Abstract
Understanding how cell adhesion proteins form adhesion domains is a key challenge in cell biology. Here, we use single-molecule atomic force microscopy (AFM) to demonstrate the force-induced formation and propagation of adhesion nanodomains in living fungal cells, focusing on the covalently anchored cell-wall protein Als5p from Candida albicans. We show that pulling on single adhesins with AFM tips terminated with specific antibodies triggers the formation of adhesion domains of 100–500 nm and that the force-induced nanodomains propagate over the entire cell surface. Control experiments (with cells lacking Als5p, single-site mutation in the protein, bare tips, and tips modified with irrelevant antibodies) demonstrate that Als5p nanodomains result from protein redistribution triggered by force-induced conformational changes in the initially probed proteins, rather than from nonspecific cell-wall perturbations. Als5p remodeling is independent of cellular metabolic activity because heat-killed cells show the same behavior as live cells. Using AFM and fluorescence microscopy, we also find that nanodomains are formed within ∼30 min and migrate at a speed of ∼20 nm·min−1, indicating that domain formation and propagation are slow, time-dependent processes. These results demonstrate that mechanical stimuli can trigger adhesion nanodomains in fungal cells and suggest that the force-induced clustering of adhesins may be a mechanism for activating cell adhesion.
Keywords: single-molecule techniques, microbial adhesion, glycoproteins, fungi
Cell adhesion is a ubiquitous feature of living cells and plays essential roles in a variety of cellular processes, including neuronal interactions, cellular communication, inflammation, and microbial infection (1–5). A growing body of evidence indicates that, in mammalian cells, the early stage of adhesion involves the formation of adhesion domains (i.e., focal adhesion complexes) composed of aggregated proteins (6, 7). A remarkable trait of adhesion domains is their ability to grow and strengthen under force (8–10). Whether such force-dependent behavior also occurs with microbial adhesins is unknown.
Adhesins in the Candida albicans Als gene family bind the pathogen to host tissues and initiate biofilm formation (11, 12). Als proteins are covalently attached to the cell wall and consist of an N-terminal Ig-like region, which initiates cell adhesion, followed by a threonine-rich region with an amyloid-forming sequence (T), a tandem repeat (TR) region that participates in cell–cell aggregation, and a stalk region projecting the molecule away from the cell surface (Fig. 1A) (13–15). Previous studies in C. albicans and in a Saccharomyces cerevisiae surface display model showed that Als5p-mediated adhesion involves a rapid cell-to-ligand adhesion step, followed by slower cell-to-cell aggregation mediated by the T and TR regions (13, 15). These kinetics suggest that, following adhesion to ligands, Als5p may undergo conformational changes that mediate cellular aggregation. Consistent with this, Rauceo et al. (16) found that, following adhesion of one region of the cell to fibronectin-coated beads, the entire surface of the cell became competent to mediate cell–cell aggregation. This led the authors to suggest a model for Als5p-mediated aggregation in which an adhesion-triggered change in the conformation of Als5p propagates around the cell surface, forming ordered adhesion domains. Whether single-molecule techniques can demonstrate the formation of Als5p adhesion domains in a live cell is the question that we address here.
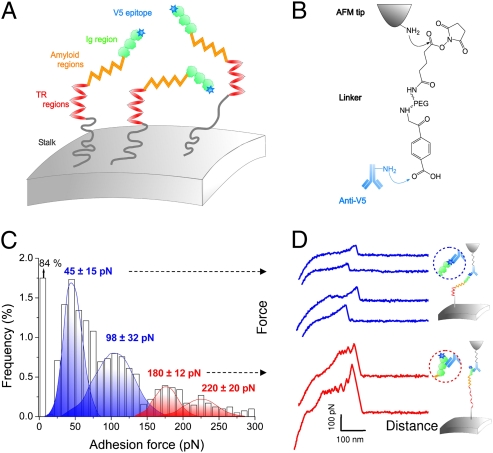
Fig. 1.
Detection and unfolding of single Als5p proteins in live cells. (A and B) Principle of the single-molecule force experiment. S. cerevisiae cells expressing Als5p proteins tagged with a V5 epitope (A) are probed, in buffer, using AFM tips terminated with anti-V5 antibodies (B). (C and D) Adhesion force histogram (n = 4,096) from four maps of 1,024 data points (C) and representative force curves (D) obtained by recording spatially resolved force curves over 1 × 1-μm areas of the cell surface using anti-V5 tips. The anti-V5 tip is capable of dual detection: the blue curves show single weak adhesion peaks reflecting recognition of the V5-tag, and the red curves feature sawtooth patterns with multiple force peaks documenting the unfolding of the entire protein via Ig binding. All curves were obtained at 20 °C using a loading rate of 10,000 pN·s−1 and an interaction time of 500 ms. Similar data were obtained using different tips and cells (10 tips, 6 cells). Als proteins expressed on S. cerevisiae exhibit the same activities as they do in C. albicans (13, 14).
Single-molecule atomic force microscopy (AFM) is a powerful tool for studying how proteins respond to mechanical forces (17–22). Stretching modular proteins such as titin (23) and tenascin (24) yields characteristic force signatures that reflect the force-induced unfolding of secondary structures (α-helices, β-sheets). AFM imaging has also visualized force-induced conformational changes in membrane proteins such as bacteriorhodopsin and aquaporin (25, 26). Yet, the use of single-molecule AFM to investigate the force-induced clustering of receptors in live cells has thus far not been documented. In this report, we demonstrate the triggering of Als5p adhesion nanodomains with force and their surface propagation across the entire cell. The results indicate that the adhesion function of Als5p is coupled to its local assembly within adhesion nanodomains, for which we propose the term “nanoadhesomes.” Comparative genomics shows similar protein design in other fungal adhesins (14), suggesting that clustering of cell adhesion proteins in response to mechanical stimuli may be a general mechanism for activating cell adhesion in eukaryotes.
Results
Dual Detection of Als5p Proteins in Live Cells.
We analyzed single Als5p proteins on yeast cells that were never exposed to mechanical force. To this end, a V5 epitope tag was inserted at the N-terminal end of full-length Als5p proteins (Fig. 1A) to allow for specific detection with AFM tips terminated with anti-V5 antibodies (Fig. 1B). Yeast cells expressing V5-tagged Als5p proteins were trapped into porous polymer membranes for in situ AFM analysis. Individual tagged proteins were detected by recording spatially resolved force curves on cell surfaces using an anti-V5 tip. As shown in Fig. 1C, 16% of the force curves featured adhesion force peaks. The corresponding adhesion force histogram displayed four maxima centered at 45 ± 15 piconewtons (pN), 98 ± 32 pN, 180 ± 12 pN, and 220 ± 20 pN. For two reasons, we attribute the ∼45 and ∼98 pN forces to the relatively weak binding of the anti-V5 antibody to the V5 epitope. First, similar forces were measured between an anti-V5 tip and a model surface functionalized with V5 epitopes (Fig. S1A). Second, the specificity of the measured interaction was confirmed by showing a substantial reduction of adhesion frequency upon injection of free epitopes on both model surfaces (Fig. S1B) and live cells (Fig. S2).
Strikingly, force curves with large adhesion peaks (∼180 and ∼220 pN) showed profiles characteristic of Als5p binding to protein ligands (Fig. 1D, red curves). Unlike the low-adhesion force curves (Fig. 1D, blue curves), these curves showed sawtooth patterns with multiple force peaks that we have previously reported as the sequential unfolding of the Als5p TR domains (22). Because (i) we used a protocol for tip modification that favors single antibody interactions and (ii) the large adhesion peaks were essentially never seen on model surfaces modified with V5 epitopes (Fig. S1), we attribute the large peaks to Als5p binding to protein ligands rather than to multiple antibody–epitope interactions. These observations lead us to conclude that the anti-V5 tip is capable of dual detection, i.e., molecular recognition of Als5p via the V5 epitope or strong multipoint attachment to ligands on the antibody (Fig. 1D, Insets).
Force-Induced Formation of Als5p Nanodomains.
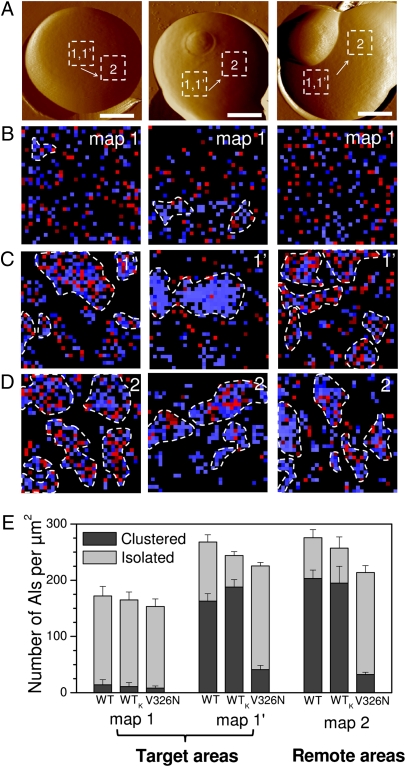
We next mapped the spatial arrangement of Als5p proteins. Adhesion force maps (32 × 32 force curves on 1 × 1-μm areas) obtained from wild-type cells (Fig. 2A) on which force was never applied revealed that the proteins were evenly distributed, without any clear evidence for clustering (Fig. 2B). Remarkably, we found that recording a second force map on the same given target area of the cell surface (Fig. 2C) led to an increase of the total protein surface density from 172 ± 16 proteins/μm2 (n = 6 maps of 1,024 data points recorded over 1 μm2) to 268 ± 13 proteins/μm2 (Fig. 2E). In addition, we observed that proteins in the second force map were no longer evenly distributed but formed clusters of 100–500 nm (Fig. 2C). As shown in Fig. 2E, the fraction of clustered molecules increased from 8% to 61%. This dramatic change in spatial organization was reproducibly observed upon recording additional maps. We note that the adhesion force histograms corresponding to consecutive maps (Fig. 2C) showed four maxima similar to those associated with primary maps (Fig. 1C).
Fig. 2.
Formation and propagation of Als5p nanodomains. (A) AFM topographic images (scale bars: 1.5 μm), in buffer, showing three different wild-type S. cerevisiae cells expressing V5-tagged Als5p proteins. (B) Adhesion force maps (1 × 1 μm) recorded with an anti-V5 tip on a given target area of the native cells, i.e., cells that were never subjected to force (map 1; recorded on the dashed squares in A). Blue and red pixels correspond to forces smaller and larger than 150 pN, respectively, and thus to V5-tagged Als5p recognition and unfolding. (C) Second adhesion force maps (1 × 1 μm) recorded on the same target area (map 1′). The heterogeneous distribution of colored pixels, which represents the detection of single Als5p, documents the formation of nanoscale clusters (highlighted by dashed lines). We define a cluster as a group of proteins containing at least 10 molecules (colored pixels) in direct contact with each other (via edges or corners). (D) Adhesion force maps (1 × 1 μm) recorded on remote areas (map 2) localized several hundred nanometers away from each other (see dashed squares in A). (E) Surface density histograms showing the number of proteins per square micrometer measured for wild-type cells (WT), for heat-killed wild-type cells (WTk), and for V326N mutant cells under different conditions: first maps were recorded in given target areas, and second maps were recorded in the same target areas or in remote areas, as marked. Darker and lighter colors represent the surface density of clustered and isolated proteins. For each condition, data obtained from minimum three maps generated on three different cells (n ≥ 3,072).
We also observed that heat-killed cells showed the same behavior as live cells (Fig. 3 A–E), the Als5p density in killed cells increasing from 153 ± 14 proteins/μm2 (n = 4 maps of 1,024 data points recorded over 1 μm2) to 244 ± 7 proteins/μm2 when recording two consecutive force maps in the same area (Fig. 2E). Because killed cells cannot increase surface expression of adhesins in response to the mechanical stimulus, we infer that the increase in active Als5p molecules and their clustering must be due to rearrangement of molecules already present on the cell surface. These results show that the force-induced clustering of Als5p is independent of cellular metabolic activity, consistent with the notion that Als5p-mediated aggregation does not involve metabolic activity (16).
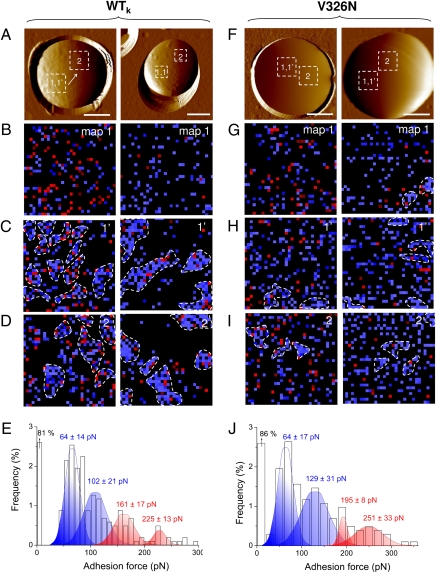
Fig. 3.
Nanodomain formation in heat-killed cells and V326N mutant cells. (A and F) AFM topographic images (scale bars: 2 μm), in buffer, showing two heat-killed (60 °C, 30 min) cells expressing wild-type Als5p (A) and two cells expressing the V5-tagged Als5p mutant protein Als5pV326N bearing a single-site mutation in the amyloid region (F). (B–D and G–I) Sets of consecutive adhesion force maps (1 × 1 μm) recorded with an anti-V5 tip on the same target areas (map 1 and map 1′), followed by third maps recorded on remote areas (map 2). (E and J) Adhesion force histograms corresponding to the data (n = 1,024) shown in map 1.
Our finding that the precise delivery of piconewton forces on the cell surface triggers the formation of Als5p nanodomains may be interpreted in two ways. An appealing explanation is that Als5p clustering results from protein redistribution triggered by force-induced conformational changes in the initially probed proteins. However, an alternative model may also be proposed in which the force applied by the tip perturbs the cell wall nonspecifically and changes the subsequent local force map, via, e.g., alteration of the carbohydrate surface layer. A series of control experiments were carried out to rule out the second model. First, we showed above that the anti-V5 tip interacts specifically with V5-tagged Als5p proteins (Fig. S1 and Fig. S2) and is able to sequentially unfold the TR domains upon stretching (22). Second, yeast cells lacking the V5-tagged Als5p proteins showed essentially no Als5p detection, and thus no clusters, even when consecutive maps were recorded (Fig. S3 A–C). Third, recording two consecutive maps on cells expressing V5-tagged Als5p proteins using a bare tip or an irrelevant antibody (i.e., anti-lysozyme) did not induce clustering or an increase in protein density (Fig. S3 D–I). Fourth, as we will see below, Als5p reorganization was abolished with a single-site mutation in the protein. Fifth, the Als5p reorganization is a slow, time-dependent process (see below for details). From these observations, we conclude that Als5p clustering results from protein redistribution triggered by force-induced conformational changes in the probed proteins. These data on live cells are reminiscent of those obtained on biomimetic model systems (9). Applying piconewton lifting forces on two contacting fluid membranes with magnetic tweezers was shown to induce the migration of mobile integrins into the contact zone and, in turn, to increased adhesion. The authors suggest that such force-induced growth of adhesion domains could play a key role in signaling events leading to mechanosensing in living cells.
Time-Dependent Propagation of Als5p Nanodomains.
Because adhesive activation of Als-expressing cells propagates around the cell surface (16), we asked whether the force-induced nanodomains can propagate across the surface by recording consecutive force maps on remote areas, i.e., areas localized several micrometers away from the first maps. Strikingly, we observed adhesion domains on remote areas (Fig. 2D) that were very similar to those observed on target areas (Fig. 2C). The same behavior was observed on many different remote areas, indicating that delivering piconewton forces on a target area eventually induces the formation of Als5p clusters across the entire cell surface.
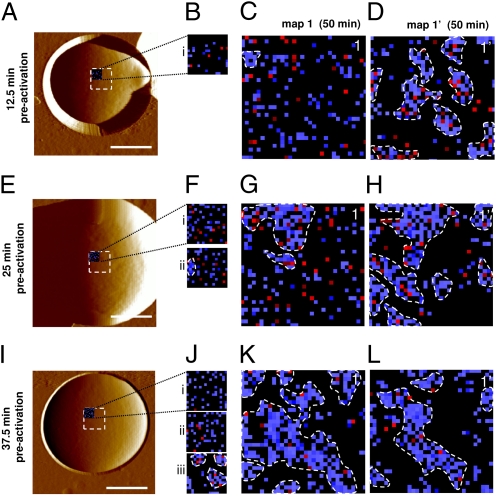
What is the time required for protein reorganization? Because clusters were essentially never seen in primary maps (see map1 in Fig. 2), we postulated that nanodomain formation and propagation are rather slow processes. To further investigate this time dependency, cells were preactivated by force probing during increasing periods of time (Fig. 4). Specifically, we recorded a sequence of small adhesion force maps on the same target areas (16 × 16 force curves on 500 × 500 nm for 12.5 min; see i, ii, and iii in Fig. 4 B, F, and J), followed by two consecutive “classical” maps (32 × 32 force curves on 1 × 1 μm for 50 min; see 1 and 1′ in Fig. 4 C, D, G, H, K, and L). Note that force curves in the small and large maps were recorded using the same approach and retraction rates and using the same lateral spacing between two curves. We found the following: (i) recording a single small map (Fig. 4B) was not sufficient to observe substantial clustering in a subsequent large map (Fig. 4C), clustering being observed only in the next large map (Fig. 4D); (ii) recording two consecutive small maps (Fig. 4F, i and ii) was able to induce clustering in the upper left corner of the next consecutive large map, but not in the other corners (Fig. 4G), revealing that 25 min of preactivation induces formation, but not propagation, of the clusters; and (iii) recording three small maps (Fig. 4J, i, ii, and iii) triggered the formation of Als5p clusters, but also their subsequent propagation because clusters were observed in the entire large map (Fig. 4K) and thus in regions that were never exposed to force. Because it takes 25 min to record the upper two corners of a large map, migration from the left to the right corner occurred within 25 min. These observations show that the formation of Als5p clusters in a 500 × 500-nm target area takes about 30 min (i.e., between 25 and 37.5 min). After the domains are formed, they propagate at a speed of ∼20 nm·min−1 (500 nm in 25 min), suggesting that propagation across the entire cell is reached within ∼6 h. Because activation kinetics from adhesion assays show faster activation (10–30 min) (16, 27), we believe that the speed of propagation in vivo is due to activation at several points on the cell surface through interaction with beads or other cells. These results lead us to conclude that domain formation and propagation are two slow, time-dependent processes. We expect that, if clustering was due to some perturbation of the cell wall by probing, then it should happen quickly. Hence, our finding of the time requirement for reorganization provides direct evidence that this process results from force-induced conformational changes in the probed proteins.
Fig. 4.
Time scale of the Als5p reorganization. (A, E, and I) AFM topographic images (scale bars: 1.5 μm), in buffer, showing three different cells expressing V5-tagged Als5p proteins. (B, F, and J) Small preactivation maps (16 × 16 force curves) recorded on 500 × 500-nm areas with an anti-V5 tip on the three native cells that were never subjected to force (maps i, ii, and iii recorded in the upper left corner of the dashed squares in A, E, and I). (C, D, G, H, K, L) Two consecutive adhesion force maps of 1 × 1 μm (map 1 and 1′) recorded on the same preactivated areas shown in B, F, and J.
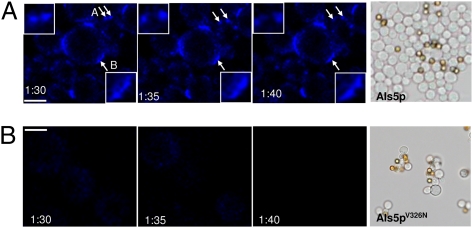
As a complement, time-lapse confocal microscopy was used with the aim of visualizing the clustering and movement of Als5p domains on shear-activated aggregated cells after staining with the amyloid-specific dye thioflavin T (Fig. 5A) (13). Consistent with AFM, adhesion domains (brightly fluorescent regions) were found to migrate (Fig. 5A, “A” arrows and Inset) and coalesce (Fig. 5A, “B” arrow and Inset) on the cell surface.
Fig. 5.
Aggregated cells expressing Als5p exhibit dynamic punctate surface fluorescence after staining with thioflavin T. Cells expressing Als5p (A) or Als5pV326N (B) were force-activated in a 1-h adhesion assay in the presence of the amyloid dye thioflavin T (100 nM). After aggregation, time-lapse confocal images were obtained at 5-min intervals. The images show migration (“A” arrows and Inset) and coalescence (“B” arrow and Inset) of punctate surface fluorescent domains. (Scale bars: 2 μm.) The right panels show results from aggregation assays of cells expressing Als5p or Als5pV326N.
Als5p Clustering Is Abolished with the V326N Single-Site Mutation.
We finally analyzed the relationship between the structure of Als5p and its clustering and adhesive properties. Als5p and other yeast adhesion proteins have conserved amyloid-forming sequences, and self-propagating amyloid formation is important for formation of robust cellular aggregates (13, 14, 16). The V326N single-site mutation in the amyloid-forming region of Als5p abrogates amyloid formation (13). Cells expressing Als5p bound to BSA-coated beads under shear and formed large cellular aggregates, whereas Als5pV326N cells adhered similarly to the beads, but formed few aggregates (Fig. 5, Right panels). Figure 3 F–J shows that Als5p clustering properties were almost completely abolished in the V326N mutant. The dynamic thioflavin T-staining regions were also absent from cells expressing the mutated protein (Fig. 5B). The minimal fluorescence was short-lived compared with that on cells expressing wild-type Als5p, implying that the spots were not self-propagating in cells expressing the V326N variant. These data show that clustering and propagation depend on a specific region of the Als5p protein.
Discussion
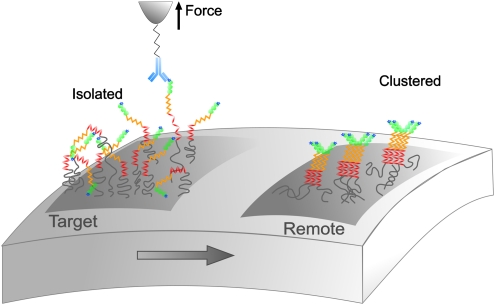
We have shown that the localized delivery of piconewton forces on living yeast cells triggers the formation and propagation of Als5p adhesion nanodomains. We suggest a two-step mechanism to explain the force-induced formation of such nanoadhesomes (Fig. 6). In the initial stage, stretching and unfolding of Als5p with the AFM tip extends conformations in which homologous hydrophobic interactions are favored. Fast hydrophobic interactions between extended tandem repeats would first promote Als5p self-associations and the recruitment of further neighboring proteins (15). Next, slower interactions would take place between the stimulated proteins, such as amyloid interactions between threonine-rich regions, and then propagate across the entire surface. Such a model is consistent with our finding that clustering requires specific interactions with Als5p and that Val326 is essential for the process. An alternative model would be that remodeling of the cell-wall polysaccharide matrix creates mobile clusters of covalently attached Als5p molecules, even in heat-killed cells. It is difficult to reconcile such rearrangement with our current understanding of cell-wall structure in yeast (28).
Fig. 6.
Proposed mechanism for the Als5p nanoadhesome. Pulling on single Als5p proteins with piconewton forces triggers the formation and propagation of Als5p clusters over the entire cell surface. Because the molecules are 70–120 nm long, they show local rotational mobility, allowing them to interact with neighboring molecules without involving free diffusion over the entire cell surface.
We also suggest that in nature C. albicans adhesion is governed by similar mechanisms. During the early stage of aggregation, local forces generated at cell–cell contacts could lead to the formation and propagation of Als domains that eventually strengthen cellular aggregation. Clustered Als5p proteins will resist larger forces than isolated proteins because of reduced diffusion of neighboring binding sites. The process in which Als5p adhesion domains strengthen yeast–yeast aggregation is reminiscent of events occurring in animal cells. A well-known example is the strong correlation between the cell adhesion activity of cadherins and their concentration within cell–cell adhesion sites (29).
In conclusion, our results demonstrate the formation and propagation of Als5p adhesion nanodomains in response to mechanical stimuli. Because force-induced activation is a common biological phenomenon (8–10), our observations suggest that force-dependent adhesin clustering may be a general mechanism for activating cell adhesion. A detailed understanding of this phenomenon would offer exciting prospects in therapeutics, e.g., for developing antimicrobial strategies.
Methods
Microorganisms and Cultures.
S. cerevisiae W3031B harboring plasmids pGK114 or pGK114V326N were grown on SC−trp plates. Mutant strain V326N was obtained by site-specific mutagenesis and was confirmed by sequencing, as will be described elsewhere (13). Two or three colonies from the SC–trp plate used as inoculum were transferred into Sc–trp medium (1.7 g/L yeast extract without amino acids and without ammonium sulfate, 1.92 g/L yeast synthetic drop-out medium supplements without trp, 5 g/L ammonium sulfate, and 20 g/L galactose). Cells were agitated at 30 °C, grown overnight, and harvested by centrifugation. They were washed three times with sodium acetate buffer and resuspended in 10 mL buffer to a concentration of ∼106 cells/mL.
Atomic Force Microscopy.
AFM measurements were performed at room temperature (20 °C) in buffered solutions (sodium acetate; pH 4.75), using a Nanoscope IV Multimode AFM (Veeco Metrology Group) and oxide-sharpened microfabricated Si3N4 cantilevers (Microlevers, Veeco Metrology Group). Cells were immobilized by mechanical trapping into porous polycarbonate membranes (Millipore), with a pore size similar to the cell size. After filtering a concentrated cell suspension, the filter was gently rinsed with buffer, carefully cut (1 × 1 cm), and attached to a steel sample puck (Veeco Metrology Group), and the mounted sample was transferred into the AFM liquid cell while avoiding dewetting. The spring constants of the cantilevers were measured using the thermal noise method (Picoforce, Veeco Metrology Group), yielding values ranging from 0.008 to 0.021 N/m. All force measurements were recorded with a loading rate of 10,000 pN/s.
AFM tips were functionalized with anti-V5 antibodies (Invitrogen) using PEG-benzaldehyde linkers as described by Ebner et al. (30). Cantilevers were washed with chloroform and ethanol, placed in a UV ozone cleaner for 30 min, immersed overnight in an ethanolamine solution (3.3 g ethanolamine in 6 mL of DMSO), washed three times with DMSO and two times with ethanol, and dried with N2. The ethanolamine-coated cantilevers were immersed for 2 h in a solution prepared by mixing 1 mg acetal-PEG-NHS dissolved in 0.5 mL chloroform with 10 μL triethylamine, washed with chloroform, and dried with N2. Cantilevers were further immersed for 5 min in a 0.1% iodine/acetone solution, washed three times in acetone, dried under N2, and then covered with a 200-μL droplet of a PBS solution containing anti-V5 (0.2 mg/mL) to which 2 μL of a 1 M NaCNBH3 solution was added. After 50 min, cantilevers were incubated with 5 μL of a 1-M ethanolamine solution to passivate unreacted aldehyde groups and then washed with and stored in PBS 10 min later. Model surfaces were functionalized with V5 epitopes using the same protocol.
Confocal Microscopy.
Aggregation assays were carried out as previously described (16), with cells activated during a 60-min adhesion assay to BSA-coated beads under shear in the presence of 100 nM thioflavin T (Sigma). Cells were observed at 5-min intervals beginning 90 min after aggregation to mimic the preobservation interval under AFM conditions. The microscope was a Nikon Eclipse 90i confocal microscope with 408 excitation and 450 emission filters.
Supplementary Material
Acknowledgments
Work at the Université Catholique de Louvain was supported by Fonds National de la Recherche Scientifique, the Université Catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté Française de Belgique (Concerted Research Action). Work at Brooklyn College was supported by National Institutes of Health Grant SC1 GM083756. Y.F.D. and D.A. are, respectively, Senior Research Associate and Research Fellow of the Fonds National de la Recherche Scientifique.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013893107/-/DCSupplemental.
References
- 1.Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- 2.Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: Signalling functions at the synapse. Nat Rev Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber C, Fraemohs L, Dejana E. The role of junctional adhesion molecules in vascular inflammation. Nat Rev Immunol. 2007;7:467–477. doi: 10.1038/nri2096. [DOI] [PubMed] [Google Scholar]
- 4.Gray-Owen SD, Blumberg RS. CEACAM1: Contact-dependent control of immunity. Nat Rev Immunol. 2006;6:433–446. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- 5.Verstrepen KJ, Reynolds TB, Fink GR. Origins of variation in the fungal cell surface. Nat Rev Microbiol. 2004;2:533–540. doi: 10.1038/nrmicro927. [DOI] [PubMed] [Google Scholar]
- 6.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 7.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 8.Bershadsky AD, Kozlov M, Geiger B. Adhesion-mediated mechanosensitivity: A time to experiment, and a time to theorize. Curr Opin Cell Biol. 2006;18:472–481. doi: 10.1016/j.ceb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Smith AS, Sengupta K, Goennenwein S, Seifert U, Sackmann E. Force-induced growth of adhesion domains is controlled by receptor mobility. Proc Natl Acad Sci USA. 2008;105:6906–6911. doi: 10.1073/pnas.0801706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc Natl Acad Sci USA. 2009;106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9:176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- 13.Otoo HN, Lee KG, Qiu WG, Lipke PN. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot Cell. 2008;7:776–782. doi: 10.1128/EC.00309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsook CB, et al. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell. 2010;9:393–404. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank AT, et al. Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot Cell. 2010;9:405–414. doi: 10.1128/EC.00235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauceo JM, et al. Global cell surface conformational shift mediated by a Candida albicans adhesin. Infect Immun. 2004;72:4948–4955. doi: 10.1128/IAI.72.9.4948-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bustamante C, Macosko JC, Wuite GJL. Grabbing the cat by the tail: Manipulating molecules one by one. Nat Rev Mol Cell Biol. 2000;1:130–136. doi: 10.1038/35040072. [DOI] [PubMed] [Google Scholar]
- 18.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 19.Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu Rev Biochem. 2008;77:127–148. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- 20.Müller DJ, Helenius J, Alsteens D, Dufrêne YF. Force probing surfaces of living cells to molecular resolution. Nat Chem Biol. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 21.Puchner EM, Gaub HE. Force and function: Probing proteins with AFM-based force spectroscopy. Curr Opin Struct Biol. 2009;19:605–614. doi: 10.1016/j.sbi.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Alsteens D, et al. Unfolding individual Als5p adhesion proteins on live cells. ACS Nano. 2009;3:1677–1682. doi: 10.1021/nn900078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 24.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 25.Müller DJ, Büldt G, Engel A. Force-induced conformational change of bacteriorhodopsin. J Mol Biol. 1995;249:239–243. doi: 10.1006/jmbi.1995.0292. [DOI] [PubMed] [Google Scholar]
- 26.Scheuring S, et al. High resolution AFM topographs of the Escherichia coli water channel aquaporin Z. EMBO J. 1999;18:4981–4987. doi: 10.1093/emboj/18.18.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaur NK, Klotz SA, Henderson RL. Overexpression of the Candida albicans ALA1 gene in Saccharomyces cerevisiae results in aggregation following attachment of yeast cells to extracellular matrix proteins, adherence properties similar to those of Candida albicans. Infect Immun. 1999;67:6040–6047. doi: 10.1128/iai.67.11.6040-6047.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez M, de Groot PWJ, Klis FM, Lipke PN. Glycoconjugate structure and function in fungal cell walls. In: Moran A, Brennan P, Holst O, von Itzstein F, editors. Microbial Glycobiology: Structures, Relevance, and Applications. Amsterdam; London; New York: Elsevier; 2009. pp. 169–183. [Google Scholar]
- 29.Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 30.Ebner A, et al. A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconjug Chem. 2007;18:1176–1184. doi: 10.1021/bc070030s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.