Abstract
Cardiac and skeletal muscle development and maintenance require complex interactions between DNA-binding proteins and chromatin remodeling factors. We previously reported that Smyd1, a muscle-restricted histone methyltransferase, is essential for cardiogenesis and functions with a network of cardiac regulatory proteins. Here we show that the muscle-specific transcription factor skNAC is the major binding partner for Smyd1 in the developing heart. Targeted deletion of skNAC in mice resulted in partial embryonic lethality by embryonic day 12.5, with ventricular hypoplasia and decreased cardiomyocyte proliferation that were similar but less severe than in Smyd1 mutants. Expression of Irx4, a ventricle-specific transcription factor down-regulated in hearts lacking Smyd1, also depended on the presence of skNAC. Viable skNAC−/− adult mice had reduced postnatal skeletal muscle growth and impaired regenerative capacity after cardiotoxin-induced injury. Satellite cells isolated from skNAC−/− mice had impaired survival compared with wild-type littermate satellite cells. Our results indicate that skNAC plays a critical role in ventricular cardiomyocyte expansion and regulates postnatal skeletal muscle growth and regeneration in mice.
Cardiogenesis is regulated in a temporally and spatially precise fashion by numerous signaling, transcriptional, and translational networks (1, 2). Epigenetic events, including posttranslational modification of histones, modulate the structure of chromatin and the accessibility of regulatory sequences to transcriptional activators and repressors. Acetylation of conserved lysine residues in histone tails by histone acetyltransferases stimulates chromatin relaxation and transcription, whereas deacetylation by histone deacetylases represses transcription (3). Methylation of lysine and arginine residues in histones affects chromatin conformation and either facilitates or inhibits transcription (4).
Smyd1 is a muscle-restricted member of a family of chromatin remodeling proteins that contains both MYND and SET domains (5). The MYND domain coordinates protein–protein interactions to recruit a corepressor complex, and the SET domain commonly functions as a methyltransferase for histones or other proteins (6, 7). Targeted deletion of Smyd1 in mice results in a failure of ventricular cardiomyocyte maturation with ventricular hypoplasia, similar to cardiac phenotypes in embryos lacking Mef2c or Hand2 (dHAND) (5, 8, 9). Smyd1 is a direct transcriptional target of Mef2c and regulates the expression of Hand2 and Irx4, all of which function in a transcriptional network to control ventricular cardiomyocyte growth and differentiation (5, 10). Because of the early embryonic lethality of Smyd1 mutant mice, its role in skeletal muscle development remains unclear in mice. Knockdown of SmyD1a and SmyD1b expression in zebrafish embryos by morpholino antisense oligos resulted in a myofibril organization defect (11). Although Smyd1 may not bind DNA, it is likely recruited to muscle-specific target genes through physical interactions with unknown DNA-binding partners.
Here, we show that the muscle-restricted isoform of the DNA-binding protein nascent polypeptide-associated complex (skNAC) is a major interacting partner of Smyd1 in the developing heart by yeast two-hybrid screening, similar to skeletal muscle (12), and that disruption of skNAC in mice results in partial embryonic lethality between embryonic day (E) 9.5 and E12.5 due to ventricular cardiomyocyte hypoplasia. The subset of skNAC mutants that survived to adulthood had reduced postnatal skeletal muscle growth. In addition, we found that skNAC-null skeletal muscle had markedly impaired regenerative capacity in response to injury. Thus, our findings demonstrate that skNAC is involved in early cardiac development and postnatal skeletal muscle growth and regeneration.
Results
skNAC Is a Muscle-Specific Partner of Smyd1 During Cardiogenesis.
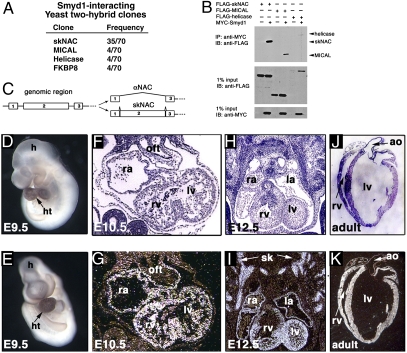
To understand the molecular mechanism by which Smyd1 regulates embryonic heart development, we constructed a yeast two-hybrid library from mouse embryonic heart (E11.5) and screened for interacting partners with full-length Smyd1 as bait. Half of the candidates isolated from the screen (35 of 70) encoded skNAC, a muscle-specific transcription factor that interacts with Smyd1 in skeletal muscle (12) (Fig. 1A). Expression of skNAC and Smyd1 is induced during skeletal myogenesis in culture, and the interaction domains in both Smyd1 and skNAC have been mapped previously (12). Several other proteins were isolated, and their interactions were confirmed by reversing prey and bait. These included a cytoplasmic protein with a flavoprotein monooxygenase domain, MICAL (4 of 70), involved in myofilament organization and actin dynamics (13); a unique protein with a helicase domain (4 of 70); and FK506 binding protein 8 (4 of 70), which regulates BCL2 (14). Each of these interactions was confirmed by coimmunoprecipitation assays (Fig. 1B).
Fig. 1.
Smyd1-interacting partners in the heart and muscle-specific expression of skNAC. (A) Frequency of Smyd1-interacting partners isolated by yeast two-hybrid screening of an embryonic heart cDNA library. (B) Immunoprecipitation (IP) of Smyd1 with anti-MYC antibodies and immunoblot (IB) with anti-FLAG antibodies are shown along with Western blots of input protein used. (C) Genomic organization of the NAC gene with splice forms of skNAC and αNAC indicated. (D and E) Whole-mount in situ hybridizations with exon 2-specific probe at E9.5 in lateral (D) and frontal views (E). (F–K) Section in situ hybridizations at indicated stages with dark-field and bright-field images. h, head; ht, heart; oft, outflow tract; ra, right atrium; la, left atrium; rv, right ventricle; lv, left ventricle; ao, aorta; sk, skeletal muscle precursor.
We focused our further studies on skNAC because of its strong interaction with Smyd1 and its muscle-specific expression, although Smyd1 interaction with the other proteins may be important as well. skNAC is generated by splicing-in of a 6-kb second exon (Fig. 1C) and encodes a transcriptional activator of the myoglobin promoter (15). The ubiquitous αNAC does not contain exon 2 but can function as a transcriptional coactivator of osteocalcin through an interaction with Jun and also acts as a positive regulator of human erythroid cell differentiation (16, 17).
Whole-mount in situ hybridization revealed high levels of skNAC expression in all four chambers of the looping heart at E9.5 (Fig. 1 D and E), but lower expression before E9.0. Section in situ hybridization showed weak expression in the heart at E8–E8.5. At E10.5 and E12.5, expression was restricted to the myocardial layers, including the compact zone, interventricular septum, and trabeculae (Fig. 1 F–I). At E12.5, high levels of expression were also observed in skeletal muscle precursors (Fig. 1I). Transcripts in the heart and skeletal muscle persisted throughout subsequent development and adulthood (Fig. 1K). The temporal and spatial expression patterns of skNAC were almost identical to those of Smyd1 (5), consistent with their physical interaction.
skNAC−/− Embryos Exhibit Ventricular Hypoplasia.
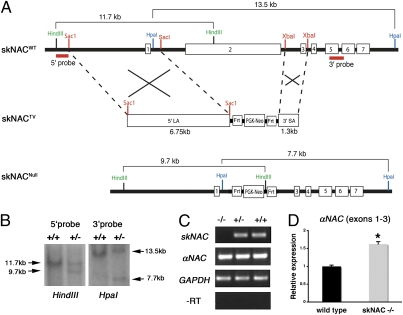
To investigate the in vivo function of skNAC, we deleted skNAC in mice. Because Northern analysis demonstrated ubiquitous αNAC expression from E7 onward, we designed a targeting vector to replace only the skNAC-specific exon 2 with the neomycin-resistance gene cassette (Neo), flanked by Flp recombinase (FRT) sites (Fig. 2A). This approach was designed to disrupt skNAC expression, while maintaining αNAC expression. Targeted ES cells were identified by Southern analysis (Fig. 2B), and chimeras were generated from two independent lines. Heterozygous mice lacking skNAC were generated either on a mixed or isogenic background and were normal. The offspring from intercrosses of heterozygous skNAC mice were genotyped before weaning. As summarized in Table S1, 17% of pups from the mixed 129S6/C57BL/6 background and 8% from the 129S6 isogenic background were homozygous, instead of the expected Mendelian ratio of 25%. This partial embryonic lethality occurred even upon deletion of Neo on the null allele by intercrossing with ubiquitous ACTb:FLPe mice (18). The absence of skNAC but maintenance of αNAC expression in homozygous mutant mice was confirmed by RT-PCR (Fig. 2C). By real-time quantitative PCR (qPCR), we found that αNAC expression was not reduced in the skNAC mutants but rather slightly up-regulated (Fig. 2D).
Fig. 2.
Targeting strategy for deletion of the skNAC-specific exon 2 in mice. (A) Organization of the skNAC wild-type allele (skNACWT) and targeting vector (skNACTV) and replacement of exon 2 by homologous recombination (skNACNull). (B) Southern analysis of genomic DNA of targeted ES cells after digestion with HindIII and HpaI and after hybridization with 5′ and 3′ probes, respectively. (C) RT-PCR analysis showing the absence of skNAC transcripts and the maintenance of αNAC transcripts in skNAC−/− embryonic heart. GAPDH is a loading control. (D) qPCR showing expression of αNAC transcripts in skNAC−/− hearts. Data are presented as mean with SE (n = 6, *P < 0.01).
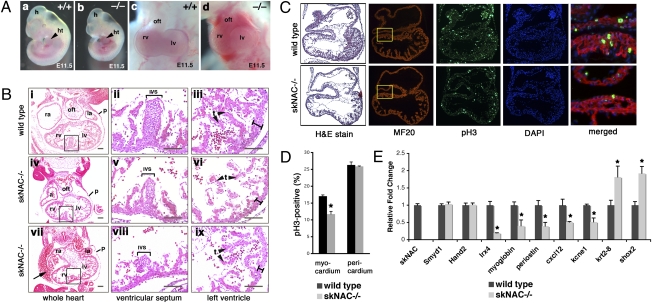
To investigate the timing and cause of prenatal death upon loss of skNAC, we examined embryos from timed pregnancies of skNAC heterozygous intercrosses. Most of the deaths occurred between E10.5 and E12.5 (Table S1), but animals that survived this period were viable and fertile. Because the phenotypes of affected skNAC−/− embryos were identical on the mixed and isogenic backgrounds, we used 129S6 isogenic mice for all subsequent experiments. The development of skNAC−/− embryos was slightly delayed, but most embryos were generally normal until E10.5. Approximately half of the embryos beyond E10.5 displayed pericardial edema and some developmental delay, typical of many mouse mutants with cardiac dysfunction (Fig. 3 Aa and Ab). From the muscle-specific expression of skNAC and maintenance of αNAC expression, this phenotype is likely secondary to hemodynamic insufficiency from cardiac dysfunction. At E11.5, pericardial hemorrhage was also frequently noted on gross inspection, suggesting breach of the myocardial wall (Fig. 3 Ac and Ad).
Fig. 3.
Cardiomyocyte defect in skNAC−/− embryos. (A) Gross morphological abnormalities in skNAC−/− embryos compared with wild type at E11.5. Lateral views and frontal views focused on the heart show hemorrhage within the pericardium. h, head; ht, heart; oft, outflow tract; rv, right ventricle; lv, left ventricle. (B) Transverse sections of wild type and mutant (×2) E11.5 embryos stained with H&E. Note blood cells in the pericardial sac of skNAC mutants (arrow). Areas for ventricular septum and left ventricle wall are shown at higher magnification. Thin myocardial wall in skNAC−/− hearts is indicated (black bar) (vi and ix). ra, right atrium; la, left atrium; p, pericardium; ivs, interventricular septum; t, trabeculae. (Scale bars, 100 μm.) (C) Histological sections of wild type and skNAC−/− E10.0 hearts stained with either H&E, MF20 antibody, anti-pH3 antibody, or DAPI. Merged image of boxed area is shown at higher magnification. (D) Percentage proliferating myocytes was determined as the number of the pH3-positive, MF20-positive cells divided by the total number of DAPI/MF20-stained nuclei per section. Values are the average of multiple sections from each of four embryos per genotype, with error bars indicating SE (*P < 0.01). (E) qPCR analysis for mRNA levels of dysregulated genes in hearts of skNAC mutants at E10.5. Data are presented as mean of each genotype (n = 3) with SE (*P < 0.01).
Histological analysis of affected skNAC-null embryos confirmed the pericardial effusion and revealed blood cells in the pericardial chamber (Fig. 3B). In skNAC−/− embryos, the myocardial wall was thinner than in wild-type littermates, the interventricular septum was poorly formed, and trabeculae were decreased and poorly organized. In agreement with the gross appearance, section in situ hybridization analysis with cardiac chamber-specific markers, such as Hand2, Hand1, and Tbx5, and markers of valve-forming regions, such as Tbx2 and TGFβ2, revealed appropriate specification of subdomains within the heart. Although skNAC expression was restricted to myocardial layers of the heart, we performed extensive histological analyses for potential vascular or placental defects in the mutant embryos, because these can also cause hemodynamic insufficiency. We did not find any abnormalities outside the heart of skNAC-null embryos. Vascular patterning appeared normal as marked by an antibody against the endothelial-specific protein PECAM.
Ventricular Hypoplasia Is Caused by Proliferation Defects.
We investigated whether the thin ventricular myocardial layer in skNAC−/− embryos was caused by a proliferation defect or by abnormal apoptosis. We examined hearts at multiple times, focusing on hearts before obvious pathologic abnormalities to avoid secondary effects. At E10.0, before evidence of pericardial edema or developmental delay, we found that the myocardium of skNAC−/− hearts had a lower percentage of phosphohistone H3 (pH3)-positive cells than wild-type littermates by counting pH3 and MF20 double-positive myocytes (Fig. 3C). Quantitative analyses of the proliferation defect showed 30% fewer proliferating myocytes in skNAC−/− than in wild-type hearts (Fig. 3D; P < 0.01). The proliferation rates in the pericardium and other parts of the embryo were similar in wild-type and mutant mice at E10.0 (Fig. 3D). TUNEL assays revealed no apoptotic cells in the hearts of either wild-type or skNAC−/− embryos.
Altered Gene Expression in the Hearts of skNAC Mutants.
To identify gene expression changes associated with the myocardial defects in skNAC−/−hearts, we performed mRNA microarray analysis of skNAC−/− and wild-type hearts at E10.5. We excluded any hearts that showed signs of slow contraction, pericardial edema, or hemorrhage to avoid gene expression changes secondary to cardiac dysfunction. Three independent biologic samples for each genotype were analyzed. Surprisingly, only 23 genes were differentially expressed in all skNAC−/− hearts, 17 of which are currently annotated genes.
We validated candidates from the microarray by qPCR (Fig. 3E). Myoglobin, a muscle-specific hemoprotein (19), is a reported direct target of skNAC (15) and was one of the most down-regulated genes in skNAC mutant hearts. Interestingly, two of the other down-regulated genes, Periostin and Cxcl12 (Sdf-1), positively regulate cardiomyocyte or endothelial proliferation in the developing heart, consistent with the proliferation defect in skNAC mutant hearts (20, 21). We reported that expression of Irx4, a ventricular-specific member of the family of transcription factors that contains an Iroquois box (22), was decreased in Smyd1 mutants (5). The observation that Irx4 was also down-regulated in the skNAC-null heart (Fig. 3E) suggests that some targets may be commonly regulated, although the Smyd1-dependent gene Hand2 (9) was unaffected in skNAC mutants, implying that it is independently regulated. Finally, we found that some genes normally expressed at low levels in differentiated working myocardium were up-regulated. These genes included the epithelial marker Krt2.8 (23) and Shox2, a transcription factor required for normal cardiac conduction system development (24), possibly indicating incomplete myocyte differentiation in skNAC mutant hearts. Many of these dysregulated genes are critical for normal cardiac physiology and could individually or collectively contribute to the cardiac dysfunction in embryos lacking skNAC.
skNAC Is Important for Postnatal Skeletal Muscle Growth and Regulates Fiber Type Specification.
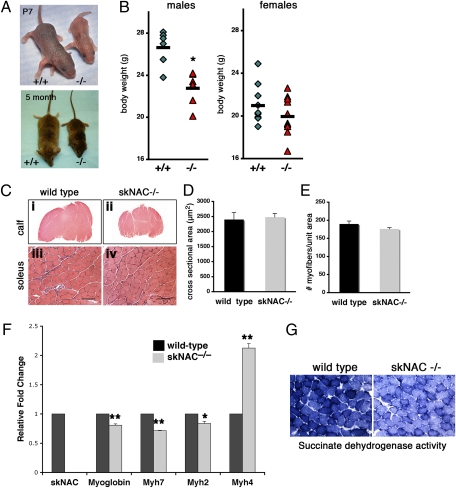
In the subset of viable skNAC−/− mice that survived postnatally, growth retardation was evident as early as the end of the first week after birth, although at the time of birth there was no obvious difference among littermates. The difference in size persisted throughout adulthood (Fig. 4A). skNAC−/− mice were smaller than gender-matched littermates at 3 mo of age, and the weight difference was statistically significant in males (P < 0.01) (Fig. 4B). skNAC−/− mice were physically active and fertile. Cardiac function was normal by echocardiography.
Fig. 4.
Defects in postnatal skeletal muscle growth in skNAC−/− mice. (A) Growth retardation of skNAC−/− mice at 1 wk (P7) and 5 mo of age. (B) Body weight of skNAC−/− mice and gender-matched littermates at 3 mo of age (male, n = 6; female, n = 11). The average is marked with a black line (*P < 0.01). (C) Reduced muscle mass in skNAC−/− mice. (i and ii) A comparison of H&E-stained sections of wild-type and skNAC−/− calf muscles shows the smaller size of skNAC−/− muscle at 3 mo of age. (iii and iv) At higher magnification, cross-sectional area of individual myofibers looks similar in wild-type and skNAC−/− soleus. (Scale bars, 100 μm.) (D and E) Quantitative analysis of myofibers in skNAC−/− or wild-type muscle. The number of myofibers in 480,000 μm2 was counted, and the cross-sectional area of individual fibers was measured by using ImageJ software. Data are presented as mean with SE (n = 3). (F) Muscle fiber type markers in soleus of skNAC−/− mice. qPCR revealed that expression of Myh4 (glycolytic, type IIb) was significantly increased in skNAC−/− muscle, whereas Myh7 (oxidative, type I) and Myh2 (oxidative, type IIa) markers were reduced. Data are presented as mean with SEs (n = 3, *P < 0.05, **P < 0.01). (G) Histochemical assay for SDH activity (blue stain) to measure oxidative potential in skNAC−/− soleus muscle. The skNAC−/− soleus demonstrated less oxidative potential than wild-type soleus. Representative samples are shown (n = 3).
Histological comparison showed less muscle mass in the calf muscles of skNAC−/− than of wild-type mice, although the sizes of individual myofibers were similar in both (Fig. 4C). Quantitative analysis of multiple animals confirmed that there was no difference in the cross-sectional area of myofibers (Fig. 4D) or the number of myofibers per unit area (Fig. 4E). The combined results suggest that the reduced muscle mass was more likely due to fewer myofibers rather than to less growth of individual myofibers.
Because myoglobin and skNAC both are highly expressed in slow oxidative skeletal muscle and myoglobin is a skNAC target (15), we determined whether loss of skNAC affected the specification of oxidative vs. glycolytic fibers of the muscle. Skeletal muscle fibers can be classified with respect to myoglobin content. Slow oxidative (type I) and fast oxidative (type IIa) fibers have large amounts of myoglobin and many mitochondria, whereas fast glycolytic (type IIb) fibers have low myoglobin content and large amounts of glycogen. Myoglobin-null mice have myofiber type switching from oxidative to glycolytic fibers (25). More of the type IIb glycolytic fiber marker Myh4 was expressed in skNAC−/− soleus muscle, and less of oxidative fiber markers, such as Myh7 (type I) and Myh2 (type IIA) (Fig. 4F). We confirmed the lower oxidative potential of skNAC−/− muscle by staining for succinate dehydrogenase (SDH) in frozen sections. SDH is located in the inner membrane of mitochondria and is responsible for oxidizing succinate to fumarate in the citric acid cycle. skNAC−/− sections showed lower oxidative potential as marked by SDH activity (Fig. 4G). These findings suggest that loss of skNAC causes myofiber type switching from oxidative to glycolytic fibers.
skNAC Is Involved in Postnatal Muscle Regeneration.
The reduced muscle mass of skNAC−/− mice was apparent early in life (Fig. 4A). Postnatal muscle growth is mainly achieved by addition of myoblasts derived from satellite cells to existing myofibers (26). Satellite cells are a population of muscle precursor cells that exist in postnatal muscle tissues. During periods of muscle growth and regeneration after injury, satellite cells reenter the cell cycle and fuse with myofibers (27, 28). Thus, we studied muscle growth defects in skNAC−/− mice during regeneration after injury.
Necrotic injury was induced in the calf muscles (gastrocnemius and soleus) of 2- to 3-mo-old skNAC−/− and wild-type mice by injection of cardiotoxin. The injuries were histologically analyzed 12 d after the injury (Fig. 5A). Wild-type muscle showed small myofibers containing centrally located nuclei, indicating normal muscle regeneration, and the overall muscle architecture was almost restored. However, regeneration of skNAC−/− muscle was significantly impaired, as evidenced by myonecrosis and infiltration of mononuclear cells. In addition, skNAC−/− muscle exhibited abnormal fat accumulation, marked by Oil Red O staining. One month after injury, regenerated wild-type muscle was indistinguishable from uninjured muscle, whereas injured skNAC−/− muscle still had many myofibers with centrally located nuclei and infiltrated mononuclear cells and adipocytes (Fig. 5B). Twelve days after the injury, numbers of cells with central nuclei were similar in skNAC−/− and wild-type muscle (Fig. 5C). Quantitative analysis of the cross-sectional area of myofibers 1 mo after the injury showed significantly smaller myofiber size in skNAC−/− muscle (Fig. 5D). These findings suggest that skNAC−/− satellite cells may be able to initiate the regeneration process but may not be able to complete the regeneration program.
Fig. 5.
Regeneration defects of skeletal muscle in skNAC−/− mice. (A) H&E staining of skeletal muscle 12 d after cardiotoxin-induced injury. Note normalizing architecture in wild-type muscle, with regenerating myofibers containing centrally located nuclei (arrowhead). skNAC-deficient muscle had signs of myonecrosis, with infiltrating mononuclear cells (arrow) and fat deposits (asterisk). Accumulation of fat in regenerating myofibers in skNAC−/− mice was marked by Oil Red O staining. (Scale bars, 100 μm.) (B) skNAC−/− muscle still had many myofibers with central nuclei (arrowhead), mononuclear infiltrating cells (arrow), and adipocytes (asterisk) 1 mo after injury; wild-type muscle was almost indistinguishable from preinjury muscle. Oil Red O staining indicates fat replacement. (Scale bars, 100 μm.) (C) Quantitative analysis of regenerating myofibers with central nuclei 12 d after the injury (n = 3). (D) Quantitative analysis of cross-sectional area of regenerated myofibers 1 mo after the injury. Myofibers of skNAC−/− mice had smaller cross-sectional area than wild-type myofibers (*P < 0.0001). (E and F) Proliferation and cell death profiles of satellite cells isolated from skNAC−/− mice. Single satellite cells isolated from the tibialis anterior muscle of skNAC−/− or control (skNAC+/−) animals were plated within a hydrogel-microwell array and monitored for 96 h by time-lapse microscopy (34). A representative graph from the three experimental replicates is displayed. Data are shown as mean with SE (*P < 0.05). (E) There was no significant difference in proliferation profiles of skNAC+/− and skNAC−/− satellite cells 96 h after plating. (F) Satellite cells from skNAC−/− mice had a higher cell death percentage than those from control skNAC+/− littermates over the 96-h period.
To determine whether skNAC plays a role in satellite cell activation and proliferation, we isolated satellite cells from the tibialis anterior muscle by FACS and monitored proliferation of individual cells for 96 h on a microwell coated with hydrogel. We found no significant difference in the proliferation profiles of control (skNAC+/−) or skNAC−/− satellite cells (Fig. 5E). However, skNAC−/− satellite cells had more cell death than control satellite cells over a 96-h period (Fig. 5F). These results suggest that skNAC is likely involved in survival and possibly differentiation of satellite cell-driven myoblasts, rather than satellite cell activation.
Discussion
This study shows that skNAC is a major Smyd1-interacting protein in the developing embryonic heart and is important for ventricular cardiomyocyte expansion in a subset of embryos. Dysregulation of numerous genes involved in cardiac proliferation and differentiation was observed in skNAC mutant hearts, many overlapping with genes altered in Smyd1 mutants. skNAC-null mice that survived to adulthood revealed a role for skNAC during postnatal skeletal muscle growth and for muscle regeneration in response to injury. Thus, in skeletal muscle, and possibly developing cardiac muscle, skNAC may regulate progressive addition or expansion of muscle cells from the pool of progenitors.
The physical interaction of skNAC and Smyd1 in the embryonic heart and the coincident expression of these two genes are consistent with their cooperative regulation of downstream events. Interestingly, Irx4 was down-regulated in skNAC−/− embryonic hearts, similar to the previous observation in Smyd1−/− mice (5). Irx4 is a member of the Iroquois homeobox transcription factor family that is expressed specifically in the ventricular chambers of the looped heart, where it activates ventricle-specific genes and represses atrial genes (22). Similarly, periostin, which promotes reentry of adult cardiomyocytes into the cell cycle by activating phosphoinositol-3 kinase, was down-regulated in skNAC mutants and may contribute to the cardiomyocyte proliferation defect (20).
skNAC is a Smyd1-interacting protein in cultured skeletal myoblasts (12) and is markedly up-regulated after skeletal muscle injury (29). Adult skeletal muscle possesses a remarkable regenerative capacity because of satellite cells, which serve as a resident pool of muscle progenitors. In rapidly growing neonatal muscle, nuclei of satellite cells and newly derived myoblasts from activated satellite cells account for approximately 30% of myofiber-associated nuclei, but after cessation of muscle growth by 3 wk of age, satellite cells are less than 5% of adult nuclei (30). The postnatal muscle growth defect we observed in skNAC−/− neonates and the failure of muscle regeneration in adults suggest a defect in the satellite cells. In addition, we also observed extensive replacement of myofibers by adipocytes during regeneration of skNAC−/− mice. Recent studies showed that brown fat cells arise in vivo from the Myf5-positive myoblast lineage by the action of a PRDM16/EBP-b transcriptional complex (31, 32). Because satellite cells from skNAC−/− mice had a higher cell death in myoblast culture system, skNAC might have a role in progenitor survival and differentiation into the muscle lineage rather than activation of satellite cells. However, the mechanism of action remains elusive. It will be important to determine whether the interaction with Smyd1, a SET domain-containing partner, is necessary for skNAC function in muscle growth and reneration.
Materials and Methods
Yeast Two-Hybrid Library Construction and Screening.
A cDNA library for yeast two-hybrid screening was constructed with 200 ng of poly(A) RNA isolated from E11.5 embryonic hearts with the MATCHMAKER Library Construction kit, according to the manufacturer's protocol (Clontech). Approximately 5 × 106 clones were screened with a full-length Smyd1 as bait; colonies that were positive for β-galactosidase activity within 1 h were selected, and inserts were amplified by PCR and sequenced.
Targeted Deletion and Generation of skNAC-Deficient Mice.
Genomic fragments surrounding exon 2 of skNAC were isolated and subcloned into a vector containing a PGK-Neo cassette, which was introduced into 129S6 ES cells as previously described (33). Two correctly targeted ES cell clones were injected into C57BL/6 blastocysts to ultimately generate skNAC−/− mice.
Cardiotoxin-Induced Muscle Injury.
The calf muscle of adult mice (2–3 mo old) were injected with 100 μL of 10 μM cardiotoxin (Calbiochem), and mice were sacrificed at specified times. Calf muscles (gastrocnemius and soleus) were dissected, mounted on 7% tragacanth, snap-frozen in isopentane cooled in liquid nitrogen, cryosectioned, and stained for histological examination.
Satellite Cell Isolation and Microwell Culture.
Isolation of satellite cells and microwell arrays from skNAC−/− and control animals were performed as previously described (34, 35). Additional detail is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Genomics Core (C. Barker, L. Ta, and Y. Hao) and Histology Core (J. Fish and C. Miller) at the Gladstone Institutes for technical support; R. Yeh for bioinformatics support and array analyses; B. Taylor for manuscript preparation; S. Ordway and G. Howard for editorial assistance; Drs. Penney Gilbert and Alessandra Sacco for expert instruction in satellite cell preparation; Karen Havenstrite for hydrogel microwell fabrication and timelapse in the H.M.B laboratory; and P. Tucker and members of our laboratory for discussion and review of the manuscript. J.A.M. was supported by the Stanford Medical Scholars Program. H.M.B. is supported by grants from the National Institutes of Health (NIH), Juvenile Diabetes Research Foundation, Muscular Dystrophy Association, Leukemia and Lymphoma Society, California Institute for Regenerative Medicine (CIRM), Stanford BioX Award, and the Baxter Foundation in Stem Cell Biology. D.S. is supported by the National Heart, Lung, and Blood Institute/NIH, CIRM, and the Younger Family Foundation. K.N.I. was a postdoctoral scholar of the CIRM. This work was also supported by NIH/National Center for Research Resources Grant C06 RR018928 (to the Gladstone Institutes).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data were deposited to the Gene Expression Omnibus database (accession no. GSE5841).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013493107/-/DCSupplemental.
References
- 1.Srivastava D. Making or breaking the heart: From lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 4.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem. 2006;2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb PD, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 6.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuikov S, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 8.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava D, et al. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 10.Phan D, et al. BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development. 2005;132:2669–2678. doi: 10.1242/dev.01849. [DOI] [PubMed] [Google Scholar]
- 11.Tan X, et al. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci USA. 2006;103:2713–2718. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sims RJ, 3rd, et al. m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J Biol Chem. 2002;277:26524–26529. doi: 10.1074/jbc.M204121200. [DOI] [PubMed] [Google Scholar]
- 13.Hung RJ, et al. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–827. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirane M, Nakayama KI. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat Cell Biol. 2003;5:28–37. doi: 10.1038/ncb894. [DOI] [PubMed] [Google Scholar]
- 15.Yotov WV, St-Arnaud R. Differential splicing-in of a proline-rich exon converts alphaNAC into a muscle-specific transcription factor. Genes Dev. 1996;10:1763–1772. doi: 10.1101/gad.10.14.1763. [DOI] [PubMed] [Google Scholar]
- 16.Akhouayri O, Quélo I, St-Arnaud R. Sequence-specific DNA binding by the alphaNAC coactivator is required for potentiation of c-Jun-dependent transcription of the osteocalcin gene. Mol Cell Biol. 2005;25:3452–3460. doi: 10.1128/MCB.25.9.3452-3460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez S, et al. NACA is a positive regulator of human erythroid-cell differentiation. J Cell Sci. 2005;118:1595–1605. doi: 10.1242/jcs.02295. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 19.Meeson AP, et al. Adaptive mechanisms that preserve cardiac function in mice without myoglobin. Circ Res. 2001;88:713–720. doi: 10.1161/hh0701.089753. [DOI] [PubMed] [Google Scholar]
- 20.Kühn B, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13:962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 21.Zhang M, et al. SDF-1 expression by mesenchymal stem cells results in trophic support of cardiac myocytes after myocardial infarction. FASEB J. 2007;21:3197–3207. doi: 10.1096/fj.06-6558com. [DOI] [PubMed] [Google Scholar]
- 22.Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161–1164. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- 23.Wald FA, Oriolo AS, Casanova ML, Salas PJ. Intermediate filaments interact with dormant ezrin in intestinal epithelial cells. Mol Biol Cell. 2005;16:4096–4107. doi: 10.1091/mbc.E05-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaschke RJ, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 25.Grange RW, et al. Functional and molecular adaptations in skeletal muscle of myoglobin-mutant mice. Am J Physiol Cell Physiol. 2001;281:C1487–C1494. doi: 10.1152/ajpcell.2001.281.5.C1487. [DOI] [PubMed] [Google Scholar]
- 26.Moss FP, Leblond CP. Nature of dividing nuclei in skeletal muscle of growing rats. J Cell Biol. 1970;44:459–462. doi: 10.1083/jcb.44.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins CA, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Munz B, Wiedmann M, Lochmüller H, Werner S. Cloning of novel injury-regulated genes. Implications for an important role of the muscle-specific protein skNAC in muscle repair. J Biol Chem. 1999;274:13305–13310. doi: 10.1074/jbc.274.19.13305. [DOI] [PubMed] [Google Scholar]
- 30.Cardasis CA, Cooper GW. An analysis of nuclear numbers in individual muscle fibers during differentiation and growth: A satellite cell-muscle fiber growth unit. J Exp Zool. 1975;191:347–358. doi: 10.1002/jez.1401910305. [DOI] [PubMed] [Google Scholar]
- 31.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kajimura S, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 34.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.