Abstract
Editing of the pre-mRNA for the DNA repair enzyme NEIL1 causes a lysine to arginine change in the lesion recognition loop of the protein. The two forms of NEIL1 are shown here to have distinct enzymatic properties. The edited form removes thymine glycol from duplex DNA 30 times more slowly than the form encoded in the genome, whereas editing enhances repair of the guanidinohydantoin lesion by NEIL1. In addition, we show that the NEIL1 recoding site is a preferred editing site for the RNA editing adenosine deaminase ADAR1. The edited adenosine resides in an A-C mismatch in a hairpin stem formed by pairing of exon 6 to the immediate upstream intron 5 sequence. As expected for an ADAR1 site, editing at this position is increased in human cells treated with interferon α. These results suggest a unique regulatory mechanism for DNA repair and extend our understanding of the impact of RNA editing.
Keywords: ADAR, nucleic acids, base excision repair, oxidative stress, DNA damage
RNA editing reactions modify, insert, or delete nucleotides and can change the coding properties of an RNA molecule (1). Deamination at C6 of adenosine (A) in RNA generates inosine (I) at the corresponding nucleotide position. Because inosine is decoded as guanosine during translation, this modification can lead to codon changes (recoding) and the introduction of amino acids into a gene product not encoded in the gene (2, 3). Adenosine to inosine editing is widespread in human cells with thousands of transcripts modified, mainly in introns and untranslated regions (4). Current estimates have the number of A to I sites in the human transcriptome at > 15,000 with the vast majority of these sites occurring in Alu repeats (5). However, hundreds of A to I sites also occur in nonrepeat sequences with at least 50 different recoding events known in human cells (6, 7). Recoding by adenosine deamination is common in the nervous system with targets including ligand-gated ion channels, voltage-gated ion channels, and G-protein coupled receptors (2, 3, 8). In several of these cases, recoding has a clear effect on the function of the protein. For instance, editing within three different codons in the message for a serotonin receptor changes an intracellular loop that interacts with G proteins and reduces the ability of the receptor to transmit signal into the cell (3). Consistent with these observations, A to I editing is required for nervous system function in metazoans (9–11). However, although both of the enzymes responsible for A to I editing in humans (ADAR1 and ADAR2) are expressed in tissues throughout the body, little is known about the effect of recoding of targets with roles outside the nervous system (7).
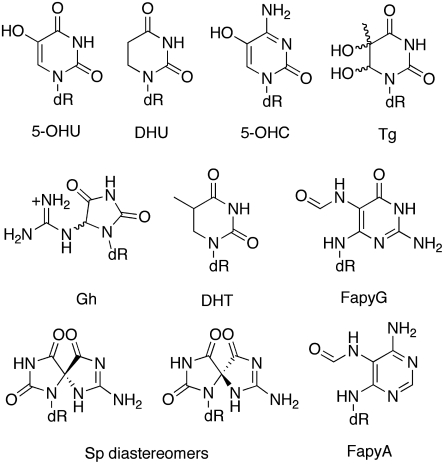
A recent whole transcriptome sequence analysis from various human tissues identified over 200 possible A to I editing sites in nonrepeat sequences, including a site predicted to cause recoding in the mRNA for the DNA repair enzyme NEIL1 (lysine 242 AAA codon edited to AIA codon for arginine) (6). NEIL1 plays a key role in the initiation of base excision repair of oxidized base lesions by catalyzing the cleavage of the N-glycosidic linkage to the 2’-deoxyribose (12). This enzyme is capable of removing a wide array of modified DNA bases including thymine glycol (Tg), 5-hydroxycytosine (5-OHC), 5-hydroxyuracil (5-OHU), dihydrothymine (DHT), dihydrouracil (DHU), the formamidopyridines (FapyG and FapyA), guanidinohydantoin (Gh), and spiroiminodihydantoin (Sp) (Fig. 1) (12–15). Oxidized base lesions arise in DNA at rates of thousands per day as a result of endogenous metabolic activity as well as from oxidative stress induced by inflammation, radiation, or toxic agents (16). The most thoroughly examined oxidized base is 8-oxo-7,8-dihydroguanine (OG) which is one of the primary substrates for the human OG glycosylase (hOGG1) but is not efficiently processed by NEIL1 (12, 14, 16). The best substrates documented for NEIL1 are the hydantoin lesions (Gh and Sp) that form from further oxidation of OG or from reactions with potent oxidants such as singlet oxygen (12, 14). These lesions have garnered much attention due to their extremely high mutagenic potential in cells which is significantly greater than OG (16). Thymine glycol is the most common pyrimidine base modification produced under oxidative stress and ionizing radiation (17). Tg is a substrate of the oxidative DNA glycosylase, NTH1, and though not miscoding, its ability to strongly block DNA replication makes it a toxic lesion in cells (17). NEIL1 is also distinct from hOGG1 and NTH1 in catalyzing both β and δ lyase reactions of the abasic site leaving a phosphate group at the 3’ end of the break (18) that allows NEIL1 to participate in an alternative AP-endonuclease independent BER pathway, in addition to short- and long-patch base excision repair (19). Moreover, NEIL1 is also unique among BER glycosylases in its activity with oxidized lesions in single-stranded, bulged and bubble DNA (12, 20). This property of NEIL1 coupled with the established interactions with proteins such as RPA, PCNA, and CSB strongly suggest its involvement in repair during replication and/or transcription (12, 20). The various features of NEIL1 suggest that this glycosylase plays central roles in facilitating repair and initiating different repair pathways dependent on the context and type of lesion encountered.
Fig. 1.
Known substrates for the base excision repair glycosylase NEIL1. Abbreviations: 5-OHU, 5-hydroxyuracil; DHU, dihydrouracil; 5-OHC, 5-hydroxycytosine; Tg, thymine glycol; Gh, guanidinohydantoin; DHT, dihydrothymine; FapyG, 2,6-diamino-4-hydroxy-5-formamidopyrimidine; Sp, spiroiminodihydantoin; and FapyA, 4,6-diamino-5-formamidopyrimidine.
The three-dimensional structure of human NEIL1 has been solved by X-ray crystallography (21). In addition, while structural data for a complex of human NEIL1 with damaged DNA has not been reported, complexes of related repair glycosylases with DNA containing damaged bases have been structurally characterized (22, 23). From an analysis of these structures, we realized the NEIL1 recoding site is located in the previously identified lesion recognition loop of this family of DNA repair enzymes (Fig. 2) (22). This observation suggested to us that RNA editing might regulate NEIL1 activity by modulating efficiency and/or specificity of damaged base removal. Regulation via RNA editing had not previously been reported for a DNA repair enzyme. Furthermore, such an effect would imply that NEIL1 repair activity is subject to regulation via pathways that control the editing enzyme responsible for recoding.
Fig. 2.
(A) Superposition of human NEIL1 structure (dark gray) with that of E. coli Fpg (green) bound to 8-oxoguanine-containing DNA (17, 19). Red open arrow indicates lesion recognition loop of Fpg. (B) Sequence alignment of Fpg/Nei family of DNA repair glycosylases indicating the position of the hNEIL1 recoding site and lesion recognition loop as identified by Imamura, Wallace, and Doublie (18, 22).
Results
The Effect of K242R Recoding on NEIL1 Activity.
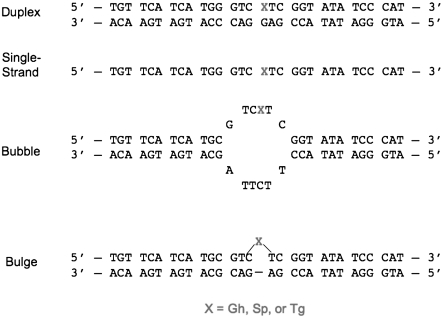
To evaluate the effect of the amino acid change on NEIL1 activity, we overexpressed and purified the protein bearing either lysine (unedited) or arginine (edited) at position 242. We analyzed the effect of the edit on the rate constants for NEIL1 removal of Tg, Gh, and the Sp1 diastereomer from single stranded DNA, duplex DNA, bulge and bubble structure DNA contexts (Fig. 1 and Fig. 3). Under conditions of multiple turnover, NEIL1 exhibits biphasic “burst” kinetics with Sp-containing substrates providing a means to accurately determine the active site fraction (14). Both enzyme forms exhibit similar active fractions indicating that the difference in amino acid at this position does not globally alter protein folding or stability needed for activity. Using the active enzyme concentration, rate constants for the glycosylase step (kg) under single-turnover conditions were measured (Table 1). We found the edited form of NEIL1 cleaves Tg when paired with G in duplex DNA 30 times more slowly than the unedited form, whereas this form reacts nearly three times faster than the unedited form with Gh in the duplex (Table 1). We also observed that when paired with A, the rate constant for Tg removal for the edited form (1.3 ± 0.1 min-1) is 40-fold reduced from that for unedited NEIL1 (53 ± 8 min-1). The superior activity of the unedited form for removal of Tg is observed in all contexts including single-strand DNA, bulge and bubble DNA contexts (Table 1). The Gh lesion is more efficiently removed by the edited form in all DNA contexts. The Sp1 diastereomer is a superb substrate for NEIL1 and is efficiently removed by both edited and unedited forms. There are small differences in processing of Sp1 by edited and unedited NEIL1; in duplex contexts, the edited form exhibits a twofold greater activity while in the bubble substrate its activity is twofold reduced. Notably, the editing reaction has altered the NEIL1 repair efficiency in a lesion-specific nature. While the edited form reacts ∼150-fold faster with Gh in the duplex substrate than with Tg, this difference is less than twofold for the unedited form. In the bubble DNA context, with both enzyme forms, Gh is preferred over Tg; however, the magnitude of the preference is ∼1,500-fold for the edited form and only 25-fold for the unedited form. Clearly, editing modulates the relative lesion specificity of base removal. The basis for the alteration in specificity and glycosylase activity caused by the conservative K to R change in the lesion recognition loop is not obvious based on the structure of the enzyme alone and awaits further detailed structural and mechanistic studies.
Fig. 3.
DNA substrates evaluated in this study.
Table 1.
Rate constants (kg)* of base removal by edited versus unedited NEIL1
| Tg† | Gh‡ | Sp1 | |||||||
| Context§ | Unedited | Edited | Ratio¶ | Unedited | Edited | Ratio | Unedited | Edited | Ratio |
| Duplex (X: G) | 76 ± 10 | 2.5 ± 0.1 | 30 | 130 ± 20 | 370 ± 40 | 0.4 | 120 ± 40 | 250 ± 20 | 0.5 |
| Single strand | 0.6 ± 0.1 | 0.02 ± 0.01 | 30 | 1.2 ± 0.1 | 2.4 ± 0.6 | 0.5 | 0.5 ± 0.1 | 0.5 ± 0.1 | 1.0 |
| Bulge | 1.4 ± 0.1 | 0.04 ± 0.02 | 35 | 5.0 ± 0.6 | 13 ± 1 | 0.4 | 1.1 ± 0.1 | 0.7 ± 0.1 | 1.6 |
| Bubble | 1.2 ± 0.1 | 0.06 ± 0.02 | 20 | 30 ± 6 | 94 ± 8 | 0.3 | 1.3 ± 0.2 | 0.6 ± 0.1 | 2.2 |
*Rate constants in min-1 measured under single-turnover conditions (20 nM substrate, 200 nM enzyme) at 37 °C. Reactions with single-strand DNA did not go to completion; slow reactions rates were determined based on initial rate rather than complete fitting of the progress curve.
†Tg paired with G. Rate constants in the same duplex paired with A for edited and unedited NEIL1 are 1.3 ± 0.1 min-1, and 53 ± 8 min-1, respectively. The ratio is 40.
‡Reactions with Gh-containing bulge and bubble substrates did not go to completion (60%–80%); however, similar relative rates for reactions that proceeded to completion were observed at 25 °C.
§See Fig. 3 for DNA structures used for NEIL1 substrates.
¶Ratio is the rate constant for unedited NEIL/rate constant for edited NEIL1.
The NEIL1 Pre-mRNA Editing Reaction.
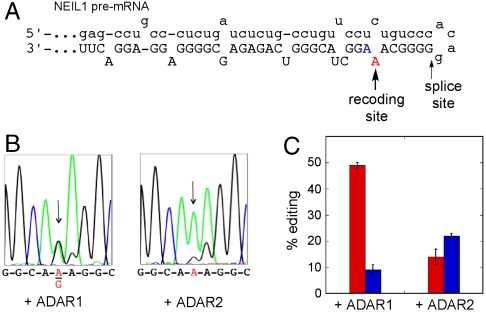
The recoding site is located in exon 6 near the intron 5/exon 6 boundary in the NEIL1 pre-mRNA. Based on in silico folding, we predicted the edited adenosine resides in an A-C mismatch in a hairpin stem formed by the pairing of the exon to the immediate upstream intronic sequence (Fig. 4) (24). The long duplex and A-C mismatch at the recoding site are features common in ADAR substrates (25). However, it is not possible to predict a priori whether ADAR1 or ADAR2 is responsible for this edit. Identification of the ADAR responsible for the NEIL1 recoding is significant because the two ADARs are themselves subject to different regulatory pathways. To determine which ADAR enzyme is responsible, we generated an RNA comprising the 200 nucleotides flanking the recoding site adenosine for in vitro editing assays. This substrate RNA was then subjected to deamination assays with overexpressed and purified human ADAR1 or ADAR2. Importantly, ADAR1 deaminates the central adenosine of the K242 AAA codon more efficiently than does ADAR2 under these conditions (49% vs. 14%) (Fig. 4 B and C). These results support ADAR1 as the editing enzyme primarily responsible for the NEIL1 recoding and implicate ADAR1 regulation in the control of NEIL1 function. In addition to the adenosine at the central position of the codon, we observed editing at the third position as well in these assays. Indeed, this nucleotide is the preferred deamination site for ADAR2 on this substrate. The third nucleotide of the K242 codon is also edited in vivo, but this edit does not cause an amino acid change (6, 26).
Fig. 4.
(A) Predicted RNA secondary structure surrounding NEIL1 recoding site. Lower case lettering indicates intron. (B) In vitro editing of a 200 nt substrate comprising the NEIL1 recoding site. (Left) Sequence of products from reaction of 1 μM human ADAR1. (Right) Sequence of products from reaction of 1 μM human ADAR2. (C) Quantification of editing efficiency of ADAR1 and ADAR2 on model RNA substrate. Red bars: central adenosine of K242 codon, Blue bars: third adenosine of K242 codon. Results are presented as an average % editing and standard deviation from three independent experiments.
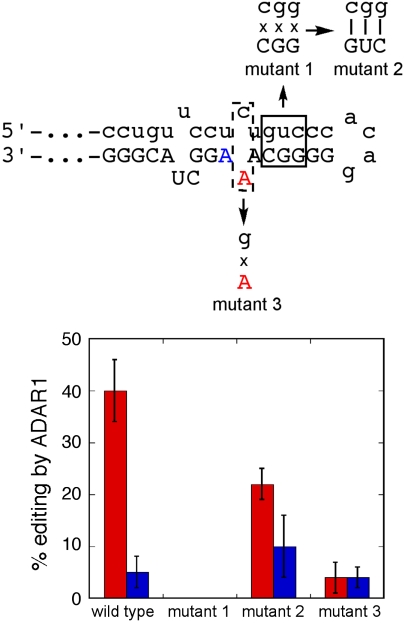
To test our secondary structure hypothesis for the RNA editing substrate, we initially introduced mutations that disrupt the predicted duplex near the editing sites by creating changes in nucleotide positions in the intron sequence that replace three pairing interactions (5′-GUC-3′•3′-CGG-5′) with three mismatches (5′-CGG-3′•3′-CGG-5′). Consistent with our prediction, this mutant RNA is not edited by ADAR1 (mutant 1, Fig. 5). Importantly, when we then make compensatory mutations that restore pairing (5′-CGG-3′•3′-GUC-5′), editing is also restored (mutant 2, Fig. 5). To test the importance of the A-C mismatch in the substrate RNA, we made a single nucleotide change that creates an A-G mismatch at this site. While editing at the third position in the K242 codon is largely unaffected by this change, the recoding site is edited poorly by ADAR1 (4%) in this RNA (mutant 3, Fig. 5). This latter result highlights the importance of the A-C mismatch in influencing editing efficiency for ADAR1 at the NEIL1 recoding site. Interestingly, the duplex secondary structure and A-C mismatch at the recoding site appear to be conserved in other vertebrate NEIL1 pre-mRNAs including from mouse, horse, and dog (Fig. S1), suggesting modulation of NEIL1 structure through RNA editing occurs in other species as well.
Fig. 5.
(Top) Mutations made to test RNA secondary structure prediction for NEIL1 recoding site. (Bottom) Quantification of editing efficiency of ADAR1 with different RNA substrates. Red bars: central adenosine of K242 codon, Blue bars: third adenosine of K242 codon. Results are presented as an average % editing and standard deviation from three independent experiments.
Changes in NEIL1 Editing in Response to Interferon.
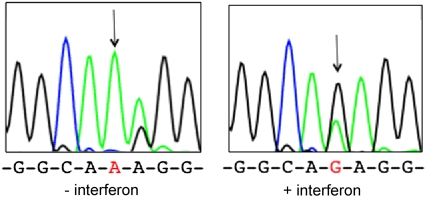
The cellular activity of ADAR1 is regulated in a variety of ways (25). To determine if NEIL1 editing is responsive to changes in ADAR1 activity, we treated U87 cells (human glioblastoma) with interferon α (IFN-α), a condition that stimulates transcription of ADAR1 p150 (27). When we amplify and sequence NEIL1 cDNA from cells with or without prior interferon treatment, we observe recoding only in the treated cells (Fig. 6). Thus, the relative amounts of edited vs. unedited NEIL1 transcripts can be regulated extracellularly. Interestingly, the third adenosine of the K242 codon is edited in the absence of interferon treatment in these cells and editing at this site is reduced by interferon. Nishikura and colleagues also reported that interferon treatment of U87 cells led to increases in editing at ADAR1 sites and decreases at ADAR2 sites on the 5-HT2cR pre-mRNA (28).
Fig. 6.
NEIL1 editing in response to IFN-α. (Left) Sequence at the recoding site in NEIL1 cDNA from U87 human glioblastoma cells cultured in the absence of IFN-α. (Right) NEIL1 cDNA sequence from U87 cells treated with IFN-α.
Discussion
While it is well established that A to I editing is necessary for creating diversity of structure for neurotransmitter receptors and ion channels, little is known about the effect this type of editing has on other protein targets. Recent whole transcriptome sequencing efforts have dramatically increased the number of known A to I editing sites that lead to codon changes in human transcripts (6, 26). Indeed, recently identified A to I recoding targets include a T cell receptor (CD6), a cyclin (cyclin 1), a tyrosine kinase (PTK2), a zinc finger protein (Znf70), and a component of the signal recognition particle (SRP9) (6, 26). However, only a small fraction of the > 50 known human A to I recoding sites have been evaluated to reveal a consequence on protein function. Here we show that an A to I editing reaction alters the glycosylase activity and damaged base specificity for the BER glycosylase NEIL1.
Modulation of the activity of a DNA repair enzyme via RNA editing represents a previously unrecognized mechanism available for regulation of DNA repair. In contrast, posttranslational modifications of DNA repair proteins are well known to modulate enzyme activity, alter interactions with protein partners and control cellular localization to permit the appropriate DNA damage response (29). Interestingly, the sequence reported in the literature for the NEIL1 protein is that of the edited form, because this was the sequence originally established for the NEIL1 cDNA (15, 30–32). Thus, all studies of lesion specificity published to date on NEIL1 have used the edited form of the protein and thus researchers should be cautioned in terms of extrapolating these results to all cellular situations, because the predominant form of NEIL1 may be different under different conditions. Moreover, both edited and unedited forms may be present under certain conditions. Both forms can be translated as we observe expression of the NEIL1 protein in untreated U87 cells (K242) (Fig. S2) and others have described overexpression of the edited form (R242) in mammalian cells (30, 33). In addition, several studies using edited NEIL1 have shown that removal of 5-OHU or FapyG by NEIL1 is enhanced by the presence of protein-binding partners (e.g., PCNA, FEN1, and CSB) (33–35). The stimulatory affects afforded by protein partners may depend on the form of NEIL1 used as well as the nature of the lesion and its nucleic acid context. Thus, the presence of edited vs. unedited forms may serve as a lesion-specific mechanism to modulate and recruit the appropriate downstream repair enzymes to coordinate the complex array of DNA damage responses (19).
The location of the site of recoding in the proposed lesion recognition loop of NEIL1 is particularly striking. Previous structural work with the bacterial Fpg glycosylase has shown that the corresponding loop becomes ordered in the presence of lesion-containing DNA; however, the manner that FapyG vs. OG lesions are recognized within this loop are completely different (23, 36). These studies illustrate the importance of flexibility in this region to adopt alternative recognition complexes to recognize a wide variety of substrates. Thus, subtle changes in the conformation and structure of this region of NEIL1 may be all that is required to shift the balance in lesion preference from one lesion to another. Interestingly, previous use of protein engineering has also shown that with only one or two mutations the substrate recognition features of some glycosylases can be altered (37). The ability to recognize a wide variety of substrates may be advantageous under normal cellular conditions where the glycosylases are patrolling the genome in search of damage; however, under conditions that produce a particularly mutagenic lesion (such as Gh), different substrate specificity may be beneficial (see below).
The ability of ADAR1 to catalyze deamination in a substrate RNA model of the NEIL1 editing site and the enhanced editing at this site observed in cells treated with IFN-α implicate ADAR1 in NEIL1 recoding. Because ADAR1 is induced in T lymphocytes and macrophages by tumor necrosis factor α (TNF-α) and IFN-γ as well as in various tissues in endotoxin-treated mice, we predict the edited form of NEIL1 would be abundant during inflammation (38). Although the full impact of NEIL1 recoding is not currently known, it is tempting to speculate that the presence of two forms of the NEIL1 enzyme, each with distinct lesion preferences and repair efficiencies, facilitates repair of DNA damage caused by the oxidative burst associated with an inflammatory response. Of note, high levels of oxidative stress have been shown to result in increased expression of NEIL1 and would be expected to favor formation of the hydantoin lesions, which are preferentially repaired by edited NEIL1 (in duplexes) (Table 1) (12). On the other hand, a recent study of lobular breast cancer progression identified ADAR1 as one of the top 5% of genes expressed in the tumor (26). Furthermore, these authors observed nearly quantitative recoding of the NEIL1 message in tumor transcripts. It is possible that prolonged hyper editing of the NEIL1 message leads to changes in the number and types of mutations that accumulate in the genome. Further studies on the consequences of NEIL1 recoding in cells expressing varying levels of ADAR1 are also warranted.
In summary, ADAR1-catalyzed editing of the NEIL1 mRNA causes the genomically encoded AAA lysine codon, corresponding to amino acid position 242 in the lesion recognition loop of the protein, to be converted to a codon for arginine. The two forms of the NEIL1 protein (edited and unedited) have distinct enzymatic properties with changes observed for both glycosylase activity and lesion specificity. Editing occurs in a hairpin duplex structure formed near the intron 5/exon 6 boundary in the NEIL1 pre-mRNA. Furthermore, NEIL1 mRNA recoding is regulated extracellularly by interferon, as predicted for an ADAR1-catalyzed reaction. These results suggest a unique regulatory mechanism for DNA repair and extend our understanding of the impact of RNA editing.
Materials and Methods
Details for mutagenesis, overexpression and purification of NEIL1, generation of lesion-containing DNA substrates, glycosylase assays, overexpression and purification of ADARs, generation of ADAR substrate RNAs, and deaminase assays can be found in SI Text.
Supplementary Material
Acknowledgments.
We thank Professor Brenda Bass (University of Utah) and Professor Susan Wallace (University of Vermont) for gifts of ADAR1 and NEIL1 expression plasmids, respectively. We also thank Ashok Bhagwat (Wayne State University) for helpful discussions. P.A.B. and S.S.D. acknowledge the National Institutes of Health for financial support in the form of Grants R01-GM061115 (P.A.B) and R01-CA090689 (S.S.D). R.A.G. was supported by training Grant T32-GM08799 from National Institute of General Medical Sciences (NIGMS)-NIH. The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or National Cancer Institute (NCI).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009231107/-/DCSupplemental.
References
- 1.Grosjean H, Benne R. Modification and editing of RNA. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- 2.Higuchi M, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 3.Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387(6630):303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 4.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLos Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barak M, et al. Evidence for large diversity in the human transcriptome created by Alu RNA editing. Nucleic Acids Res. 2009;37:6905–6915. doi: 10.1093/nar/gkp729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 7.Pullirsch D, Jantsch MF. Proteome diversification by adenosine to inosine RNA-editing. RNA Biol. 2010;7:1–8. doi: 10.4161/rna.7.2.11286. [DOI] [PubMed] [Google Scholar]
- 8.Hoopengardener B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 9.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 10.Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. A-to-I pre-mRNA editing in drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 12.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-Diamino-4-hydroxy-5-formamidopyrimidine and 4,6-Diamino-5-formamidopyrimidines from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 14.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair. 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 16.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 17.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 18.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem Rev. 1998;98:1221–1262. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 19.Hazra TK, et al. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair. 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci USA. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamura K, Wallace SS, Doublie S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J Biol Chem. 2009;284:26174–26183. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 24.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maydanovych O, Beal PA. Breaking the central dogma by RNA editing. Chem Rev. 2006;106:3397–3411. doi: 10.1021/cr050314a. [DOI] [PubMed] [Google Scholar]
- 26.Shah SP, et al. Mutational evolution in a lobular breast tumor profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 27.Patterson JP, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W, Wang Q, Kanes SJ, Murray JM, Nishikura K. Altered RNA editing of serotonin 5-HT2C receptor induced by interferon: implications for depression associated with cytokine therapy. Mol Brain Res. 2004;124:70–78. doi: 10.1016/j.molbrainres.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by posttranslational modification. DNA Repair. 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takao M, et al. A backup glycosylase in Nth1 knock-out mice is a functional Nei (Endonuclease VII) homologue. Proc Natl Acad Sci USA. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 31.Hazra TK, et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morland I, et al. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegde ML, et al. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J Biol Chem. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dou H, et al. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 35.Muftouglu M, et al. Cockanye syndrome Group B stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J Biol Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coste F, et al. Structural basis for the recognition of FapydG lesions (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by Formamidopyrimidine-DNA glycosylase. J Biol Chem. 2004;279:44074–44083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- 37.O’Brien PJ. Catalytic promiscuity and the divergent evolution of DNA repair enzymes. Chem Rev. 2006;106:720–752. doi: 10.1021/cr040481v. [DOI] [PubMed] [Google Scholar]
- 38.Yang JH, et al. Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology. 2003;109:15–23. doi: 10.1046/j.1365-2567.2003.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.