Abstract
The development of cancer drug resistance is a persistent clinical problem limiting the successful treatment of disseminated malignancies. However, the molecular mechanisms by which initially chemoresponsive tumors develop therapeutic resistance remain poorly understood. Error-prone translesional DNA synthesis (TLS) is known to underlie the mutagenic effects of numerous anticancer agents, but little is known as to whether mutation induced by this process is ultimately relevant to tumor drug resistance. Here, we use a tractable mouse model of B-cell lymphoma to interrogate the role of error-prone translesional DNA synthesis in chemotherapy-induced mutation and resistance to front-line chemotherapy. We find that suppression of Rev1, an essential TLS scaffold protein and dCMP transferase, inhibits both cisplatin- and cyclophosphamide-induced mutagenesis. Additionally, by performing repeated cycles of tumor engraftment and treatment, we show that Rev1 plays a critical role in the development of acquired cyclophosphamide resistance. Thus, chemotherapy not only selects for drug-resistant tumor population but also directly promotes the TLS-mediated acquisition of resistance-causing mutations. These data provide an example of an alteration that prevents the acquisition of drug resistance in tumors in vivo. Because TLS also represents a critical mechanism of DNA synthesis in tumor cells following chemotherapy, these data suggest that TLS inhibition may have dual anticancer effects, sensitizing tumors to therapy as well as preventing the emergence of tumor chemoresistance.
Keywords: DNA polymerase, cancer, chemotherapy, relapse
The development of acquired chemoresistance is a persistent clinical problem limiting the successful treatment of disseminated malignancies. Tumors that relapse following initial treatment frequently are refractory to subsequent administration of the initial drug regimen as well as to distinct sets of chemotherapeutics. Although a number of key pathways have been implicated in resistance to conventional chemotherapeutics, including enhanced drug efflux, increased drug metabolism, drug inactivation, enhanced DNA repair, and defects in apoptosis programs (1, 2), the mechanisms by which tumors develop drug resistance-causing mutations remains unclear.
At its core, acquired chemoresistance represents the emergence of subpopulations of drug-resistant tumor cells, a phenomenon rooted in the inherent genetic heterogeneity of the tumor itself. This heterogeneity may occur as a consequence of tumor genetic instability, a process known to underlie tumor development in numerous malignancies. Alternatively, cancer therapy itself may promote mutation and subsequent chemoresistance in relapsed tumors. Support for the latter hypothesis comes from several observations. First, conventional chemotherapeutics can be highly mutagenic (3). In fact, considerable work has gone into highlighting the mutagenic properties of platinum-based and other DNA adduct-forming chemotherapeutics as well as the genes that act in the cellular response to these toxins (4–6). Second, patients treated with conventional chemotherapies show significantly increased incidence of secondary malignancies, a phenomenon specifically tied to the mutagenic potential of genotoxic agents (7). Finally, recent tumor genome-sequencing studies have shown exceptionally high mutation frequency in relapsed malignancies (8). However, there is little evidence to implicate therapy-induced mutation directly, as opposed to the outgrowth of cells with rare preexisting mutations, as a major contributor to drug resistance.
A fundamental principle of mutagenesis is that most mutations induced by DNA-damaging agents result from the action of specialized DNA polymerases carrying out translesion synthesis (TLS) across from DNA lesions (3, 9). In eukaryotes, three genes whose products play a critical role in mutagenesis were identified first in a screen for Saccharomyces cerevisiae mutants that displayed a “reversionless” phenotype, i.e., exhibited a reduced frequency of mutations after UV irradiation (10, 11). The products of the REV1, REV3, and REV7 genes act together in a mutagenic branch of TLS that is responsible for most mutations induced by UV light and chemical mutagens (3, 9). The human orthologs of these same genes, REV1, REV3L, and REV7 (MAD2B), are similarly required for most of the mutagenesis induced by exposure to DNA-damaging agents such as UV light and by chemical mutagens such as benzo(a)pyrene diol epoxide and cisplatin (12–16). Rev1, a member of the Y family of TLS DNA polymerases, has both a dCMP transferase activity that contributes to the bypass of certain lesions and a second important role as a scaffolding protein that associates with several translesion DNA polymerases, including DNA polymerase ζ (Polζ) (3, 17, 18). Rev3 is the catalytic subunit of Polζ, a member of the B family of DNA polymerases, whereas Rev7 is the auxiliary subunit.
In this study, we present in vivo evidence showing that acquired resistance to the front-line chemotherapeutic cyclophosphamide (CTX) in a mouse model of B-cell lymphoma arises as consequence of the mutagenic TLS DNA polymerases copying over lesions caused by the chemotherapeutic agent. In doing so, we provide a link between drug-induced mutation and resistance to the mutagenic drug in a relevant physiological setting. Given the widespread use of CTX and related compounds in the clinic, our results, combined with results showing drug sensitization to lung adenocarcinomas by TLS inhibition (see ref. 19), suggest a rationale for TLS inhibition as an adjuvant therapy for DNA adduct-forming chemotherapeutics.
Results
Suppression of Translesion DNA Synthesis Sensitizes B-Cell Lymphomas to Cisplatin in Vivo.
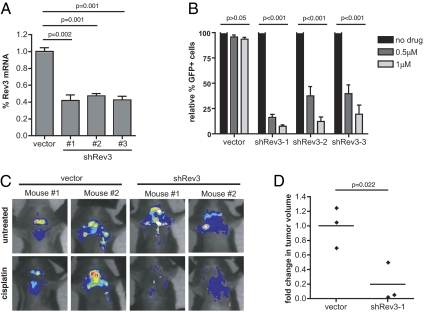
In a companion article (19), we show that suppression of the translesion polymerase Polζ (Rev3L/Rev7) can sensitize intrinsically chemoresistant lung adenocarcinomas to cisplatin. Using a well-established preclinical model of Burkitt's lymphoma, the Eμ-myc mouse (20), we sought to determine whether Rev3L depletion could further sensitize chemoresponsive tumors to cisplatin-based chemotherapy. Three distinct shRNAs targeting Rev3L were expressed from retroviral vectors, and the level of Rev3L transcript was assessed by quantitative PCR following transduction of target lymphoma cells (Fig. 1A). As an initial in vitro validation step, Rev3L shRNAs were tested for their ability to promote cisplatin sensitivity in a highly sensitive GFP competition assay. In this assay, GFP is used as a surrogate marker for the presence of an shRNA, and the impact of gene suppression is determined by the relative change in the percent of GFP-positive cells following treatment. In this context, all Rev3L shRNA-infected cells showed significantly depleted GFP percentages relative to cells transduced with a control vector (Fig. 1B).
Fig. 1.
Rev3 depletion sensitizes B-cell lymphomas to cisplatin in vitro and in vivo. (A) Quantitative RT-PCR (n ≥ 3) confirmation of target mRNA suppression using three distinct shRNAs targeting Rev3 in retrovirally transduced Eμ-myc; p19arf−/− lymphoma cells. Retroviral vectors coexpressed shRNAs and a GFP marker, and pure populations of infected cells were isolated by GFP sorting before RT-PCR. (B) Naïve lymphoma cell populations were transduced to an infection efficiency of 40–50% with Rev3 shRNAs, treated with cisplatin (0.5 and 1.0 μM), and monitored using GFP-based flow cytometry for changes in the relative percentage of shRNA-containing (GFP-positive) cells. n ≥ 3 for all samples. (C) Representative pseudocolored images showing the tumor burden in four individual mice (two control and two Rev3-knockdown mice) treated with cisplatin for 24 h. (D) Quantification of relative changes in tumor volume in control and Rev3-knockdown tumors before and 24 h after cisplatin treatment. n = 3 individual mice in each group. All quantified data shown represent the mean ± SD. P values were determined using Student's t tests.
We then injected pure populations of GFP-sorted control and Rev3L-deficient lymphoma cells into syngeneic recipient mice and allowed palpable tumors to form (∼2 wk). Upon tumor presentation, mice were treated with a single 10-mg/kg dose of cisplatin and monitored using in vivo GFP imaging. Although all lymphomas were sensitive to cisplatin, mice bearing Rev3L-deficient tumors exhibited a significantly more rapid reduction in GFP-positive tumor cells than seen in treated control mice, with tumor regression occurring within 24 h following treatment (Fig. 1 C and D). Thus, Rev3L suppression can sensitize cells acutely to the cytotoxic effects of cisplatin in vivo in a lymphoma model as well as in a model of an intrinsically chemoresistant model of non-small cell lung cancer (19).
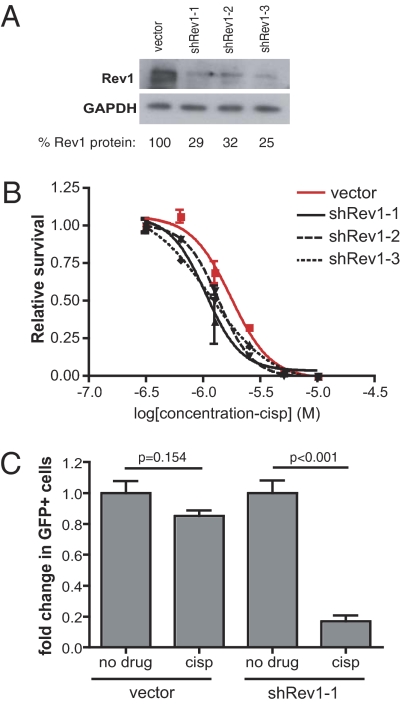
Because Rev1 also plays a key role in preventing cisplatin cytotoxicity and DNA damage-induced mutagenesis, we extended our analysis by similarly designing and testing three unique shRNA vectors targeting Rev1 and observed suppressed Rev1 protein expression in transduced cell populations by Western blot (Fig. 2A). We then subjected these cells to rigorous dose–response experiments to examine the effect of Rev1 suppression in the context of cisplatin treatment. Comparison of best-fit regression curves revealed significantly lower EC50 values in all three Rev1 shRNA populations as compared with vector control cells (Fig. 2B; shRev1-1: P = 0.0039; shRev1-2: P = 0.0076; shRev1-3: P = 0.0035). Importantly, when examined using a GFP competition assay in vivo, partially transduced cells expressing the most potent Rev1 shRNA exhibited a markedly robust negative selection in response to cisplatin, whereas vector control cells displayed a similar percentage of GFP-positive cells before and after treatment (Fig. 2C).
Fig. 2.
Rev1 depletion sensitizes B-cell lymphoma to cisplatin in vivo. (A) Western blot confirmation of Rev1 suppression in Rev1 shRNA-expressing lymphoma cell populations. Quantitation was performed on the combined intensity of both Rev1 bands. (B) Cisplatin dose–response curves in cells expressing normal (vector, red) or impaired (shRev1, black) levels of Rev1 protein (shRev1-1: P = 0.0039; shRev1-2: P = 0.0076; shRev1-3: P = 0.0035). n = 3 replicates per dose per sample. P values were determined using an F-test comparison of EC50 values derived from best-fit nonlinear regression curves. (C) Mice harboring partially transduced lymphoma cell transplants were treated with 8 mg/kg cisplatin for 24 h. Shown is the percentage of GFP-positive cells in mice treated with either cisplatin (cisp) or vehicle (PBS; no drug) alone. P values were determined using Student's t tests.
Rev1 Suppression Limits Cyclophosphamide-Induced Mutagenesis and Acquired Drug Resistance in Vitro.
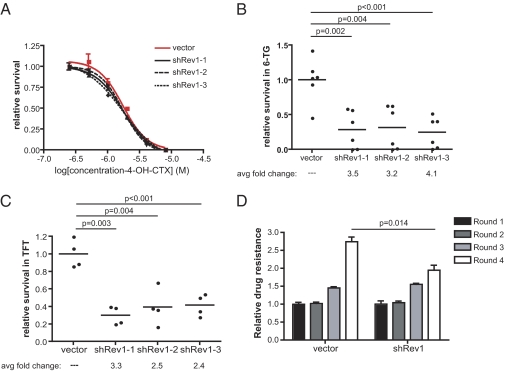
Although cisplatin serves as a front-line therapy for numerous malignancies, including testicular, ovarian, and lung cancer, the standard of care for many hematopoietic malignancies typically features alkylating rather than platinum-based chemotherapeutic agents. In particular, CTX is the front-line treatment for a wide range of lymphoma subtypes, either as a single agent or in combination with other chemotherapeutics. CTX is a nitrogen mustard alkylating agent that, like cisplatin, forms highly toxic intrastrand crosslinks between guanine nucleotides that impede normal DNA replication (21–23). We chose to evaluate the role of Rev1 in mediating the response of our lymphoma cells to CTX (i) because of its central role in mutagenesis (3, 9), (ii) because of its key role in interacting with Polζ and other TLS DNA polymerases (3, 17, 18), and (iii) because it has been implicated in the replication-dependent repair of a nitrogen-mustard–like interstrand crosslink in a Xenopus cell-free system (24). Using the same set of three Rev1-targeting shRNAs described in the context of cisplatin therapy, we examined the effect of Rev1 depletion on the acute response to increasing doses of CTX. Although statistical comparison of the resulting data revealed a slight but significant difference for two of three Rev1-deficient survival curves compared with the control curve (Fig. 3A; shRev1-1: P = 0.035; shRev1-2: P = 0.121; shRev1-3: P = 0.009), we observed no meaningful shifts in either the calculated EC50 (shRev1-1: P = 0.2555; shRev1-2: P = 0.2209; shRev1-3: P = 0.1062) or the hillslope (shRev1-1: P = 0.6663; shRev1-2: P = 0.7827; shRev1-3: P = 0.2187) values. Thus, Rev1 suppression promotes only limited sensitization of cultured lymphoma cells to CTX. This finding is reminiscent of prior observations, which showed that the loss of mutagenic TLS function has little effect on cell survival in response to agents such as UV light and benzo(a)pyrene diol epoxide (18). Notably, however, the same studies also documented a significant decrease in drug-induced mutation in response to DNA-damaging agents in TLS-deficient cells.
Fig. 3.
Rev1 suppression inhibits CTX-induced mutagenesis. (A) CTX dose–response curves in control and Rev1-knockdown cells (shRev1-1: P = 0.2555; shRev1-2: P = 0.2209; shRev1-3: P = 0.1062). n = 3 replicates per dose in each sample. P values were determined using an F-test comparison of EC50 values derived from best-fit nonlinear regression curves. (B) A graph showing the relative survival of CTX-treated (4 μg/mL 4-OH-cyclophosphamide for 1 h) control and Rev1-knockdown Eμ-myc lymphoma cells following exposure to 1 μM 6TG for 1 wk. (C) A graph showing the relative survival of CTX-treated (4 μg/mL 4-OH-cyclophosphamide for 1 h) control and Rev1-knockdown L5178Y-TK+/− mouse lymphoma cells following exposure to10 μM triflorothymidine (TFT). (D) A graph showing the relative response of control and Rev1-knockdown Eμ-myc lymphoma cells to multiple rounds of CTX treatment in vitro. In each case lymphoma cell viability was determined 48 h following exposure to 1.3 μg/mL 4-OH-cylophosphamide. Relative drug resistance was determined by normalizing viability measurements to those observed in round 1. P values shown in B–D were determined using Student's t tests.
To examine the role of Rev1-dependent TLS in CTX-induced mutations, we performed two complementary and classically defined mutagenesis assays in the presence and absence of Rev1 suppression. Notably, these assays were carried out in liquid culture, because hematopoietic malignancies are not amenable to more conventional colony-based assays. In the first setting, we used CTX-induced mutagenesis at the hprt locus to serve as a readout of relative mutagenic burden. Briefly, cells were exposed to 4 μg/mL CTX for 1 h, cultured for 2 wk, and then challenged with 6-thioguanine (6TG) to select for mutants with impaired hprt gene function. Because hprt function is required for 6TG-mediated toxicity, this assay allows for the quantitation of CTX-induced hprt mutation. As shown in Fig. 3B, Rev1 deficiency reduced the frequency of 6TG-resistant variants by 3.2- to 4.1-fold compared with control-infected cells. To confirm this observation in a related context, we made use of a mouse lymphoma cell line (L5178Y) that is heterozygous at the thymidine kinase (TK) locus. Because TK activity is necessary for the cytotoxic effects of the thymidine analog triflorothymidine, these cells provide a highly sensitized setting for selection of CTX-induced mutations in the wild-type TK allele. Using the same set of three Rev1 shRNA vectors described above, we generated three Rev1-knockdown L5178Y lymphoma cell populations and tested their relative mutagenicity in response to CTX. In agreement with the hprt experiments performed in our Eμ-myc lymphoma cells, Rev1 suppression in L5178Y cells reduced the mutagenic burden by 2.4- to 3.3-fold relative to control cells (Fig. 3C). Thus, the effects of Rev1 on survival and mutagenesis are largely separable in this context, because Rev1 shRNAs fail to sensitize lymphoma cells to CTX treatment significantly but potently inhibit CTX-induced mutagenesis.
A fundamental question in cancer chemotherapy is whether genotoxic drugs can induce mutations that promote tumor chemoresistance. Given the importance of Rev1 in CTX-induced mutagenesis, we sought to determine whether Rev1 depletion could inhibit the development of CTX resistance in treated tumor cells. To investigate this question, we treated a fixed number (1 × 106 cells/mL) of control or shRev1-expressing Eμ-myc lymphoma cells with a fixed dose (∼EC70 in control samples) of CTX, assayed for cell survival at 48 h, and allowed the cells to recover for an additional 5 d, at which point we initiated a subsequent round of chemotherapy. To chart the evolution of drug resistance in a given cell population over time, we normalized survival data recorded during each round of therapy to the initial values collected during the first round of treatment. As expected, control cell populations became progressively resistant to repeated CTX exposure (Fig. 3D, Left). In contrast, Rev1-deficient cells displayed a diminished resistance profile in the final round of treatment (Fig. 3D, Right).
Rev1 Deficiency Inhibits the Acquisition of CTX Resistance in Vivo.
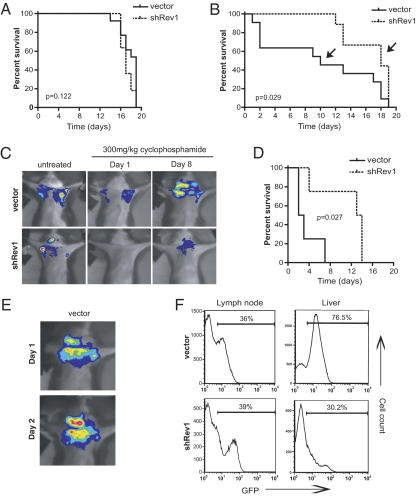
The preceding experiments strongly suggest that, in cultured cells, Rev1-dependent mutagenesis can actively promote chemotherapeutic resistance. However, the relevance of this mutagenesis to tumor relapse and drug resistance has not been investigated. To examine whether Rev1 deficiency similarly could delay the development of chemoresistant tumors in vivo, we injected GFP-sorted control and Rev1-deficient Eμ-myc lymphoma cells into syngeneic recipient mice and allowed palpable tumors to form. We then treated tumor-bearing mice with 30 mg/kg CTX and monitored tumor burden using in vivo GFP imaging. At this dose, we did not observe any difference between control and Rev1-deficient transplants with respect to acute tumor regression or time to tumor relapse (Fig. 4A). Thus, consistent with our cell culture data, Rev1 deficiency fails to promote CTX sensitivity in vivo.
Fig. 4.
Rev1 depletion improves CTX-based chemotherapy in vivo. (A) Kaplan–Meier survival curves of tumor-bearing mice treated with a single dose of CTX (300 mg/kg; n = 13 vector control, n = 11 shRev1). Day 0 represents the day of drug administration. At disease relapse, individual tumors were harvested and reinjected into new recipient mice for additional drug treatment. (B) Kaplan–Meier survival curves of control and shRev1 transplant-bearing mice challenged with a second round of CTX chemotherapy (n = 11 vector control, n = 9 shRev1). As in A, tumors were harvested at relapse and reinjected into naive recipient mice. (C) In vivo GFP imaging showing representative mice from each experimental group (indicated with arrows in B). (D) Round 3 Kaplan–Meier survival data (n = 4 vector control, n = 4 shRev1). P values for all survival studies were determined using log-rank curve comparison tests. (E) GFP imaging of a highly drug-resistant vector control tumor treated with CTX. (F) GFP histograms of dissociated whole tissue harvested from tumor-bearing mice. The percentages represent the proportion of control vector or shRev1 lymphoma cells present in the indicated tissue.
To examine the role of Rev1 in the evolution of tumor chemoresistance, we harvested tumors from individual mice at relapse, resorted tumors for GFP-positive lymphoma cells, and reinjected sorted tumor cells into syngeneic recipient mice for a second round of therapy. Following tumor transplantation and regrowth, a subset of control tumors was no longer sensitive to CTX (Fig. 4B). Mice bearing these tumors showed continued lymphoma and the development of terminal disease shortly after treatment. Strikingly, we observed a complete absence of this class of drug-resistant tumors following suppression of Rev1, with all recipient mice showing sustained periods of tumor regression and enhanced overall survival. To extend these findings further, we subjected control and Rev1-knockdown tumors to a third round of treatment. In this setting, three of four Rev1-deficient tumors still retained a pronounced sensitivity to CTX treatment, whereas all control tumor recipients showed little or no tumor-free survival (Fig. 4 C–E). Of note, drug-resistant control tumors also were significantly more aggressive than their Rev1-deficient counterparts, showing perivascular infiltration into nonhematopoietic organs such as the liver and lung (Fig. 4F). Thus, the emergence of tumor drug resistance is coincident with the acquisition of additional tumor growth characteristics, probably because of the high mutational load present in treated TLS-proficient cells.
Discussion
Chemotherapeutic intervention rarely results in complete tumor eradication. More frequently, tumors exhibit varying degrees of response and ultimately relapse with more aggressive, drug-resistant phenotypes. It has been postulated that tumor mutation rate is one of a few critical determinants of the clinical resistance of a variety of human cancers (25). To this end, mathematical models have been proposed to suggest that evolving drug-resistant tumor subpopulations emerge under the selective pressure of drug exposure (26). However, an added layer of complexity is introduced when one considers the intrinsically mutagenic properties of the therapy itself, an effect that greatly compounds any preexisting mutagenic tendencies inherent in a given tumor. Using a genetically tractable and highly dynamic model of B-cell lymphoma, we show that by impairing mutagenic translesion DNA synthesis tumors not only are sensitized to relevant chemotherapies but also are partially protected from the consequences of mutagenic chemotherapies that do not succeed in killing target cells.
A treatment strategy based on pairing a DNA-damaging chemotherapeutic agent such as CTX or cisplatin with a drug that inhibits the mutagenic TLS pathway could be very powerful, because it could reduce significantly the rate at which cells acquire chemoresistance. In vitro studies of cultured human cell lines have shown that suppressing either Rev1 or Rev3L reduces the rate of emergence of cisplatin resistance (14, 15), so there is reason to think that similar effects would be seen for cisplatin and other DNA-damaging chemotherapeutics in clinically relevant contexts. Such a strategy might be additionally effective because DNA-damaging agents such as cisplatin and the alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine not only introduce lesions into DNA but also induce the expression of Rev3L (14, 27). Because increased REV3L expression has been shown to promote resistance to cisplatin (28), a drug that inhibits the Rev1/3/7-dependent mutagenic TLS pathway would suppress the acquisition of drug-resistant mutations.
It is possible that mutagenic TLS polymerases may play a role in cancer causation as well as in acquired resistance. Recent sequencing of the genomes of cancer cell lines and of a lung cancer has shown the presence of 20,000–50,000 mutations (29–31). The majority of the mutations are inferred to have been caused by a lesion in the DNA, and the nature of the mutations, predominantly base-pair substitutions and small insertions and deletions, resemble those known to be introduced during mutagenic TLS (3, 9). Rev1 also has been implicated in the development of carcinogen-induced lung cancer (32). Normal levels of TLS DNA polymerases, together with the large amounts of DNA damage from exogenous agents such as smoking or sunlight, might be sufficient to account for the many mutations observed in tumors. However, the rate of mutagenesis also might be increased by elevated expression of TLS DNA polymerases as cancer progresses. Such elevated expression has been reported for advanced-stage gliomas (28) and colorectal adenocarcinoma, in which loss of both mismatch repair and p53 increases the levels of expression of Rev1 and REV3L by 10-fold and 20-fold, respectively (33). Furthermore, mutations or conditions that alter the complex web of protein–protein interactions that control the access of TLS DNA polymerases to primer termini (34, 35) also could increase the rate of both spontaneous and induced mutagenesis. For example, the Rev1-257Ser single-nucleotide polymorphism has been suggested as a risk factor for lung adenocarcinoma and squamous cell carcinoma, and homozygous Rev1-373Ser status is associated with an increased risk for cervical carcinoma (36).
Tumors that do have higher levels of mutagenic TLS activity, such as later-stage gliomas (28) or mismatch repair-defective, p53−/− colorectal adenocarcinomas (33), might be particularly susceptible to the sensitizing and antimutagenic effects of a drug targeting the mutagenic TLS pathway. Components of the mutagenic TLS system also can be tumor suppressors, because their loss results in increased chromosome instability in cells that can tolerate TLS deficiency (37, 38). A tumor lacking a component of the TLS system would not benefit from the chemotherapeutic strategy we are proposing but might be susceptible to drugs that inhibit other DNA-repair or tolerance pathways. In fact, combination therapies that exploit similar DNA-repair deficiencies, including the use of poly (ADP-ribose) polymerase inhibitors in breast cancer 1/2 (BRCA1/2)-deficient tumor cells or inhibitors of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) in ataxia telangiectasia mutated (ATM)-deficient cells (39–41), have gained increasing traction as synthetic lethal strategies for cancer treatment.
Methods
Cell Culture, Retroviral Vectors, and Chemicals.
Eμ-myc B-cell lymphoma and L5178Y/TK−/− lymphoma cells were cultured in B-cell medium (45% vol/vol DMEM/45% Iscove's Modified Dulbecco's Medium/10% FBS, supplemented with 2 mM l-glutamine and 5 μM β-mercaptoethanol). shRNA constructs were designed and cloned as previously described (42). Sequences (5′–3′) targeted by shRNAs are as follows: shRev3-1: TTTACTACAGATACCATGCTG; shRev3-2: TATCTTTATAAGCTGCTCCTG; shRev3-3: TACAGTTATACAAATATCCTA; shRev1-1: GCGGAGGAATTGAGAAATCTA; shRev1-2: AAACAGTGTTGCTAGCAGGCTA; shRev1-3: CCTCCGGGAACAAATAGAACAA. The vector used coexpressed GFP under the control of the SV40 promoter and is identical to the published MSCV/LTRmiR30-SV40-GFP (LMS) vector. Cisplatin (Calbiochem) and 4-OH-cyclophosphamide (Toronto Research Chemicals) were dissolved in DMSO to make 1,000–2,000× stock solutions and were diluted (0–15 μM and 0–8 μg/mL final concentration, respectively) in fresh medium containing cells at the time of treatment. For in vivo studies, cisplatin (8–10 mg/kg) and CTX (300 mg/kg) were dissolved in a 0.9% NaCl solution, protected from light, and immediately injected intraperitoneally into tumor-bearing mice.
Quantitative RT-PCR and Western Blotting.
For quantitative real-time PCR, total RNA was isolated after retroviral infection and puromycin selection. Quantitative RT-PCR was performed using SYBR green on a BioRad thermal cycler. Primer sequences are available upon request. For Western blotting, cell lysates were prepared in lysis buffer [1% sodium deoxycholine, 0.1% SDS, 1% Triton-X, 10 mM Tris-HCl (pH 8.0), 140 mM NaCl] for 10 min, cleared for 15 min at 18,000 × g and then mixed with 5× SDS sample buffer. Proteins then were run on a 10% SDS/PAGE gel, transferred to PVDF (Millipore), and detected with the following antibodies: anti-Rev1 (1:50; a kind gift from Neils De Wind's laboratory) and anti-GAPDH (1:10,000; Santa Cruz).
Mutagenesis Assays.
Retrovirally transduced cells were cultured initially for a minimum of 2 wk in medium containing hypoxanthine, aminopterin, and thymidine (HAT) to remove preexisting hprt- and tk- mutants from the experimental population. Cells then were split into fresh medium (without HAT) 24 h before treatment with cisplatin. Target cells then were mutagenized with 8 μg/mL 4-OH-cyclophosphamide for 1 h, allowed to recover, and passaged for an additional 10 d (in the absence of HAT) to stabilize any induced mutations. Mutagenized cells then were split onto fresh 10-cm feeder plates in medium containing either 6TG (Eμ-myc lymphoma) or triflorothymidine (TFT, L5178Y lymphoma) to select for variants with impaired hprt or tk function, respectively. Cell viability was determined by flow cytometry after 1 wk of selection.
In Vitro Viability Assays and FACS.
For short-term viability assays, cells were seeded in triplicate (6 × 103 per well) in 96-well plates and treated as indicated with cisplatin. After 48 h of treatment, cell viability was measured using Cell-Titer-Glo (Promega) on an Applied Biosystems microplate luminometer. Long-term viability assays were performed by initially treating 4 × 105 lung adenocarcinoma cells with 15 μM cisplatin for 24 h. Four days following treatment, cells were split 1:20 onto a fresh 10-cm plate and allowed to form colonies for ∼10 d. To visualize colonies, plates were washed with 0.05% ethidium bromide (in 50% EtOH) for 10–15 s and imaged using a UV-gel box/camera. Images were processed and colonies were counted using freely available ImageJ software. All flow cytometry was performed using Becton-Dickinson FACScan or MoFlo flow cytometers. Cell death was detected by propidium iodide (PI) incorporation (0.05 mg/mL), and dead cells were excluded from GFP analysis. Live cells were sorted using GFP coexpression as a marker of cell transduction.
In Vivo Transplantation and Imaging.
Syngeneic C57BL6/J female recipient mice were i.v. injected (via the tail vein) with 4 million lymphoma cells and were monitored until palpable tumors formed (∼14 d). Upon tumor presentation, mice were administered either 8–10 mg/kg cisplatin or 300 mg/kg CTX and were monitored until the indicated time points, at which time mice were killed and tumor material collected, if necessary. Mice subjected to live in vivo GFP imaging were immobilized using isoflurane anesthesia and were imaged/analyzed using a NightOwl (Berthold Technologies) imaging platform.
Acknowledgments
We thank members of the Hemann and Walker laboratories for helpful advice and discussions. K.X. and G.C.W. are supported by Grant ES015818 from the National Institute of Environmental Health Sciences and Grant P30 ES002109 from the Center of Environmental Health Sciences (MIT). G.C.W. is an American Cancer Society Research Professor. M.T.H. is a Rita Allen Fellow and the Latham Family Career Development Assistant Professor of Biology and is supported by National Institutes of Health Grant RO1 CA128803. J.D. is supported by an MIT Department of Biology training grant and a Ludwig Center Graduate Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.R.L. is a guest editor invited by the Editorial Board.
References
- 1.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: A link between cancer genetics and chemotherapy. Cell. 2002;108:153–164. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg E, et al. DNA Repair and Mutagenesis. Washington, DC: American Society of Microbiology; 2005. [Google Scholar]
- 4.Ahmad S. Platinum-DNA interactions and subsequent cellular processes controlling sensitivity to anticancer platinum complexes. Chem Biodivers. 2010;7:543–566. doi: 10.1002/cbdv.200800340. [DOI] [PubMed] [Google Scholar]
- 5.Beck DJ, Brubaker RR. Mutagenic properties of cis-plantinum(II)diammino-dichloride in Escherichia coli. Mutat Res. 1975;27:181–189. doi: 10.1016/0027-5107(75)90077-9. [DOI] [PubMed] [Google Scholar]
- 6.Zwelling LA, Bradley MO, Sharkey NA, Anderson T, Kohn KW. Mutagenicity, cytotoxicity and DNA crosslinking in V79 Chinese hamster cells treated with cis- and trans-Pt(II) diamminedichloride. Mutat Res. 1979;67:271–280. doi: 10.1016/0165-1218(79)90021-1. [DOI] [PubMed] [Google Scholar]
- 7.Feig SA. Second malignant neoplasms after successful treatment of childhood cancers. Blood Cells Mol Dis. 2001;27:662–666. doi: 10.1006/bcmd.2001.0436. [DOI] [PubMed] [Google Scholar]
- 8.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemontt JF. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence CW, Das G, Christensen RB. REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet. 1985;200:80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 12.Gibbs PE, et al. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc Natl Acad Sci USA. 2000;97:4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, et al. hREV3 is essential for error-prone translesion synthesis past UV or benzo[a]pyrene diol epoxide-induced DNA lesions in human fibroblasts. Mutat Res. 2002;510:71–80. doi: 10.1016/s0027-5107(02)00253-1. [DOI] [PubMed] [Google Scholar]
- 14.Wu F, Lin X, Okuda T, Howell SB. DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 2004;64:8029–8035. doi: 10.1158/0008-5472.CAN-03-3942. [DOI] [PubMed] [Google Scholar]
- 15.Okuda T, Lin X, Trang J, Howell SB. Suppression of hREV1 expression reduces the rate at which human ovarian carcinoma cells acquire resistance to cisplatin. Mol Pharmacol. 2005;67:1852–1860. doi: 10.1124/mol.104.010579. [DOI] [PubMed] [Google Scholar]
- 16.Cheung HW, et al. Inactivation of human MAD2B in nasopharyngeal carcinoma cells leads to chemosensitization to DNA-damaging agents. Cancer Res. 2006;66:4357–4367. doi: 10.1158/0008-5472.CAN-05-3602. [DOI] [PubMed] [Google Scholar]
- 17.Washington MT, Carlson KD, Freudenthal BD, Pryor JM. Variations on a theme: Eukaryotic Y-family DNA polymerases. Biochim Biophys Acta. 2010;1804:1113–1123. doi: 10.1016/j.bbapap.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 19.Doles J, et al. Rev3 suppression sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1011409107. 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams JM, Cory S. Oncogene co-operation in leukaemogenesis. Cancer Surv. 1992;15:119–141. [PubMed] [Google Scholar]
- 21.Schärer OD. DNA interstrand crosslinks: Natural and drug-induced DNA adducts that induce unique cellular responses. ChemBioChem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 22.Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutat Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- 23.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Räschle M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawyers CL. Calculated resistance in cancer. Nat Med. 2005;11:824–825. doi: 10.1038/nm0805-824. [DOI] [PubMed] [Google Scholar]
- 26.Komarova NL, Wodarz D. Drug resistance in cancer: Principles of emergence and prevention. Proc Natl Acad Sci USA. 2005;102:9714–9719. doi: 10.1073/pnas.0501870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Y, Yang J, Zhu F, Xu F. Response of REV3 promoter to N-methyl-N’-nitro-N-nitrosoguanidine. Mutat Res. 2004;550:49–58. doi: 10.1016/j.mrfmmm.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, et al. REV3L confers chemoresistance to cisplatin in human gliomas: The potential of its RNAi for synergistic therapy. Neuro-oncol. 2009;11:790–802. doi: 10.1215/15228517-2009-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 32.Dumstorf CA, Mukhopadhyay S, Krishnan E, Haribabu B, McGregor WG. REV1 is implicated in the development of carcinogen-induced lung cancer. Mol Cancer Res. 2009;7:247–254. doi: 10.1158/1541-7786.MCR-08-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin X, Howell SB. DNA mismatch repair and p53 function are major determinants of the rate of development of cisplatin resistance. Mol Cancer Ther. 2006;5:1239–1247. doi: 10.1158/1535-7163.MCT-05-0491. [DOI] [PubMed] [Google Scholar]
- 34.Lehmann AR, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 35.Chun AC, Jin DY. Ubiquitin-dependent regulation of translesion polymerases. Biochem Soc Trans. 2010;38:110–115. doi: 10.1042/BST0380110. [DOI] [PubMed] [Google Scholar]
- 36.He X, et al. REV1 genetic variants associated with the risk of cervical carcinoma. Eur J Epidemiol. 2008;23:403–409. doi: 10.1007/s10654-008-9251-5. [DOI] [PubMed] [Google Scholar]
- 37.Wittschieben JP, et al. Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res. 2010;70:2770–2778. doi: 10.1158/0008-5472.CAN-09-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA polymerase zeta causes chromosomal instability in mammalian cells. Cancer Res. 2006;66:134–142. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 39.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 40.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 41.Jiang H, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–1909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]