding the influence of culture medium on growth and protein expression of Xac.
Background
Advances in genome sequencing have resulted in the description of a huge number of genes; some of these genes are hypothetical. However, the functions of many of these genes and the relationship among the many proteins that constitute the proteome of a cell or tissue cannot be easily determined. In addition, about half of the predicted proteins have no inferable functions, especially in microorganisms [1,2].
Many approaches, such as two-hybrid systems and proteomics, have been used to describe genes that are effectively translated into proteins for systems of interest without considering transcriptomics or other methodologies that focus on RNA expression. Although they are complementary techniques, proteomics has the advantage of looking directly at the protein level, as gene and protein levels are not always equivalent due to processing and regulation of gene expression at both the transcriptional and translational levels [3].
Despite the large number of proteomic studies, particularly for bacteria, there are few reports about the genus Xanthomonas. Xanthomonas consists of mainly phytopathogenic bacteria that infect a wide variety of economically important plants, such as brassicas, rice, citrus, vine, cassava, pepper, and tomato. Most proteomic analyses involving organisms of the Xanthomonadaceae family have studied Xanthomonas campestris pv. campestris [4-7], due to the completion of its genome sequencing in 2002 [8] and its ability to infect Arabidopsis thaliana, a model plant for genetic studies [9].
Xanthomonas citri subsp. citri (Xac = Xanthomonas axonopodis pv. citri) [10,11], the causal agent of citrus canker, a disease that affects most commercial citrus cultivars and causes extensive damage to citriculture around the world [12,13], is also a good target for proteomic studies. In addition, its complete genome has been sequenced [8] and compared with those from other strains [14-16].
Few studies have focused on Xac proteins, but instead have examined specific genes involved in pathogenicity and adaptation mechanisms [17,18].
This work reports our analysis of the global protein expression profile of Xac using two distinct conditions of in vitro growth. First, we used a rich medium, Nutrient Broth, which is usually used for the cultivation of microorganisms. We also used Tryptone Sucrose broth, TSE, enriched with glutamic acid. TSE is a version of TSA medium [19] without agar and was chosen due to the high growth potential of Xanthomonas in this medium compared to NB medium. The two compounds in TSE medium, sucrose and glutamic acid, were individually determined as being fundamental to the growth process and are also considered important in mimicking the plant environment [20,21].
We aimed to compare the proteome of Xanthomonas grown in NB medium as opposed to TSE medium to understand the differences in the profile of the growth curve. We verified that the presence of sucrose and glutamic acid induced for the expression of genes involved directly or indirectly with pathogenicity.
These two proteomes were analyzed by two-dimensional liquid chromatography-tandem mass spectrometry (2D-LC-MS/MS) to improve protein coverage [22-24].
Results
Proteomic analysis from Xac under two distinct conditions
The Xanthomonas citri subsp. citri was grown at 28°C in NB or TSE media. The cells were collected at an optical density (OD600) corresponding to log phase (Figure 1). The protein extracts were proteolyzed, and the tryptic peptides were separated into 100 fractions in the first dimension (see Additional file 1), followed by nLC-MS/MS investigation. Approximately 119,000 MS/MS peptide spectra for NB and 100,000 MS/MS peptide spectra for TSE were collected and analyzed. The MS/MS spectra led to the identification of 1,162 proteins from bacteria grown in NB and 1,261 proteins from bacteria grown in TSE (see Additional file 2).
Figure 1.
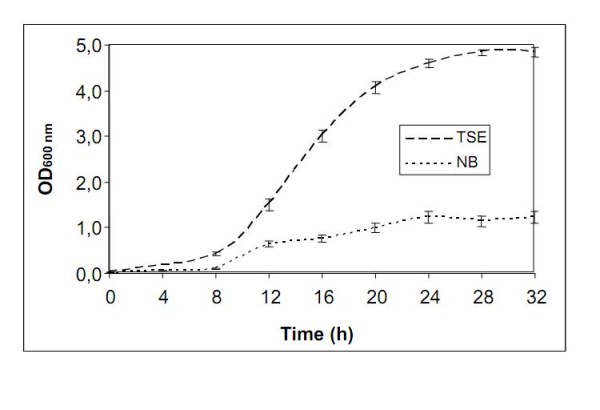
Growth curve profiles of Xanthomonas citri subsp. citri in TSE and NB media. Bacteria were grown at 28°C in Nutrient Broth (NB - 5 g/L peptone and 3 g/L meat extract) or Tryptone Sucrose Broth (TSE - 10 g/L tryptone, 10 g/L sucrose and 1 g/L sodium glutamate) media with shaking at 200 rpm for 16 hours.
Data validation
The reversed sequence database was used as a filter for incorrect or ambiguous peptides. Using the same (Methods Section) parameters for the Xac database, the search of all of the MS/MS spectra returned 5% false positive identifications on the reversed sequence databank, showing good reliability for the method.
Proteome × Genome
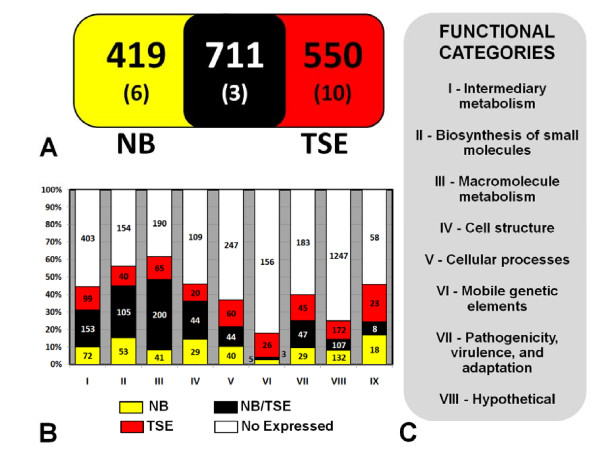
This approach provided high protein coverage, and the results were compatible with those obtained from genome analysis (see Additional file 3 and Figure 2). The total number of proteins found in the present study (1,702) corresponds to almost 40% of all of the proteins predicted by the genome, which is equivalent to that of E. coli, the most comprehensive proteome [25]. The identified Xac proteins were classified according to their annotated function (Table 1). A majority of the proteins were classified as hypothetical proteins (413) or related to metabolism (830): categories I (intermediary metabolism), II (metabolism of small molecules) and III (macromolecule metabolism).
Figure 2.
Proteomic analysis of Xanthomonas citri subsp. citri under non-infecting conditions. (A) A comparison of identified proteins from Xac grown on NB culture medium and Xac grown on TSE culture medium (* denotes plasmid's protein). (B) Comparison of the Xac proteins identified in this study and the genomic prediction per categories (C). For clarity, proteins with undefined functions are excluded from the chart. The red color represents Xac proteome in TSE, yellow color represents Xac proteome in NB, black represents protein detected in both media and white refers to proteins not detected.
Table 1.
Comparison of genome with proteomic analysis from Xanthomonas citri subsp. citri.
| Categories | Genome | Proteome | Prot/Gen % | |
|---|---|---|---|---|
| I | Intermediary metabolism | 727 | 324 | 45 |
| II | Biosynthesis of small molecules | 352 | 198 | 56 |
| III | Macromolecule metabolism | 496 | 308 | 62 |
| IV | Cell structure | 202 | 95 | 48 |
| V | Cellular processes | 391 | 145 | 37 |
| VI | Mobile genetic elements | 190 | 44 | 23 |
| VII | Pathogenicity, virulence and adaptation | 304 | 125 | 41 |
| VIII | Hypothetical | 1658 | 413 | 25 |
| IX | ORFs with undefined category | 107 | 49 | 46 |
| Total | 4427 | 1702 | 39 | |
The number of proteins detected in this study independent of growth conditions is compared with the total expected by the genome
Intermediary and small molecule metabolism
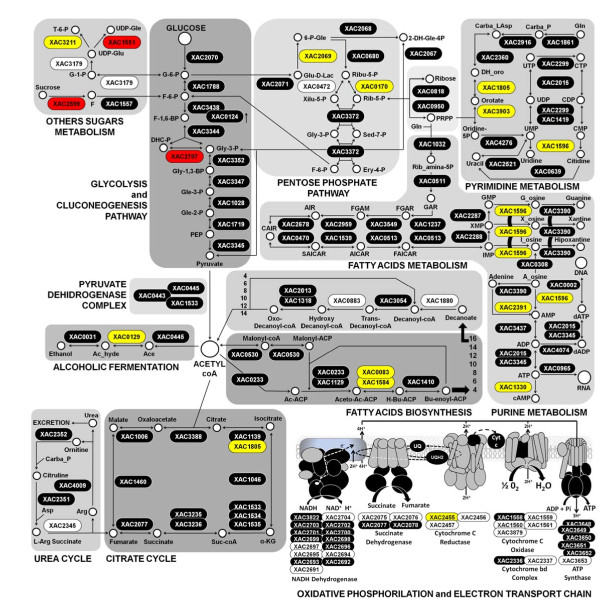
The Xac genome contains 727 and 352 predicted genes classified in intermediary metabolism and biosynthesis of small molecule categories, respectively [8]. We identified 324 proteins assigned to the intermediary metabolism category, 106 of which are related directly to energy metabolism (51%, 106 out of 209 in the genome), involving the complete glycolysis/gluconeogenesis, citrate cycle (TCA cycle), pentose phosphate pathway, other sugar metabolism, urea cycle, pyrimidine and purine biosynthesis, and fatty acid synthesis and degradation pathways (Figure 3). We also observed 23 response regulators (sensor, regulator and hybrid-proteins), 9 complete and 13 incomplete two-component systems, all proteins required for polyamine biosynthesis (XAC0484, XAC3924 and XAC3923) and 42 proteins involved in nucleotide biosynthesis (out of 51 predicted by genome).
Figure 3.
The protein expression profile of Xanthomonas citri subsp. citri according to energy production. The pathways illustrated here are almost complete. The expressed proteins in TSE are represented in red boxes, the expressed proteins in NB are represented in yellow boxes, black boxes represent proteins detected in both media and white boxes represent non-expressed or non-detected proteins.
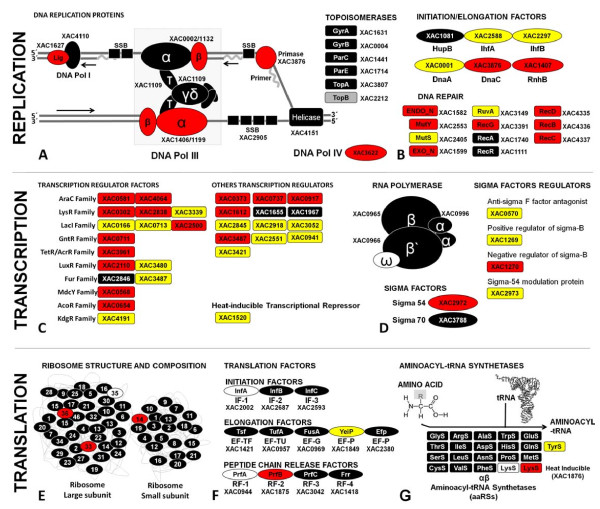
Macromolecule metabolism
We detected 308 of the 496 proteins (62%) predicted by the genome to be involved in macromolecule metabolism. Several of these expressed proteins are involved in the metabolism of DNA (53%, 71 out of 135), RNA (53%, 110 out of 209) or protein (66%, 103 out of 157) and include initiation, elongation, transcription, sigma and translation factors (Figure 4). In addition, ribosomal proteins as well as aminoacyl-tRNA synthetases to all tRNA were also expressed, consistent with the expected number of genes (54 ribosomal proteins = genome) (Figure 4E). These processes show the potential for Xac replication, in accordance with its growth curve profile (Figure 1).
Figure 4.
Distribution of identified proteins of Xanthomonas citri subsp. citri involved in replication, transcription and translation process. In total, 163 proteins (9.6% of the proteome) are involved in these processes. The expressed proteins in TSE are represented in red boxes, the expressed proteins in NB are represented in yellow boxes, black boxes represent proteins detected in both media and white boxes represent non-expressed or non-detected proteins.
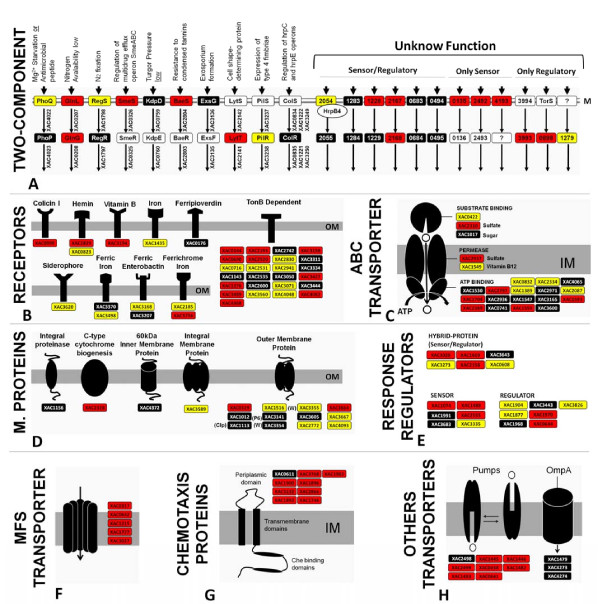
Cell structure
Similar to other bacterial proteomes, a large number of proteins involved in cell structure and cellular processes, such as surface proteins, were detected in the Xac proteome. In particular, we identified proteins involved in pilus synthesis, corresponding to genes XAC3098-3102 (the complete cluster) and subunits pilB, A, C, Y1 and X from distinct clusters. Among the members of this category, we also found 12 distinct outer membrane proteins, 5 pumps and 3 OmpA proteins (Figure 5).
Figure 5.
Distribution of identified proteins of Xanthomonas citri subsp. citri related to membrane, transport and gene regulation. In total, 154 proteins (9.1% of the proteome) are involved in these processes. The expressed proteins in TSE are represented in red boxes, the expressed proteins in NB are represented in yellow boxes, black boxes represent proteins detected in both media and white boxes represent non-expressed or non-detected proteins.
Cellular processes
In this study, 145 proteins of this category were found; among them, 17 are related to cell division (66%, 17 out of 27 of genome), 105 are related to transport (37%, 105 out of 281) and 4 distinct proteins are related to osmotic processes (XAC0304, XAC0604, XAC2915, and XAC0888).
Mobile genetic elements
In this class, 44 proteins were found, of which 12 proteins have phage-related functions and are prophages, 12 have plasmid-related functions and 20 have transposon- and intron-related functions.
Pathogenicity, virulence and adaptation
Proteins involved in adaptation (63%, 15 out of 24 in the genome) were also identified, as well as proteins related to pathogenicity. Although hrp gene expression is generally induced in plants or under certain in vitro conditions [26], four protein constituents of the type III secretion system apparatus and two proteins related to hypersensitivity were found in the Xac proteome during growth in TSE medium: HrcN (XAC0412), HrpF (XAC0394), HrcU (XAC0406) and HrcV (XAC0405), XopAE (XAC0393) and a PthA isoform was found in the two protein extracts (NB medium). The expression of proteins for both type II (XAC3534/35 - D/N, XAC3537 - L and XAC3544 - E subunits) and type IV secretion systems (XAC2614-XAC2623) was also verified.
Expression of hypothetical genes
In an attempt to find potential functions for the hypothetical proteins expressed by Xac, the BLASTp program was used to search for sequence similarities [27]. A BLAST search for homologous proteins in Xanthomonas campestris pv. vesicatoria returned 209 proteins with identity > 34%, coverage > 49% and e-values less than or equal to 10-19 (see Additional file 4). Examples of proteins previously proposed as hypothetical are XAC3997, XAC2966, XAC3898 and XAC1756, which now are categorized as proteins with putative functions, such as ABC transporter permease, BolA superfamily transcriptional regulator, membrane-bound metalloendopeptidase and PhoH-like protein, respectively.
ORFs with undefined category
A considerable number of proteins (49) without a specific category were identified in this work. These proteins were identified in both of the two treatments, suggesting that the proteins play an important role in metabolism. For example, the products of XAC2529 and XAC3245 (RhsD proteins) may have a putative function involved in pathogenesis or virulence due the cellular location. This protein was found in bacterial outer membrane or vesicles secreted together with proteins secreted by the Type III secretion system in other species of Xanthomonas [7]. Accordingly, Laia and coworkers [28] reported that a mutant for XAC3245 gene in Xac causes significant reduction of necrosis during the plant infection process.
Discussion
In the present work, the proteomic analysis of Xac cultivated in non-infecting conditions revealed the presence of 1,702 proteins, corresponding to 39% of all of the proteins predicted by the genome. A good correlation with the genome was observed (Figure 2), except for the class of mobile genetic elements, which exhibits more intense activity under stress conditions [29,30]. In addition to regulation factors, proteins involved in the pathways of DNA replication, transcription and translation are associated with active growth.
Xac expresses almost every protein involved in energy metabolism pathways, such as the TCA cycle, pentose pathway, fermentation, biosynthesis and degradation of fatty acids, glycolysis/gluconeogenesis, and purine and pyrimidine biosynthesis (Figure 3). These observations might be explained by the localization of these proteins because the detected products are mainly cytosolic proteins. It is not surprising that a large proportion of the identified proteins belong to energy metabolism. Nonetheless, this work is the first large-scale analysis for an organism of the Xanthomonadaceae family.
The presence and detection of the transcription factors, sigma factors, ribosomal subunits, translation factors and aminoacyl-tRNA synthetases (Figure 4) illustrate the good agreement between the genome and proteome of Xac under the investigated conditions.
A large number of membrane constituents were detected, including 22 ABC transporters and other response regulators. In addition, 154 proteins related to these processes were found, corresponding to 9.1% of the proteome. In addition, nine sets of two-component systems were completely identified (sensor + regulator) (Figure 5A), with two related to nitrogen metabolism (XAC02907/0208 - GlnLG and XAC1798/1797 - RegSR) and the others (e.g., XAC2054 and XAC2055) with unknown function. Alegria and coworkers [31] described the interaction between XAC2054 and the product of the hrpB gene, which was also found in the proteome, suggesting an association with the type III secretion system. The TonB-dependent receptors (25 of 46 annotated), which participate in the uptake of iron or siderophores, were also highly expressed. The significant expression levels of these receptors and 13 more specific iron receptors demonstrate the ability of Xac to take up these compounds, which is fundamental to the growth of microorganisms, and some are related to pathogenicity [32].
Xac expressed a large number of components related to cell structure. These proteins play an important role during bacterial infection and host adhesion. Examples of proteins in this group include type IV fimbriae and pilus proteins, which are important to the processes of colonization and biofilm formation. The co-expression of hemagglutinin protein, which is coded by the gene XAC1815, corroborates this physiological condition, which is fundamental to pathogen adaptation in the plant tissue with consequent induction of pathogenicity [33]. In addition, proteins involved in different cellular processes were detected, including a large set of proteins related to cell division and a larger number of transporter and chemotaxis proteins (Figure 5). These proteins are mainly involved in adaptation to culture conditions.
Mobile genetic elements represented about 3% of the Xac proteome, a minor group of the detected proteins. This result was expected because these genes are generally related to survival response mechanisms, such as antibiotic resistance and virulence [34]. In this study, Xac was grown on privileged media under non-stressing conditions. The genes corresponding to the identified proteins, such as transposases (XAC1872 and XAC3233), were located mainly in the chromosome.
Additional experiments are needed to investigate the appropriate conditions for the expression of these genes, which have significant importance for genome plasticity and evolution [35].
The expression of genes related to pathogenicity was surprising, especially in TSE medium (Additional file 5). Among these proteins, members of secretory systems are the most investigated [36-39].
The type III secretion system is a generally conserved mechanism found in both plant and animal pathogens for the export, secretion and delivery of specific proteinaceous effector molecules (virulence or pathogenicity factors) directly or indirectly into host cells in a contact-dependent manner. The type II secretion system is involved with the secretion of cell wall-degrading enzymes, such as carbohydrate esterases and proteases.
Genes encoding components of the secretion apparatus are not constitutively expressed but activated in planta and in minimal media mimicking the environmental conditions present in the plant apoplast. Some proteins related to secretory systems were identified in this work; these proteins were detected only during bacterial growth priority on TSE medium. NB medium has been suggested as a more appropriate medium to compare the differential expression of proteins in infectious non-inducing conditions. Furthermore, a higher level of xanthan gum production has been observed in TSE culture (data not shown), which is associated with virulence factors [40]. Recently, new genes involved in the pathogenesis process have been reported [28], and the author showed that expression of these genes is dependent on the plant-pathogen interaction. Among these genes, the XAC0340 product was the only one detected in TSE media, which could be associated with the presence of specific components in this medium, such as sucrose or glutamic acid.
Sucrose has been described as an inducer of hrpF gene expression in Xanthomonas campestris pv vesicatoria, in this case by modifying the culture medium MM1 with additional amino acids that have sulfur in their composition [20]. Glutamic acid has been reported to potentiate the growth of Xanthomonas, particularly in the case of Xanthomonas fuscans B, which is consistent with the growth curves. Quorum sensing due to the high density of cells would stimulate the expression of genes related to the type III secretion system, as described for enteropathogens [41,42] and phytopathogens [43,44]. These results support the idea that NB is a more appropriate medium for control experiments.
One goal of this study was to provide experimental annotation of predicted genes. The products of these genes may have important roles in bacterial development. A total of 209 of the 413 hypothetical proteins have homologs in Xanthomonas campestris pv. vesicatoria, which allowed us to hypothesize their function (see Additional file 4).
Finally, this proteomic analysis also provided information regarding the influence of culture medium on the growth and protein expression of Xac. Figure 1 shows the more effective growth in TSE medium, reflected in the larger number of proteins expressed in this condition. As shown in Figure 2A, a significant number of proteins were exclusively detected under each condition. These proteins probably depend directly on the components of each medium. Therefore, the choice of medium can be crucial in developing a control experiment.
Conclusions
This report is the first presentation of a proteome map of the most aggressive strain of Xanthomonas pathogen of all orange varieties, Xac [16]. We used a 2D-LC-MS/MS strategy to characterize this proteome. Protein mixtures were digested with proteases, and the resulting peptides were separated by multidimensional liquid chromatography and analyzed sequentially by MS/MS, resulting in very high protein coverage.
We were able to identify proteins with a variety of functions covering most cellular processes, including regulatory and signal transduction proteins, even though these proteins are normally present in small amounts in the cell.
The complete set of energy metabolism pathways identified in this study denotes the ability of Xanthomonas to convert carbon sources from other biomolecules of fundamental importance for metabolism, as shown in Figure 3, and which helps to explain the profile of the growth curve (Figure 1). In addition to this metabolic energy profile, annotated genes related to metabolic function of macromolecules were determined in this study. Initiation factors for replication, transcription, and translation and sigma factors are shown in Figure 4. However, the receptors and membrane transporters identified in this study drew the most attention. The quantity and diversity of our results support the growth dynamics, colonization and spread of Xanthomonas in plant tissues. The two-component systems and response regulators, membrane receptors closely related to the uptake of iron and membrane transporters (MFS, ABC, OMPs and other pumps), are also interesting. Many of these proteins were annotated by sequence homology, but their functions have not been characterized in Xanthomonas, as shown in Figure 5.
This platform for proteomics, in corroboration with other tools for functional analysis of the genome of Xac, like transcriptomics [45], mutatomics [28] or two-hybrid systems [37,38], will enable a more integrated understanding of metabolic pathways, essential for the adaptation of pathogens in plant tissue. Additional studies directed toward the development of biotechnological tools to combat the pathogen may help to reduce worldwide crop losses due to the genus Xanthomonas.
Methods
Bacterial strains, growth and cell lysis
The Xanthomonas citri subsp. citri strain 306 used both in the genome project [8] and in this study was obtained from the culture collection of plant pathogenic bacteria of IAPAR (Instituto Agronômico do Paraná, PR, Brazil). This strain was grown at 28°C in Nutrient Broth (Difco™ NB - 5 g/L peptone and 3 g/L meat extract) or Tryptone Sucrose Broth (TSE - 10 g/L tryptone, 10 g/L sucrose and 1 g/L sodium glutamate) media with shaking at 200 rpm for 16 hours. The cells were collected at an optical density (OD600) corresponding to the log phase.
The extraction of total proteins was performed as described by Mehta & Rosato [17]. The cells were collected by centrifugation, and the pellet was washed in phosphate buffer (1.24 g/L K2HPO4; 0.39 g/L KH2PO4; 8.8 g/L NaCl, pH 7.2). Next, the pellet was suspended in 0.75 mL of extraction buffer (0.7 M sucrose; 0.5 M Tris-HCl, pH 7; 30 mM HCl; 50 mM EDTA; 0.1 M KCl and 40 mM DTT) and incubated for 15 min at room temperature. The same volume of phenol was added; after 15 min of agitation, the suspension was centrifuged at 10000 g and 4°C for 3 min, and the supernatant was recovered. This procedure was repeated twice.
The proteins were precipitated with five volumes of 0.1 M ammonium acetate in methanol, and the precipitate was washed once with 80% acetone. Protein concentrations were estimated by the Bradford method (Bio-Rad, Hercules, CA, USA). A lysate sample containing 1 mg of protein was denatured by adding 50 μL of buffer (8 M urea and 25 mM NH4HCO3, pH 8.0), reduced with 10 mM DTT at 37°C for 1 h, and alkylated with 50 mM iodoacetamide in the dark for 30 min. The urea concentration was reduced to 1 M by dilution. A 20-μg sample of trypsin (Promega, modified sequencing grade) (1:50) was added to digest the proteins at 37°C overnight. Neat formic acid was added to stop the digestion.
Strong cation exchange (SCX) chromatography
Tryptic peptides were fractionated by strong cation exchange chromatography. The separation was performed on an SP-Sepharose HP™ column (GE Healthcare) using the AKTA PRIME™ chromatography system (GE Healthcare). The column was washed with buffer A (25 mM NH4HCO3, pH 3.0), and the peptides were eluted with buffer B (25 mM NH4HCO3, 500 mM KCl, pH 3.0) at a flow rate of 1 mL per minute. Buffer B concentrations of 10%, 20%, 25% and 30% were used to displace fractions during SCX chromatography. Fractions (about 100) were collected at 1 min intervals and concentrated by vacuum centrifugation to produce a final volume of 100 μL.
The nLC-MS/MS System
The SCX fractions were loaded into a Waters CapLC™ system (Waters, Milford, MA). The digested proteins were desalted in-line using a Waters Opti-Pak C18 trap column. The sample injection volume was typically 10 μL, and the LC was performed using a NanoEase C18™ 150 mm × 75 μm column (Waters, Milford, MA) with elution (0.6 μL/min) using a linear gradient (10 to 50%) of acetonitrile containing 0.1% formic acid (solution B).
Electrospray tandem mass spectra were recorded using a Q-Tof quadrupole/orthogonal acceleration time of flight spectrometer (Waters, Milford, MA) interfaced with the CapLC™ capillary chromatograph. The ESI voltage was set at 3500 V using a metal needle, the source temperature was 100 ºC and the cone voltage was 100 V. The instrument control and data acquisition were conducted by a MassLynx data system (Version 4.0, Waters) and experiments were performed by scanning from a mass-to-charge ratio (m/z) of 200 to 2000 using a scan time of 1 s applied during the entire chromatographic process. The mass spectra corresponding to each signal from the Total Ion Current (TIC) chromatogram were averaged, allowing an accurate molecular mass determination. Exact mass MS/MS is achieved automatically using the Q-Tof's LockSpray™ (Waters, Milford, MA). The reference ion used was the mono charged ion of Rifampicin at m/z 823.4130.
Database Searching
All data were processed using the ProteinLynx Global server (version 2.0, Waters). Proteins were identified by correlation of tandem mass spectra and Xanthomonas axonopodis pv. citri str. 306 genome data bank available at NCBI, using the MASCOT™ software (Matrix Science, version 2.1). One missed cleavage per peptide was allowed and an initial mass tolerance of 0.05 Da was used in all searches. Cysteines were assumed to be carbamidomethylated and variable modification of methionine (oxidation) was allowed. To evaluate the false positive rate of this approach, a reversed sequence databank (a database in which the sequences have been reversed) containing the same number of proteins in the Xac database was constructed.
Abbreviations
Xac: Xanthomonas citri subsp. citri; nLC: capillary liquid chromatography; nLC-MS/MS: capillary liquid chromatography/tandem mass spectrometry; SCX: strong cation exchange chromatography; 2D LC-MS/MS: two-dimensional liquid chromatography/tandem mass spectrometry; NB: Nutrient Broth; TSE: Tryptone Sucrose broth enriched with glutamic acid.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MRS, APF and RMF carried out the proteomics experiments. MRS, LMM and FCG were responsible for bioinformatic analysis and have been involved in drafting the manuscript. FCG has made substantial contributions to analysis and interpretation of data. RM, JCFO, MIT and JAF made substantial contributions to the study conception and design and critically revised the manuscript for intellectual content. All authors edited the manuscript and approved the final version.
Supplementary Material
Strong cation exchange chromatography. Tryptic peptides were fractionated by strong cation exchange chromatography. Fractions (about 100) were collected at 1 min intervals and concentrated by vacuum centrifugation to produce a final volume of 100 μL. Figure A shows the chromatogram of peptides from Xac proteins detected in TSE medium and Figure B shows the chromatogram of peptides from Xac proteins detected in NB medium.
Proteins identified of Xanthomonas citri subsp. citri. Complete list of all proteins expressed in both conditions available, categorized according annotation.
Map of identified proteins in TSE × NB conditions. The comparison between expressed products of Xac show more regions of continuous genes in TSE medium, showing that NB expressed minor number of proteins. The red color represents Xac proteome in TSE, yellow color represents Xac proteome in NB, black represents protein detected in both media and white refers to proteins not detected. Gray boxes represent hypothetical or conserved hypothetical proteins and green boxes show the proteins expressed only in TSE medium
Functions assigned to some hypothetical proteins. A BLAST search for homologous proteins in Xanthomonas campestris pv. vesicatoria returned 209 proteins with identity > 34%, coverage > 49% and e-values less than or equal to 10-19.
List of proteins related to pathogenicity or virulence exclusively detected in TSE medium.
Contributor Information
Márcia R Soares, Email: marcia@iq.ufrj.br.
Agda P Facincani, Email: agdapf1@yahoo.com.br.
Rafael M Ferreira, Email: marini64@gmail.com.
Leandro M Moreira, Email: lmmorei@gmail.com.
Julio CF de Oliveira, Email: juliunifesp@yahoo.com.br.
Jesus A Ferro, Email: jesus@fcav.unesp.br.
Maria IT Ferro, Email: mitferro@fcav.unesp.br.
Rogério Meneghini, Email: rogerio.meneghini@bireme.org.
Fábio C Gozzo, Email: fabio@iqm.unicamp.br.
Acknowledgements
We acknowledge the Laboratório Nacional de Luz Síncroton (LNLS) for the use of its Mass Spectrometry facility. M.R. Soares received a post-doctoral scholarship from FAPESP (Fundação de amparo à pesquisa do estado de São Paulo). Part of this work was financed by FAPESP Grant (04/02006-7).
References
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Conserved hypothetical' proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004;32:5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Gygi SP. Proteomics: the Move to Mixtures. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- Watt SA, Wilke A, Patschkowski T, Niehaus K. Comprehensive analysis of the extracellular proteins from Xanthomonas campestris pv. campestris B100. Proteomics. 2005;5:153–167. doi: 10.1002/pmic.200400905. [DOI] [PubMed] [Google Scholar]
- Chung WJ, Shu HY, Lu CY, Wu CY, Tseng YH, Tsai SF, Lin CH. Qualitative and comparative proteomic analysis of Xanthomonas campestris pv. campestris 17. Proteomics. 2007;7:2047–2058. doi: 10.1002/pmic.200600647. [DOI] [PubMed] [Google Scholar]
- Andrade AE, Silva LP, Pereira JL, Noronha EF, Reis FB Jr, Bloch C Jr, Santos MF, Domont GB, Franco OL, Mehta A. In vivo proteome analysis of Xanthomonas campestris pv. campestris in the interaction with the host plant Brassica oleracea. FEMS Microbiol Lett. 2008;281:167–174. doi: 10.1111/j.1574-6968.2008.01090.x. [DOI] [PubMed] [Google Scholar]
- Sidhu VK, Vorholter FJ, Niehaus K, Watt SA. Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol. 2008;8:87. doi: 10.1186/1471-2180-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LMC. et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- Goodman HM, Ecker JR, Dean C. The genome of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1995;92:10831–10835. doi: 10.1073/pnas.92.24.10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad NW, Postnikova E, Lacy GH, Sechler A, Agarkova I, Stromberg PE, Stromberg VK, Vidaver AK. Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv malvacearum (ex smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and "var. fuscans" of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst Appl Microbiol. 2005;28:494–518. doi: 10.1016/j.syapm.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Schaad NW, Postnikova E, Lacy G, Sechler A, Agarkova I, Stromberg PE, Stromberg VK, Vidaver AK. Emended classification of xanthomonad pathogens on citrus. Syst Appl Microbiol. 2006;29:690–695. doi: 10.1016/j.syapm.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Stall RE, Civerolo EL. In: Xanthomonas campestris pv. citri: cause of citrus canker. Swings JG, Civerolo EL, editor. Chapman and Hall, London; 1993. pp. 48–50. [Google Scholar]
- Gottwald TR, Graham JH. In: Compendium of Citrus Diseases. Timmer LW, Garnsey SM, Graham JH, editor. Am Phytopathol Soc, São Paulo; 2000. pp. 5–7. [Google Scholar]
- Moreira LM, de Souza RF, Almeida NF Jr, Setubal JC, Oliveira JC, Furlan LR, Ferro JA, da Silva AC. Comparative genomics analyses of citrus-associated bacteria. Annu Rev Phytophathol. 2004;42:163–184. doi: 10.1146/annurev.phyto.42.040803.140310. [DOI] [PubMed] [Google Scholar]
- Moreira LM, De Souza RF, Digiampietri LA, Da Silva ACR, Setubal JC. Comparative analyses of Xanthomonas and Xylella complete genomes. Omics. 2005;9:43–76. doi: 10.1089/omi.2005.9.43. [DOI] [PubMed] [Google Scholar]
- Moreira LM, Almeida NF Jr, Potnis N, Digiampietri LA, Adi SS, Bortolossi LC, da Silva AC, da Silva AM, de Moraes FE, Oliveira JC. et al. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics. 2010;11:238. doi: 10.1186/1471-2164-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Rosato YB. A simple method for in vivo expression studies of Xanthomonas axonopodis pv. citri. Curr Microbiol. 2003;47:400–403. doi: 10.1007/s00284-003-4051-3. [DOI] [PubMed] [Google Scholar]
- Alegria MC, Souza DP, Andrade MO, Docena C, Khater L, Ramos CH, da Silva AC, Farah CS. Identification of new protein-protein interactions involving the products of the chromosome- and plasmid-encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri. J Bacteriol. 2005;187:2315–25. doi: 10.1128/JB.187.7.2315-2325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, MaGbanua MM, White FF. Identification of two novel hrp-associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae. J Bacteriol. 2000;182:1844–53. doi: 10.1128/JB.182.7.1844-1853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte R, Bonas U. A Xanthomonas Pathogenicity Locus Is Induced by Sucrose and Sulfur-Containing Amino Acids. Plant Cell. 1992;4:79–86. doi: 10.1105/tpc.4.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Han SW, Bartley LE, Ronald PC. Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proc Natl Acad Sci USA. 2006;103:18395–400. doi: 10.1073/pnas.0605508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Wu CC, MacCoss MJ, Howell KE, Yates JR. A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- Taoka M, Yamauchi Y, Shinkawa T, Kaji H, Motohashi W, Nakayama H, Takahashi N, Isobe T. Only a small subset of the horizontally transferred chromosomal genes in Escherichia coli are translated into proteins. Mol Cell Proteomics. 2004;3:780–787. doi: 10.1074/mcp.M400030-MCP200. [DOI] [PubMed] [Google Scholar]
- Bonas U. Hrp genes of phytopathogenic bacteria. Curr Top Microbiol Imunol. 1994;192:79–98. doi: 10.1007/978-3-642-78624-2_4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Laia ML, Moreira LM, Dezajacomo J, Brigati JB, Ferreira CB, Ferro MI, Silva AC, Ferro JA, Oliveira JC. New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaptation revealed by a transposon-based mutant library. BMC Microbiol. 2009;9:12. doi: 10.1186/1471-2180-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy Pierre, Gasperi Giuliano, Christian Biémont, Claude Bazin. Stress and transposable elements: co-evolution or useful parasites? Heredity. 2000;85:101–106. doi: 10.1046/j.1365-2540.2000.00751.x. [DOI] [PubMed] [Google Scholar]
- Grandbastien MA. Activation of plant retrotransposons under stress conditions. Trends in Plant Science. 1998;3:181–187. doi: 10.1016/S1360-1385(98)01232-1. [DOI] [Google Scholar]
- Alegria MC, Docena C, Khater L, Ramos C, da Silva AC, Farah CS. New protein-protein interactions identified for the regulatory and structural components and substrates of the type III Secretion system of the phytopathogen Xanthomonas axonopodis pathovar citri. J Bacteriol. 2004;186:6186–97. doi: 10.1128/JB.186.18.6186-6197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denancé N, Vasse J, Lauber E, Arlat M. Plant carbohydrate scavenging through tonb-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One. 2007;2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One. 2009;4:e4358. doi: 10.1371/journal.pone.0004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL. Identification of genomic islands in six plant pathogens. Gene. 2006;374:134–141. doi: 10.1016/j.gene.2006.01.029. [DOI] [PubMed] [Google Scholar]
- Tobes R, Pareja E. Bacterial repetitive extragenic palindromic sequences are DNA targets for Insertion Sequence elements. BMC Genomics. 2006;7:62. doi: 10.1186/1471-2164-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier O, Van den Ackerveken G, Bonas U. HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol Microbiol. 2000;38:828–838. doi: 10.1046/j.1365-2958.2000.02173.x. [DOI] [PubMed] [Google Scholar]
- Alegria MC, Docena C, Khater L, Ramos CH, da Silva AC, Farah CS. New protein-protein interactions identified for the regulatory and structural components and substrates of the type III secretion system of the phytopathogen Xanthomonas axonopodis pv. citri. J of Bacteriol. 2004;186:6186–6197. doi: 10.1128/JB.186.18.6186-6197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegria MC, Souza DP, Andrade MO, Docena C, Khater L, Ramos CH, da Silva AC, Farah CS. Identification of new protein-protein interactions involving the products of the chromosome- and plasmid-encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri. J Bacteriol. 2005;187:2315–2325. doi: 10.1128/JB.187.7.2315-2325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano JR, Collmer A. TYPE III SECRETION SYSTEM EFFECTOR PROTEINS: Double Agents in Bacterial Disease and Plant Defense. Annu Rev Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- Lu GT, Ma ZF, Hu JR, Tang DJ, He YQ, Feng JX, Tang JL. A novel locus involved in extracellular polysaccharide production and virulence of Xanthomonas campestris pathovar campestris. Microbiology. 2007;157:737–746. doi: 10.1099/mic.0.2006/001388-0. [DOI] [PubMed] [Google Scholar]
- Henke JM, Bassler BL. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol. 2004;186:3794–805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V, Mellies JL, Nguyen W, Shin S, Kaper JB. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999;96:15196–201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Coulthurst SJ, Pritchard L, Hedley PE, Ravensdale M, Humphris S, Burr T, Takle G, Brurberg MB, Birch PR. et al. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 2008;4:e1000093. doi: 10.1371/journal.ppat.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MN, Yang CH, Barak JD, Jahn CE, Charkowski AO. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J Bacteriol. 2005;187:639–48. doi: 10.1128/JB.187.2.639-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira LM, Almeida NF Jr, Potnis N, Digiampietri LA, Adi SS, Bortolossi LC, da Silva AC, da Silva AM, de Moraes FE, Oliveira JC. et al. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics. 2010;11:238. doi: 10.1186/1471-2164-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strong cation exchange chromatography. Tryptic peptides were fractionated by strong cation exchange chromatography. Fractions (about 100) were collected at 1 min intervals and concentrated by vacuum centrifugation to produce a final volume of 100 μL. Figure A shows the chromatogram of peptides from Xac proteins detected in TSE medium and Figure B shows the chromatogram of peptides from Xac proteins detected in NB medium.
Proteins identified of Xanthomonas citri subsp. citri. Complete list of all proteins expressed in both conditions available, categorized according annotation.
Map of identified proteins in TSE × NB conditions. The comparison between expressed products of Xac show more regions of continuous genes in TSE medium, showing that NB expressed minor number of proteins. The red color represents Xac proteome in TSE, yellow color represents Xac proteome in NB, black represents protein detected in both media and white refers to proteins not detected. Gray boxes represent hypothetical or conserved hypothetical proteins and green boxes show the proteins expressed only in TSE medium
Functions assigned to some hypothetical proteins. A BLAST search for homologous proteins in Xanthomonas campestris pv. vesicatoria returned 209 proteins with identity > 34%, coverage > 49% and e-values less than or equal to 10-19.
List of proteins related to pathogenicity or virulence exclusively detected in TSE medium.