Abstract
Malignant tumors result from the accumulation of genetic alterations in oncogenes and tumor suppressor genes. Much less is known about the genetic changes in benign tumors. Seborrheic keratoses (SK) are very frequent benign human epidermal tumors without malignant potential. We performed a comprehensive mutational screen of genes in the FGFR3-RAS-MAPK and phosphoinositide 3-kinase (PI3K)-AKT pathways from 175 SK, including multiple lesions from each patient. SK commonly harbored multiple bona fide oncogenic mutations in FGFR3, PIK3CA, KRAS, HRAS, EGFR, and AKT1 oncogenes but not in tumor suppressor genes TSC1 and PTEN. Despite the occurrence of oncogenic mutations and the evidence for downstream ERK/MAPK and PI3K pathway signaling, we did not find induction of senescence or a DNA damage response. Array comparative genomic hybridization (aCGH) analysis revealed that SK are genetically stable. The pattern of oncogenic mutations and X chromosome inactivation departs significantly from randomness and indicates that spatially independent lesions from a given patient share a clonal relationship. Our findings show that multiple oncogenic mutations in the major signaling pathways involved in cancer are not sufficient to drive malignant tumor progression. Furthermore, our data provide clues on the origin and spread of oncogenic mutations in tissues, suggesting that apparently independent (multicentric) adult benign tumors may have a clonal origin.
Seborrheic keratoses (SK) are one of the most common benign human tumors. Their incidence increases with age and old individuals can have many lesions (Fig. S1A), occasionally displaying a linear distribution; little is known about their etiology (1). SK are classified in three histological subtypes (acanthotic, hyperkeratotic, and adenoid; Fig. S1B) and at the microscopic level, they are similar to epidermal nevi (EN), a benign skin lesion that generally occurs at birth or in young children and that also has no malignant potential. EN can present as single lesions, as multiple lesions, or in association with more complex phenotypes (2). Because of their anatomical distribution, it has long been proposed that EN arise as a result of mosaicism (3); a proportion of EN harbor somatic embryonic mutations in the gene coding for fibroblast growth factor receptor (FGFR3) and for the p110α subunit of phosphoinositide 3-kinase (PIK3CA) (4–6).
Oncogenic mutations in one of these genes have also been found in a fraction of SK (6–8). These findings in benign epidermal tumors are similar to those reported in benign melanocytic tumors: The V600E BRAF hotspot mutation—originally identified in malignant melanoma—is also frequently present in benign melanocytic nevi (9) and NRAS mutations have been identified in congenital melanocytic nevi. A distinctive difference between the benign epidermal and melanocytic tumors is that the latter harbor a variable risk of malignant transformation (10), whereas SK and EN have no malignant potential. Therefore, SK and EN represent unique opportunities to analyze the biological significance of oncogenic mutations in benign tumors.
FGFR3 encodes a transmembrane receptor tyrosine kinase that is a target of both germ-line and somatic mutations. Activating FGFR3 mutations have been found in the extracellular, juxta-transmembrane, and kinase domains. Germ-line mutations result in skeletal syndromes associated with shortened long bones, occur only in a few codons, and display a good genotype–phenotype association (11) whereas somatic FGFR3 mutations—often at the same hotspots—have been identified in several human cancers including multiple myeloma (12) and urothelial carcinoma (13). Extracellular domain mutations commonly lead to an unpaired Cys, allowing spontaneous receptor homodimerization, increased tyrosine phosphorylation, and ligand-independent receptor activation (14). Cytoplasmic domain hotspot mutations cause conformational changes in the activation loop that increase the kinase activity. The level of activation correlates with the severity of the skeletal phenotype (14). FGFR3 signaling involves the activation of Shc, phospholipase-Cγ, STAT1, Gab1, and FRS2α. The docking proteins FRS2α and FRS2β mediate activation of the RAS-MAPK and phosphoinositide 3-kinase (PI3K)-AKT pathways (15). Therefore, the identification of mutations in PIK3CA in SK and EN was in agreement with the involvement of the PI3K-AKT pathway in the development of benign skin tumors. Activation of ERK1/2 has been observed in FGFR3 mutant keratinocytes (16).
We have proposed that mutations in additional genes involved in the RTK-RAS-MAPK or the PI3K-AKT pathways could be involved in SK not harboring mutations in FGFR3 and PIK3CA (8). Among the candidates are RAS and PTPN11 (protein tyrosine phosphatase, nonreceptor type 11). Oncogenic HRAS can activate the PI3K-AKT pathway (17). Genes coding for proteins involved in the MAPK pathway are mutated in the germ line of patients with Costello (HRAS), LEOPARD (PTPN11), or Noonan syndrome (KRAS, PTPN11), which are often associated with a skin phenotype (18). Additional genes involved in the PI3K pathway that are mutated in cancers are AKT and the tumor suppressors PTEN and TSC1. Whereas activating germ-line mutations in AKT may be embryonic lethal, PTEN germ-line mutations result in Cowden's disease, an inherited cancer syndrome displaying also a skin phenotype (19). TSC1 codes for hamartin and inhibits the mTOR pathway through the TSC1/2 complex that inactivates Rheb (20). Germ-line TSC1 mutations cause the autosomal dominant tuberous sclerosis syndrome and somatic TSC1 mutations have been identified in bladder cancer (21).
Herein, we provide a comprehensive analysis of oncogenes and tumor suppressor genes of the FGFR3-RAS-MAPK and the PI3K-AKT pathway in human SK. We show a high prevalence of oncogenic mutations in FGFR3, PIK3CA, AKT1, KRAS, HRAS, and EGFR. Many of these lesions display two simultaneous oncogenic mutations. In contrast, TSC1 and PTEN mutations are uncommon in SK. Unlike in benign melanocytic lesions, we find no evidence for senescence playing a major role in halting tumor progression in SK. Finally, the mutational pattern—together with X chromosome inactivation analysis—strongly suggests that spatially distinct, apparently independent, lesions share a clonal relationship.
Results
We collected 175 SK from 25 Spanish and German patients and analyzed mutations in genes of the FGFR3-RAS-MAPK and PI3K-AKT pathways. A summary of the results and detailed information on patients and mutations are shown in Fig. 1A and Datasets S1 and S2.
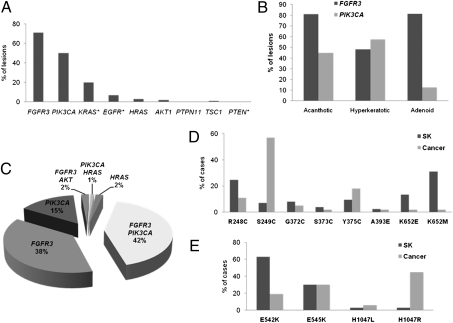
Fig. 1.
Mutational analysis of SK. (A) Prevalence of mutations in genes coding for proteins in the FGFR3-RAS-MAPK and PI3K-AKT pathways (*genes analyzed only in a subgroup of samples). (B) FGFR3 and PIK3CA mutation prevalence according to histological subtype. (C) Compound mutational genotype of SK. (D) Comparison of the spectrum of FGFR3 mutations in SK and in human cancers (COSMIC database). (E) Comparison of the spectrum of PIK3CA mutations in SK and in human cancers (COSMIC database).
Analysis of Oncogenic Mutations in the FGFR3-RAS-MAPK and PI3K-AKT Pathways.
FGFR3 hotspot mutations were detected in 71% (124/175) of SK. Mutation rate was similar among Spanish and German patients (68% vs. 79%); kinase domain mutations were more common among Spanish cases (P = 0.01). The most frequent amino acid substitutions were K652M (31%), R248C (25%), and K652E (13%). Two lesions harbored two FGFR3 mutations each. FGFR3 mutation rate was unrelated to age of appearance or total SK number. FGFR3 mutations were more frequent in acanthotic and adenoid SK than in hyperkeratotic SK (P < 0.0001 and P < 0.05, respectively) (Fig. 1B). PIK3CA hotspot activating mutations were detected in 50% (80/159) of SK with a higher prevalence among German patients (72% vs. 41%; P < 0.001); the mutation spectrum was similar in both groups. The E542K mutation was the most frequent substitution (63%), followed by E545K (30%).
HRAS mutations occurred in 3% (5/160) of the lesions (four G12D and one Q61K). PTPN11 hotspot mutations were not detected in 144 SK analyzed. The AKT1 E17K hotspot mutation was identified in 2% (3/173) of SK. Overall, 79.4% (139/158) of informative SK had mutations in one gene or more and 45% of them had oncogenic mutations in two genes at the same time (Fig. 1C). FGFR3 and HRAS mutations were mutually exclusive, as were PIK3CA and AKT1 mutations, as in human cancers (22, 23).
The mutations found in FGFR3, PIK3CA, and HRAS in SK have been proven to be oncogenic (24–26) and to be involved in a wide variety of human cancers (http://www.sanger.ac.uk/genetics/CGP/cosmic/). However, the mutational spectrum of FGFR3 in SK was different from that of malignant tumors, especially bladder cancer, where FGFR3 is the most commonly mutated oncogene (13, 27, 28). When comparing SK with malignant tumors included in the COSMIC database, the S249C mutation was more common in cancer (53.9% vs. 7.1%, P < 10−7) whereas mutations in codons 652 and 248 were more common in SK (44.4% vs. 4.9% and 24.6% vs. 10.5%, respectively; P < 10−7 and P < 10−5) (Fig. 1D). Importantly, germ-line K652M and R248C mutations are associated with severe forms of dwarfism and high FGFR3 constitutive kinase activity (14). Regarding PIK3CA, helical domain mutations were more common in SK than in cancer: The E542K mutation frequency was 63% in SK vs. 19% in malignant tumors, whereas the H1047R kinase domain mutation was significantly less common in SK than in cancer (3% vs. 45%) (P < 0.0001) (Fig. 1E).
Analysis of Tumor Suppressor Genes TSC1 and PTEN.
One of 128 SK (1%) revealed a somatic point mutation (c.G1528C, D510H) in TSC1. The D510H semiconservative substitution has BLOSUM 62 (29) and Grantham (30) pathogenicity scores of −1 and 81, respectively. Upon transfection of wild-type and D510H TSC1 cDNA into 97-1 and HCV29 cells, levels of the mutant protein were ≈70% lower than those of wild-type protein. Low TSC1 levels would result in reduced TSC1–TSC2-dependent inhibition of mTOR activity and increased mTOR signaling (31). No difference in RNA splicing of exons 14–17 was detected between wild-type and mutant RNA from this lesion. Therefore, this mutation is of uncertain functional relevance. Somatic PTEN mutations were not found in 30 SK analyzed.
Screening for Additional Oncogenic Mutations.
To identify additional genes involved in SK, we searched for hotspot mutations in 14 SK lacking mutations in the primary screen, using the OncoCarta Panel v1.0. Nine SK harbored KRAS mutations and one had a R108K EGFR mutation, previously reported in brain tumors and shown to be oncogenic in vitro (32). The OncoCarta Panel identified five and one additional mutations in PIK3CA and FGFR3, respectively. There were no mutations in the remaining genes. We sequenced KRAS exon 2 in 45 additional SK: 20% (12/59) of SK revealed a codon 12 mutation (seven G12D, three G12V, and two G12C). Altogether, at least one oncogenic mutation was detected in 156/175 (89%) SK.
Genomic Alterations in SK.
We used array CGH to assess genomic stability. Two SK from each of 10 patients and the corresponding germ-line leukocyte DNA were used. In 7/20 lesions, gains at 9q34.2 were identified that were not consistently confirmed by quantitative PCR. We found an increased signal for Y chromosome markers in two SK from patient G5. Quantitative PCR showed that the SK contained a normal dosage of Y chromosome genes whereas a loss of Y chromosome sequences in mosaic occurred in blood. Overall, SK are genomically stable.
Expression of Senescence, Proliferation, and Apoptosis Markers in SK.
The occurrence of strongly activating mutations in FGFR3 in SK, the presence of multiple oncogenic mutations in genes participating in the most common pathways involved in cancer, and the benign nature of SK suggest the activation of mechanisms suppressing tumor progression. Therefore, we assessed proliferation, apoptosis (DNA damage response, DDR), and whether a senescence program was activated (33, 34).
Proliferation in the basal cell layer was similar in SK and normal skin; by contrast, 72% of SK showed Ki67 reactivity in suprabasal layers vs. 0% of normal skin samples (P < 0.001) (Fig. 2). Ki67 expression was significantly higher in FGFR3 wild-type vs. mutant lesions (P = 0.033). By contrast, pHH3 immunoreactivity was not significantly different in normal skin and in SK. To get insight into how SK differ from malignant tumors, we analyzed proliferation in a panel of 38 squamous cell carcinomas (SCC). Ki67 staining was higher in SK than in SCC (P = 0.016) whereas phospho-histone H3 (pHH3) staining was much higher in the latter than in SK (P < 0.001). There was no significant difference in staining for activated caspase 3 in normal skin and in SK.
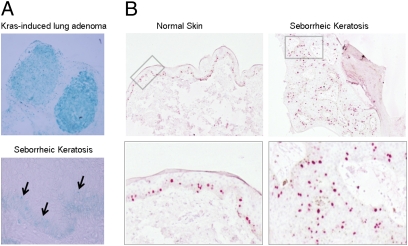
Fig. 2.
β-Galactosidase activity and proliferation in normal skin and SK. (A) β-Galactosidase staining in KRAS-induced mouse lung adenoma as a control (Upper) and in SK (Lower). There was no evidence of β-galactosidase activity in most of the SK lesions analyzed. (B, Upper) Ki67 staining of normal skin and SK. A significant increase in Ki67-positive cells was found in the suprabasal layers of the epidermis in SK. (Lower) A higher magnification of the indicated region is shown.
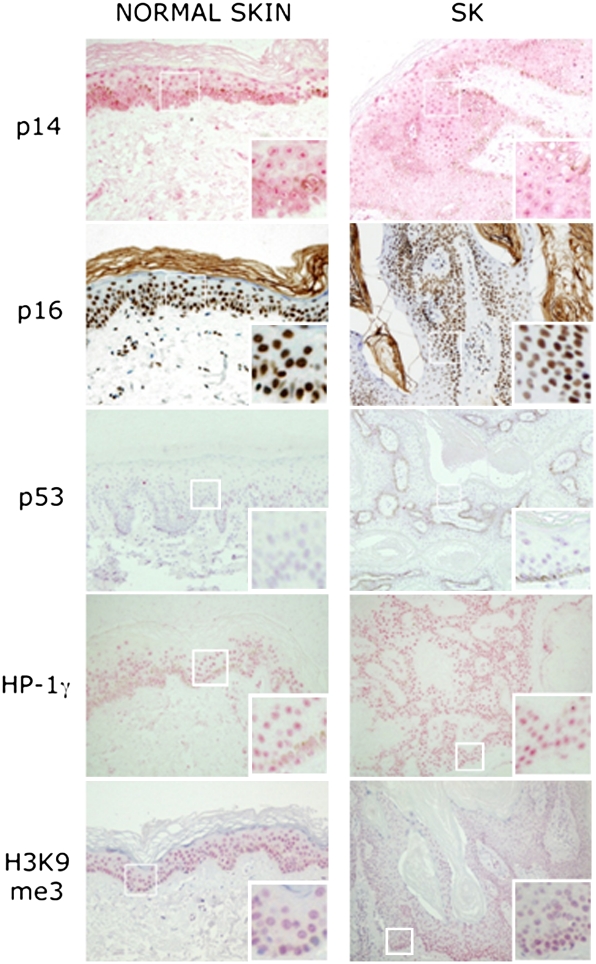
To analyze the activation of senescence, pH 6.0 β-galactosidase activity and expression of senescence markers (p14, p16, p53, Dec1, HP-1γ, and H3K9me3) were assessed (35). Weak β-galactosidase activity was observed in the basal layer in 5/49 lesions (Fig. 2), the remaining cases being negative. Expression of p14, p16, p53, and H3K9me3 was similar in normal skin and SK, regardless of mutational status (Fig. 3). When stratified by mutation, Dec1 and HP-1γ expression was significantly lower in FGFR3 mutant vs. wild-type lesions. These findings are in agreement with the observation that expression of 53BP1 and γ-H2AX, markers of DDR, was similar in normal skin and in SK.
Fig. 3.
Immunohistochemical analysis of senescence-associated markers. Senescence marker expression was similar in normal skin and SK. Insets show a higher magnification of the indicated areas.
RAS-MAPK and PI3K-AKT-mTOR Signaling in SK.
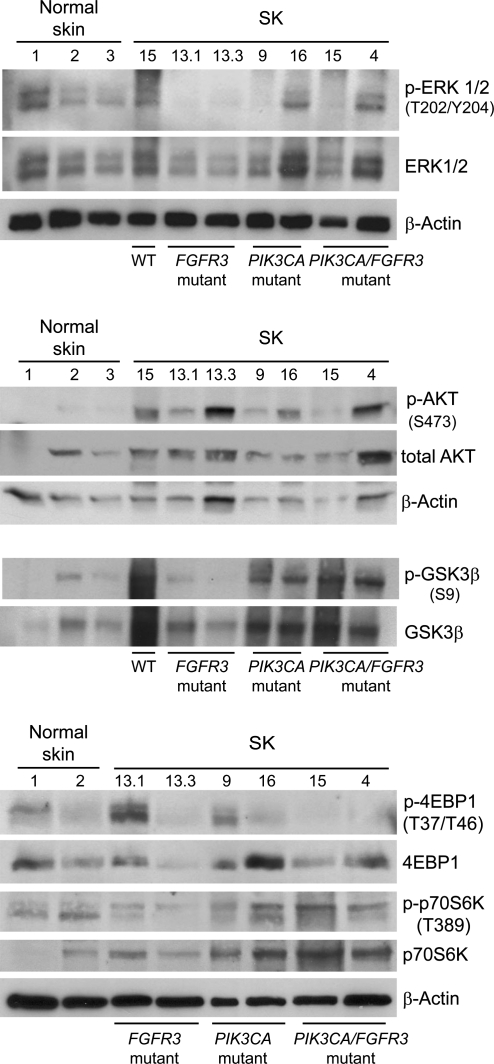
The above findings raise the question of whether oncogenic mutations activate the corresponding signaling pathways. We compared the expression of p-ERK1/2, p-AKT (Ser473), p-GSK3β (Ser9), p-4EBP1 (Thr37/Thr46), and p-p70S6 kinase (Thr389) in normal epidermis and in SK with diverse mutational patterns, using Western blotting and immunohistochemistry. The levels of phosphorylated proteins varied substantially. Overall, p-AKT levels were more clearly up-regulated in SK than p-ERK, with highest levels found in PIK3CA mutant samples (Fig. 4 and Fig. S2). In agreement with this, strong phosphorylation of GSK3β Ser9, an AKT substrate, was found. However, p-4EBP1 levels were low in PIK3CA mutant SK. Immunohistochemical analysis showed a clear-cut increase in p-ERK1/2 in SK compared with normal skin (Fig. S3).
Fig. 4.
ERK/MAPK and PI3K pathway signaling in SK compared with normal skin: Western blotting analysis of p-ERK1/2, p-AKT, p-GSK3β, and p-4EBP1. Increased ERK phosphorylation was commonly observed in SK. A very high increase in p-AKT was consistently found regardless of the mutational genotype; in agreement with this, GSK3β phosphorylation was commonly increased. In contrast, p-4EBP1 levels were not consistently elevated in samples with PIK3CA mutations.
Analysis of the Clonal Relationship of Multicentric Intraindividual SK.
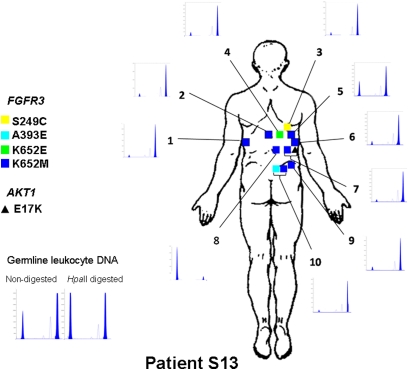
The prevalence and pattern of mutations found in multiple SK from a given patient revealed a remarkable deviation from independence (Dataset S2). For example, all SK analyzed from patients S7 (n = 7), S13 (n = 10), G1 (n = 5), G2 (n = 5), and G4 (n = 5) harbored an FGFR3 mutation; the combined probability of this occurring in the five patients is 3.8 × 10−5. As for PIK3CA, all SK from patients G1 (n = 5), G7 (n = 5), and G8 (n = 7) were mutated whereas all lesions from patients S5 (n = 9) and S13 (n = 10) were wild type; the combined probability for this distribution is 1.7 × 10−7. Regarding the specific amino acid substitution, the three FGFR3 mutant lesions from patient S5 displayed the K652M mutation; 8/10 lesions from patient S13 harbored the FGFR3 K652M mutation and all were PIK3CA wild type; 5/7 lesions from patient S2 harbored the PIK3CA E542K mutation yet showed diverse FGFR3 mutations. Similar patterns of mutational aggregation were found in other patients. Detailed maps of SK location and genotype were generated; Fig. 5 and Fig. S4 show the maps of patients S2, S5, and S13. In patient S2, SK harboring the E542K mutation were found either in close proximity (at 5 cm from each other) or at distant locations (i.e., in the left arm and in the right thigh). In patient S5, 3 SK harboring the FGFR3 K652M mutation were located in the back whereas other neighboring lesions were FGFR3 wild type. In patient S13, the 8 SK harboring the K652M mutation were located in the back.
Fig. 5.
Distribution and genotypes of SK in patient 13. All 10 lesions sampled harbored a mutation in FGFR3 and all of them were wild type for PIK3CA. Eight of the lesions harbored the same K652M mutation; lesion no. 7 harbored an additional E17K AKT mutation and lesion no. 10 an additional A393E FGFR3 mutation. X chromosome inactivation analysis shows that normal germ-line leukocyte DNA displays a balanced distribution of X chromosome inactivation whereas all but one of the SK displayed a common X chromosome inactivation pattern. The probability of this distribution occurring by chance is estimated to be 1.9 × 10−7.
To assess clonality and explore the possibility that SK harboring two mutations might contain two independent cell populations, we used the human androgen receptor (HUMARA) assay to determine X chromosome inactivation in lesions from eight heterozygous females. Three different samples of normal skin from patient S6 showed lack of clonality. By contrast, 28/30 SK with a single mutation and 18/19 SK with two or more mutations harbored selective inactivation of one X chromosome, supporting their clonal nature (9/19 expected vs. 1/19 observed, for nonclonal lesions, P = 0.01). Patient S13 was most informative because eight lesions shared the same FGFR3 mutation: Seven of them, located at 5–10 cm distance from one another, displayed inactivation of the same X chromosome (Fig. 5). The probability that these are independent events is 3.8 × 10−8. These findings provide strong evidence that apparently distinct SK share a clonal origin.
Discussion
This is one of the most detailed mutational analyses of cancer genes in benign human tumors. Because of the ease of skin sampling, we were able to study a large number of lesions per individual and to assess their genetic makeup and clonal relationship. The skin is an excellent model to study such questions due to the visibility and accessibility of the lesions; yet, the emerging paradigms may apply to other tissues. The major findings of this study are (i) multiple oncogenic mutations occur in benign lesions with no malignant potential, (ii) downstream signaling can be demonstrated, (iii) we find no evidence that a senescence program is activated, and (iv) apparently independent lesions display shared mutational and X chromosome inactivation patterns, raising questions about their phylogenetic relationship and the mechanisms involved in the appearance of multicentric synchronous tumors.
It has been proposed that at least 5–7 mutations are required for cancer development (36) and whole-genome sequencing studies have shown that breast, colon, pancreatic, and brain tumors harbor 48–101 putative pathogenic mutations (37, 38). However, little is known about the number and types of mutations present in benign tumors. Here we show that bona fide activating mutations in oncogenes participating in two of the major pathways involved in human cancer (39) occur frequently and commonly together in SK. The notion that nonneoplastic cells can harbor genetic mutations is not novel (9). However, the finding that multiple mutations in oncogenes occurring at the hotspots associated with human cancer lead to downstream signaling, but no malignant potential, is provocative. The same genes mutated in SK are mutated in urothelial cell carcinoma (i.e., FGFR3, PIK3CA, AKT1, HRAS, and KRAS) and the prevalence of mutations is similar in both sites, suggesting shared biological properties in these stratified epithelia. Nevertheless, the types of mutations differ: In the case of SK, FGFR3 tyrosine kinase domain mutations are much more common in the benign skin lesions. This finding is somewhat surprising because—in the germ line—these mutations are associated with the most severe bone phenotype (40) and in vitro they lead to highest constitutive kinase activity, suggesting strongest oncogenic potential (14). It is not currently possible to determine whether the distinct mutational spectrum found in SK and urothelial cell carcinoma results from differences in the mutational mechanisms involved, DNA repair, or tissue-specific mutation effects. As for PIK3CA, the SK-associated mutations occur more commonly in the helical than in the kinase domain, unlike in most cancers. Helical domain mutations also predominate in urothelial cell carcinoma (41). It has been proposed that gain-of-function derived from mutations in the helical domain requires interaction of p110α with RAS-GTP and is less affected by binding of the regulatory subunit p85, unlike with kinase domain mutants that do not require RAS-GTP (42). All lesions harboring RAS mutations had either wild-type or helical domain PIK3CA mutations. Intriguingly, in breast tumors, helical domain mutations have been associated with a more aggressive in vivo metastatic phenotype in mice (43) and with less favorable clinical features (44).
The selective occurrence of FGFR3 and PIK3CA mutations with an apparently more pro-oncogenic behavior in SK prompted us to examine cell proliferation and the activation of a senescence program. Regarding proliferation, we found that Ki67 expression was higher in SK than in normal skin in the suprabasal layers of the epidermis, yet pHH3 levels were much lower than would be expected on the basis of the Ki67 labeling. These observations, together with the low mitotic index characteristic of SK, suggest that oncogene-initiated epidermal cells are primed to enter the cell cycle but do not display a high proliferation rate. Senescence can be induced by activating mutations in oncogenes, as well as by loss of tumor suppressors, oxidative stress, and persistent DNA damage, and it has emerged as a potent tumor suppressor mechanism (45). Senescence markers have been shown to be activated in several preneoplastic lesions in humans such as melanocytic nevi (46), neurofibromas (47), and prostate intraepithelial neoplasia (48). We used an extensive panel of markers to examine senescence in SK but find no evidence supporting that it plays a major role protecting SK from the effects of oncogenic mutations. One mechanism through which oncogenes activate senescence is the induction of a DDR (49). We looked for evidence of DDR by comparing γ-H2AX foci and 53BP1 expression in SK and normal skin and found no significant differences. The lack of detectable p53 expression also suggests that neither DDR nor TP53 mutations occur in SK. In mouse models of melanoma, the BRAF V600E mutation induces development of senescent nevi and an additional mutation in the PTEN suppressor leads to rapidly progressive metastatic melanoma (50). BRAF and PTEN participate in the RAS-MAPK and PI3K-AKT pathways, as do FGFR3 and PIK3CA, raising the issue of gene-specific functions, tissue-specific functions, or a distinct role of oncogenes vs. tumor suppressors in driving cancer progression.
To assess whether the mutated oncogenes activate downstream signaling, we analyzed phosphorylation of relevant substrates in the MAPK and PI3K pathways. We found a strong activation of AKT in SK as well as evidence for negative feedback loops, or pathway cross-talk, restraining the effect of oncogenic mutations, as suggested by the low levels of phosphorylation of 4EBP1. These events likely contribute to tumor suppression and to the absence of a senescence response.
Importantly, we did not find evidence of alterations in the tumor suppressors TSC1, PTEN, and TP53 in SK, suggesting that this category of genes plays a more fundamental role in driving progression to malignant tumors. In agreement with this notion, SK were genomically stable. Altogether these findings suggest that the genomic architecture of tumors, rather than the mutations in oncogenes, is an essential determinant of tumor progression. This proposal is also in agreement with the finding that germ-line FGFR3 mutations, or somatic embryonic mutations in FGFR3 and PIK3CA, do not impose a significantly increased cancer risk in patients with short bone syndromes or epidermal nevi, respectively (11, 51). A role for FOXN1 as a regulator that is downstream of FGFR3 that determines a benign vs. a malignant phenotype has recently been proposed (52).
The analysis of multiple, anatomically distinct, lesions from the same patient has also provided insights into important issues such as the patterns of mutations found in tumors, their origin, and clonal relatedness. FGFR3 and PIK3CA showed highly skewed mutational patterns: Certain individuals showed a greater tendency than others to present with mutations in one of these genes and specific mutations were more common than expected by chance. There is some precedent supporting the notion of individual susceptibility to mutation development in the case of JAK2 in leukemia (53, 54). Recently, a genome-wide association study in patients with bladder cancer showed that a SNP located in 4p16, in the vicinity of FGFR3, is associated with somatic FGFR3 mutations in tumors (55). However, this study did not address the issue of multicentricity that we analyzed in detail here. The finding of the same mutation in closely distributed, but distinct, SK raises the notion that they might be clonally related. The clonal patch size in the skin has been estimated as being 2 mm2 (56). The X chromosome inactivation pattern found in SK not only confirms that SK are clonal, as reported (57), but also indicates either preferential inactivation of one X chromosome—providing an advantage for cell growth—or the shared origin of apparently independent lesions. Both issues merit further studies in the context of multicentric synchronous or metachronous tumors. Because in EN somatic embryonic mutations in FGFR3 and PIK3CA occur in mosaicism, it is conceivable that the mutations found in SK of aged individuals might also have occurred early in life, spread through the skin in a radial manner, remained “silent” until adulthood, and then have led to benign tumors grown. A precedent for a role of somatic embryonic mutations in tumors is best established in childhood leukemia: this malignancy occurs only in a fraction of children in whom tumor-causing translocations are found at birth (58, 59). These questions are important to understand the kinetics of acquisition of tumor-related mutations, their significance when found in healthy individuals, and the requirements for disease development.
Because the skin is “right before our eyes,” it is possible to follow the natural history of tumors better than at any other site. Our findings provide unique notions on the mechanisms limiting malignant transformation in response to oncogene activation and on the origin of apparently independent neoplasms. As occurs in the skin, similar mechanisms may operate in internal tissues for which such analyses are more difficult to perform.
Materials and Methods
Detailed information is provided in SI Materials and Methods.
Patients and Samples.
The material was collected at the Departments of Dermatology at Hospital del Mar (Barcelona) and the University Clinic, Regensburg, Germany. Informed consent was obtained according to the guidelines of the local Ethics Committees and the Declaration of Helsinki.
Mutational Analyses.
FGFR3 mutations were assessed using a SNaPshot assay (4). PIK3CA, HRAS, and PTPN11 mutations were assessed by Sequenom using specifically designed assays. The remaining assays are described in SI Materials and Methods.
X Chromosome Inactivation Analysis.
The HUMARA allele-specific PCR was used.
β-Galactosidase Staining and Immunohistochemistry.
Sections of 50 fresh frozen SK were stained using a senescence β-galactosidase staining kit (Cell Signaling Technology). Lung adenoma from KRasV12 mice (33) was used as a control. Immunohistochemical assays for markers of proliferation, senescence, apoptosis, and signaling pathways are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Alfaro, N. del Pozo, C. Guerrero, E. Herschberger, L. Kuenzel, T. Lobato, F. Mercadillo, D. Pastor, and T. Schifferstein for excellent technical support; M. Torres and Centro Nacional de Genotipado (CEGEN)-Santiago for help with the Sequenom analysis; M. Serrano, S. Chanock, and J. Valcárcel for valuable discussions and comments; M. Collado (CNIO, Madrid, Spain) for reagents and discussions; and L. Sánchez and the CNIO Immunohistochemistry Core Facility for valuable contributions. This study was partially supported by Grants SAF2007-60860 and Consolíder ONCOBIO from Ministerio de Ciencia e Innovación (Madrid) (to F.X.R.), Deutsche Forschungsgemeinschaft Grant HA 5531/1-1 and Deutsche Dermatologische Gesellschaft/Arbeitsgemeinschaft Dermatologische Forschung Research Grant 2007 (to C.H.), Grant PI04/1728 from Fondo de Investigación Sanitaria (to R.M.P.), Cancer Research UK Grant C6228/A5433 (to M.A.K. and J.E.B.), European Union-7FP Grant 201663-UROMOL (to N.M. and F.X.R.), and a grant from Asociación Española Contra el Cáncer (to F.X.R.). M.M. was recipient of a Sara Borrell Fellowship from the Instituto de Salud Carlos III, Madrid.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 20599.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008365107/-/DCSupplemental.
References
- 1.Hafner C, Vogt T. Seborrheic keratosis. J Dtsch Dermatol Ges. 2008;6:664–677. doi: 10.1111/j.1610-0387.2008.06788.x. [DOI] [PubMed] [Google Scholar]
- 2.Happle R, Rogers M. Epidermal nevi. Adv Dermatol. 2002;18:175–201. [PubMed] [Google Scholar]
- 3.Happle R. Mosaicism in human skin. Understanding the patterns and mechanisms. Arch Dermatol. 1993;129:1460–1470. [PubMed] [Google Scholar]
- 4.Hafner C, et al. Mosaicism of activating FGFR3 mutations in human skin causes epidermal nevi. J Clin Invest. 2006;116:2201–2207. doi: 10.1172/JCI28163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernández S, et al. Fibroblast growth factor receptor 3 mutations in epidermal nevi and associated low grade bladder tumors. J Invest Dermatol. 2007;127:1664–1666. doi: 10.1038/sj.jid.5700705. [DOI] [PubMed] [Google Scholar]
- 6.Hafner C, et al. Oncogenic PIK3CA mutations occur in epidermal nevi and seborrheic keratoses with a characteristic mutation pattern. Proc Natl Acad Sci USA. 2007;104:13450–13454. doi: 10.1073/pnas.0705218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logié A, et al. Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum Mol Genet. 2005;14:1153–1160. doi: 10.1093/hmg/ddi127. [DOI] [PubMed] [Google Scholar]
- 8.Toll A, Real FX. Somatic oncogenic mutations, benign skin lesions and cancer progression: Where to look next? Cell Cycle. 2008;7:2674–2681. doi: 10.4161/cc.7.17.6523. [DOI] [PubMed] [Google Scholar]
- 9.Pollock PM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 10.Krengel S, Hauschild A, Schäfer T. Melanoma risk in congenital melanocytic naevi: A systematic review. Br J Dermatol. 2006;155:1–8. doi: 10.1111/j.1365-2133.2006.07218.x. [DOI] [PubMed] [Google Scholar]
- 11.Vajo Z, Francomano CA, Wilkin DJ. The molecular and genetic basis of fibroblast growth factor receptor 3 disorders: The achondroplasia family of skeletal dysplasias, Muenke craniosynostosis, and Crouzon syndrome with acanthosis nigricans. Endocr Rev. 2000;21:23–39. doi: 10.1210/edrv.21.1.0387. [DOI] [PubMed] [Google Scholar]
- 12.Chesi M, et al. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappellen D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 14.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 15.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Hafner C, et al. FGFR3 mutation affects cell growth, apoptosis and attachment in keratinocytes. Exp Cell Res. 2010;316:2008–2016. doi: 10.1016/j.yexcr.2010.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberger G, Meien S, Kutsche K. Oncogenic HRAS mutations cause prolonged PI3K signaling in response to epidermal growth factor in fibroblasts of patients with Costello syndrome. Hum Mutat. 2009;30:352–362. doi: 10.1002/humu.20855. [DOI] [PubMed] [Google Scholar]
- 18.Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: Novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- 19.Liaw D, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 20.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 21.Hornigold N, et al. Mutation of the 9q34 gene TSC1 in sporadic bladder cancer. Oncogene. 1999;18:2657–2661. doi: 10.1038/sj.onc.1202854. [DOI] [PubMed] [Google Scholar]
- 22.Carpten JD, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 23.Jebar AH, et al. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24:5218–5225. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 24.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernard-Pierrot I, et al. Oncogenic properties of the mutated forms of fibroblast growth factor receptor 3b. Carcinogenesis. 2006;27:740–747. doi: 10.1093/carcin/bgi290. [DOI] [PubMed] [Google Scholar]
- 26.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 27.Hernández S, et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol. 2006;24:3664–3671. doi: 10.1200/JCO.2005.05.1771. [DOI] [PubMed] [Google Scholar]
- 28.van Rhijn BW, et al. Molecular grading of urothelial cell carcinoma with fibroblast growth factor receptor 3 and MIB-1 is superior to pathologic grade for the prediction of clinical outcome. J Clin Oncol. 2003;21:1912–1921. doi: 10.1200/JCO.2003.05.073. [DOI] [PubMed] [Google Scholar]
- 29.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 31.Kwiatkowski DJ, Manning BD. Tuberous sclerosis: A GAP at the crossroads of multiple signaling pathways. Hum Mol Genet. 2005;14(Spec No 2):R251–R258. doi: 10.1093/hmg/ddi260. [DOI] [PubMed] [Google Scholar]
- 32.Lee JC, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–476. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- 35.Collado M, et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 36.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 37.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavormina PL, et al. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- 41.López-Knowles E, et al. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res. 2006;66:7401–7404. doi: 10.1158/0008-5472.CAN-06-1182. [DOI] [PubMed] [Google Scholar]
- 42.Zhao L, Vogt PK. Hot-spot mutations in p110alpha of phosphatidylinositol 3-kinase (pI3K): Differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle. 2010;9:596–600. doi: 10.4161/cc.9.3.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang H, et al. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase. Cancer Res. 2009;69:8868–8876. doi: 10.1158/0008-5472.CAN-09-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalinsky K, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–5059. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 45.Collado M, Serrano M. Senescence in tumours: Evidence from mice and humans. Nat Rev Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michaloglou C, et al. BRAF E600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 47.Courtois-Cox S, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majumder PK, et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer Cell. 2008;14:146–155. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 50.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Happle R. Epidermal nevus syndromes. Semin Dermatol. 1995;14:111–121. doi: 10.1016/s1085-5629(05)80006-9. [DOI] [PubMed] [Google Scholar]
- 52.Mandinova A, et al. A positive FGFR3/FOXN1 feedback loop underlies benign skin keratosis versus squamous cell carcinoma formation in humans. J Clin Invest. 2009;119:3127–3137. doi: 10.1172/JCI38543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kilpivaara O, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41:455–459. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olcaydu D, et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet. 2009;41:450–454. doi: 10.1038/ng.341. [DOI] [PubMed] [Google Scholar]
- 55.Kiemeney LA, et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat Genet. 2010;42:415–419. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaturvedi V, Chu MD S, Carrol BS M, Brenner BS JW, Nickoloff BJ. Estimation of size of clonal unit for keratinocytes in normal human skin. Arch Pathol Lab Med. 2002;126:420–424. doi: 10.5858/2002-126-0420-EOSOCU. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura H, et al. Clonal nature of seborrheic keratosis demonstrated by using the polymorphism of the human androgen receptor locus as a marker. J Invest Dermatol. 2001;116:506–510. doi: 10.1046/j.1523-1747.2001.01289.x. [DOI] [PubMed] [Google Scholar]
- 58.Greaves M. Pre-natal origins of childhood leukemia. Rev Clin Exp Hematol. 2003;7:233–245. [PubMed] [Google Scholar]
- 59.Wiemels JL, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.