Abstract
The v-Crk oncogene product consists of two protein interaction modules, a Src homology 2 (SH2) domain and an SH3 domain. Overexpression of CrkI, the cellular homolog of v-Crk, transforms mouse fibroblasts, and elevated CrkI expression is observed in several human cancers. The SH2 and SH3 domains of Crk are required for transformation, but the identity of the critical cellular binding partners is not known. A number of candidate Crk SH3 binding proteins have been identified, including the nonreceptor tyrosine kinases c-Abl and Arg, and the guanine nucleotide exchange proteins C3G, SOS1 and DOCK180. The aim of this study is to determine which of these are required for transformation by CrkI. We found that shRNA-mediated knockdown of C3G or SOS1 suppressed anchorage-independent growth of NIH-3T3 cells overexpressing CrkI, while knockdown of SOS1 alone was sufficient to suppress tumor formation by these cells in nude mice. Knockdown of C3G was sufficient to revert morphological changes induced by CrkI expression. By contrast, knockdown of Abl family kinases or their inhibition with imatinib enhanced anchorage-independent growth and tumorigenesis induced by Crk. These results demonstrate that SOS1 is essential for CrkI-induced fibroblast transformation, and also reveal a surprising negative role for Abl kinases in Crk transformation.
Keywords: SH3 domain, SH2/SH3 adaptor, Abl family kinase, SOS1, C3G
Introduction
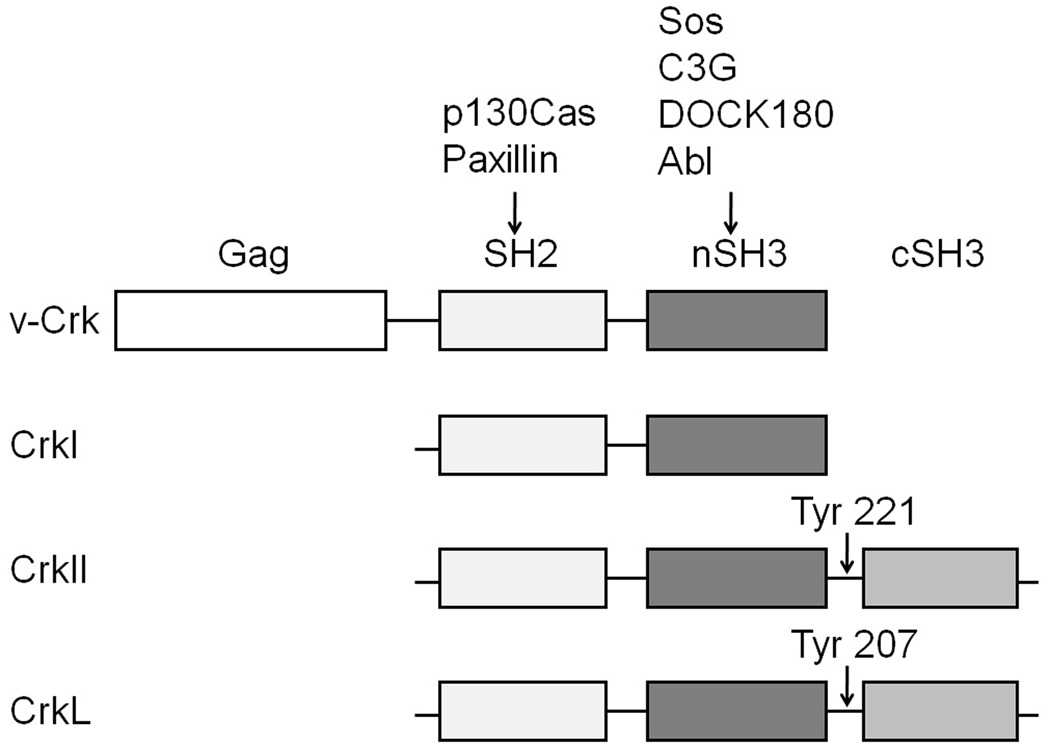
v-Crk was first identified as the oncogene product of the avian sarcoma virus CT10 (Mayer et al., 1988). It consists of a viral Gag portion fused with an SH2 and SH3 domain derived from the endogenous c-Crk gene. Two forms of the cellular homolog of v-Crk, termed CrkI and CrkII, are generated by alternative mRNA splicing (Matsuda et al., 1992; Reichman et al., 1992). CrkI, like v-Crk, consists of one SH2 and one SH3 domain (nSH3), while CrkII has an additional C-terminal SH3 domain (cSH3) (Fig. 1). The closely related CrkL gene encodes a protein with the same overall domain structure as CrkII (ten Hoeve et al., 1993). Crk acts as an adaptor protein in cell signaling, mediating protein-protein interactions via its SH2 domain (which binds to tyrosine phosphorylated peptides) and SH3 domains (which bind proline-rich peptide motifs). Thus Crk serves to couple tyrosine kinase-mediated signals to downstream effectors, such as small G proteins, in signaling pathways that regulate cell transformation, adhesion, migration, phagocytosis, differentiation, proliferation, and apoptosis (Feller, 2001).
Fig. 1.
Domain structures of Crk family adaptor proteins. v-Crk is a fusion protein consisting of N-terminal viral Gag sequences fused to cellular CrkI. CrkI consists of one SH2 and one SH3 domain (nSH3), while CrkII has an additional C-terminal SH3 domain (cSH3). Domain structure of CrkL is similar to CrkII. Tyr 221 and 207 can be phosphorylated by Abl family tyrosine kinases, leading to inhibition of binding activity.
Expression of v-Crk induces transformation of both chicken embryo fibroblasts (CEFs) and mouse fibroblasts (NIH-3T3) (Greulich and Hanafusa, 1996; Mayer et al., 1988). Overexpression of wild type (wt) CrkI induced cell transformation in rat 3Y1 cells, and subcutaneous injection of those cells into nude mice caused tumor formation (Matsuda et al., 1992). Overexpression of CrkL and CrkII also induced transformation, albeit relatively weakly, in rodent fibroblasts (Iwahara et al., 2003; Matsuda et al., 1992; Senechal et al., 1998). The binding activity of the SH2 and SH3 domains is required for Crk-induced transformation (Iwahara et al., 2003; Mayer and Hanafusa, 1990; Senechal et al., 1998). Elevated expression of CrkI has been found in different types of human cancers, including lung adenocarcinoma and glioblastoma, and its expression level is correlated with malignant features in these tumors (Miller et al., 2003; Takino et al., 2003; Wang et al., 2007). Furthermore, siRNA-mediated knockdown of Crk expression suppressed the transformed phenotype of Crk-expressing tumor cell lines, such as glioblastoma KMG4 and ovarian cancer MCAS (Linghu et al., 2006; Wang et al., 2007). Overexpression of miRNA-126, which targets the 3'UTR of Crk, can inhibit adhesion, migration, and invasion of non-small cell lung carcinoma cell lines (Crawford et al., 2008). These results suggest a role for Crk in human cancer.
More than two decades after the discovery of v-Crk, the precise mechanism of Crk-induced transformation remains elusive, despite the identification of many Crk SH2 and SH3 binding partners. Little is known, for example, about which of these interaction partners actually play key roles in the transforming activity of Crk. In fibroblasts, the two most prominent Crk SH2 binding proteins are p130Cas (Sakai et al., 1994a; Sakai et al., 1994b) and paxillin (Birge et al., 1993). Both of these are multi-domain scaffold proteins that localize to focal adhesions and serve as platforms for the assembly of multi-protein complexes that regulate cell migration, cell adhesion and cell survival (Deakin and Turner, 2008; Defilippi et al., 2006). p130Cas has been implicated in Crk transformation by several studies (Iwahara et al., 2004; Nievers et al., 1997; Riggins et al., 2003). The most prominent Crk nSH3-binding proteins are C3G, a guanine-nucleotide exchange factor (GEF) for Rap1 and R-Ras; DOCK180, a GEF for Rac; SOS1, a GEF for Ras and Rac; and the nonreceptor tyrosine kinases c-Abl and Arg (Feller et al., 1994; Hasegawa et al., 1996; Matsuda et al., 1994; Ren et al., 1994; Tanaka et al., 1994; Wang et al., 1996). Previous studies have suggested that C3G plays a role in Crk transformation, and that a Crk/C3G/R-Ras/JNK signaling pathway is activated in v-Crk transformed cells (Mochizuki et al., 2000; Tanaka et al., 1997). The PI 3-kinase/Akt pathway has also been shown to be activated in v-Crk transformed CEFs and NIH-3T3 cells. Furthermore, the small G proteins Ras and Rho have also been implicated in Crk transformation (Greulich and Hanafusa, 1996; Iwahara et al., 2003), and overexpression of SOS or H-Ras can enhance v-Crk induced activation of Akt (Akagi et al., 2002; Akagi et al., 2000; Stam et al., 2001).
In order to elucidate the role of Crk nSH3 binding proteins in CrkI-induced cell transformation, short hairpin RNAs (shRNA) were used to knock down the expression of the candidate Crk effectors C3G, DOCK180, SOS1, and Abl family kinases. These experiments were performed in NIH-3T3 cells overexpressing CrkI, which were transformed as measured by anchorage-independent growth in soft agar and tumorigenicity in athymic nude mice. We found that both C3G and SOS1 are required for maximal CrkI-induced anchorage independent growth, while only SOS1 expression was required for tumorigenicity in vivo. Furthermore, we found that Abl family kinases play a surprising negative regulatory role in suppressing CrkI transformation. This work provides new insight into the signal transduction pathways involved in Crk transformation, and raises new concerns about the use of tyrosine kinase inhibitors for therapy of human cancers that may be driven by Crk overexpression.
Results
Establishment of Crk SH3 binding protein knockdown cell lines overexpressing CrkI
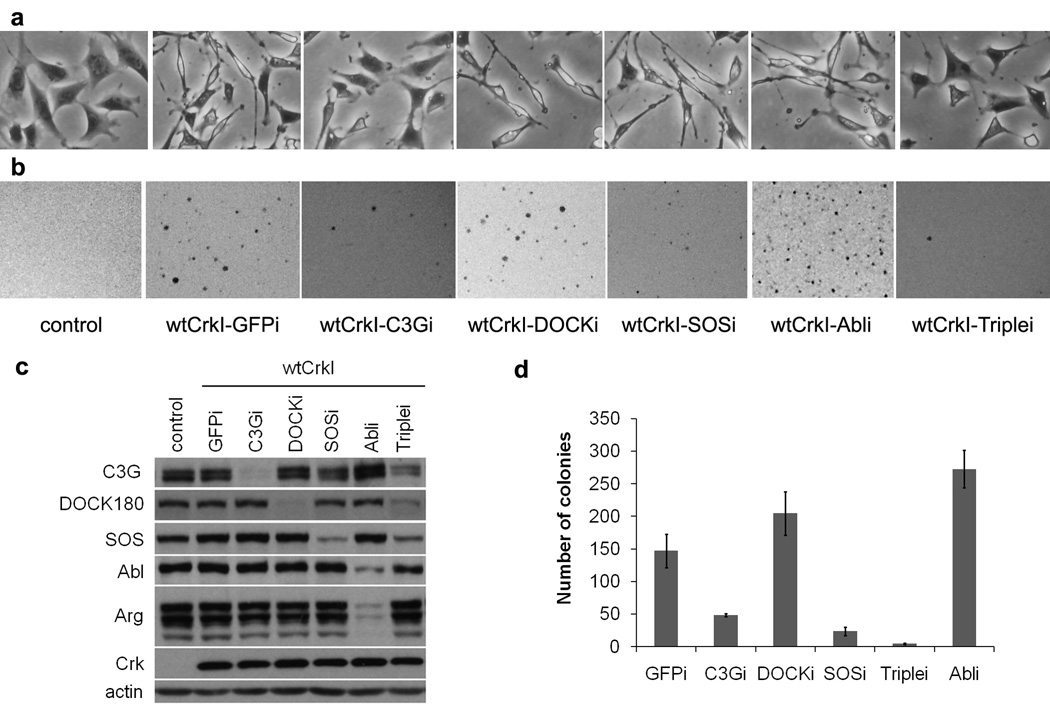
Because Crk family proteins do not transform rodent fibroblasts very efficiently (Iwahara et al., 2003), we first established a pool of mouse fibroblasts stably overexpressing wt human CrkI. NIH-3T3 cells were infected with a retrovirus carrying the human CrkI cDNA and infected cells were subjected to drug selection. CrkI-transformed NIH-3T3 cells were spindle-shaped, with a narrow and elongated cytoplasm, in contrast to the flattened, well-spread morphology of normal cells (Fig. 2a). The CrkI-overexpressing cells grew as colonies in soft agar, demonstrating anchorage-independent growth, while under the same conditions few if any colonies were formed by cells infected with control retrovirus (Fig. 2b). We then took the approach of knocking down candidate nSH3-binding proteins in these cells to assess the role of each in Crk transformation. We used retrovirus-mediated expression of short hairpin RNAs (shRNA) targeting mouse C3G, DOCK180, SOS1, Abl and Arg to establish CrkI-transformed NIH-3T3 cell lines lacking one or more nSH3 binding protein. These candidates were chosen because their binding to nSH3 (Feller et al., 1994; Matsuda et al., 1996; Ren et al., 1994) and their signaling activities are well documented. Endogenous levels of targeted Crk SH3 binding proteins were significantly decreased in the knockdown cell lines (Fig. 2c).
Fig. 2.
Morphology and anchorage-independent growth of CrkI-transformed NIH-3T3 cells lacking different Crk SH3 binding proteins. NIH-3T3 cells were infected with retrovirus expressing wt human CrkI (wtCrkI) or control virus. Infected cells were subjected to puromycin drug selection, and infected with retrovirus producing shRNAs targeting different Crk SH3 binding proteins. shRNA targeting GFP was used as control. Infected cells were subjected to hygromycin drug selection, and then used for transformation assays. (a) Morphology in monolayer culture. Magnification, x40. (b, d) Soft agar colony assay. (b) Photographs were taken 4 weeks after plating. Representatives of three independent experiments are shown. (d) Quantification of the number of colonies. Values represent the average of three independent experiments; error bars represent S.E.M. (c) Knockdown efficiency was determined by immunoblotting analysis using the appropriate antibody, indicated on left. GFPi, SOSi, C3Gi, DOCKi, Abli stand for knockdown of GFP, SOS1, C3G, DOCK180 and Abl family proteins (Abl & Arg) respectively. Triplei stands for triple knockdown of C3G, DOCK180 and SOS1. All cell lines overexpress wt human CrkI (wtCrkI) except control. GFP was targeted by shRNA in both control and wtCrkI-GFPi cell lines.
Morphologic alteration and anchorage-independent growth of knockdown cell lines
We found that the distinctive morphology of CrkI-transformed cells was strongly suppressed by shRNA-mediated knockdown of C3G; the knockdown cells displayed a flat shape similar to control NIH-3T3 (Fig. 2a). Morphological reversion of CrkI-transformed cells was not observed in the other single knockdown cell lines. However, simultaneous knockdown of three Crk SH3 binding proteins (C3G, DOCK180 and SOS1; hereafter referred to as triple knockdown) similarly reverted cells to a more normal morphology. This suggests that, among the Crk SH3 binding proteins tested, C3G is critical for the dramatic morphological changes caused by CrkI overexpression in fibroblasts.
Next we compared the transforming activities of CrkI-transformed cells and their knockdown derivatives by soft-agar colony formation assay. Anchorage independent growth in agar suspension is a hallmark of fibroblast transformation, and correlates with tumorigenicity in vivo. In this assay, the transforming activities of C3G or SOS1 knockdown cell lines were significantly impaired compared with parental CrkI-transformed cells (Fig. 2b,d). The number of colonies formed by C3G or SOS1 knockdown cell lines were about 1/3 and 1/5 of those formed by CrkI-transformed cells, respectively. Very few colonies were formed by the triple knockdown cell line, suggesting the effects of SOS1 and C3G knockdown were additive. Surprisingly, the number of colonies actually increased by approximately two-fold when the Abl family kinases Abl and Arg were knocked down. The DOCK180 knockdown cells showed a slight but statistically insignificant (P = 0.18) increase in colony number compared with parental CrkI-transformed cells. These results indicate that both SOS1 and C3G are important for the anchorage-independent growth of CrkI-transformed cells, and suggest a surprising negative role for Abl family kinases in CrkI-induced transformation.
Tumor formation in nude mice
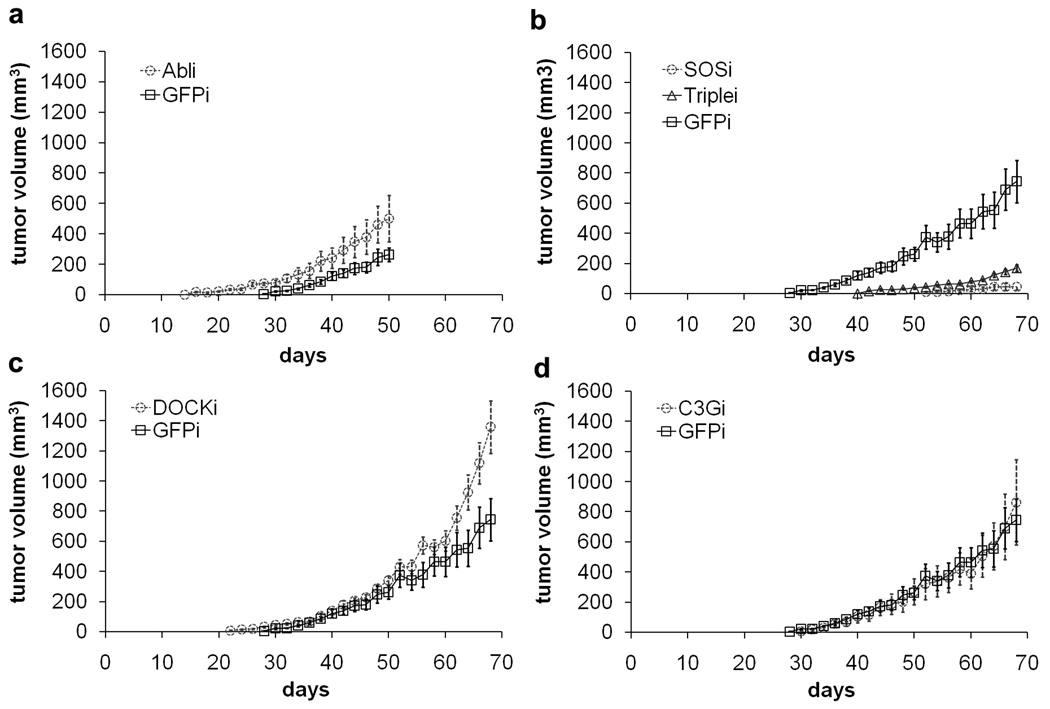
In order to better examine the in vivo transforming activities of different knockdown cell lines, we tested their ability to form tumors in athymic nude mice. Mice injected with CrkI-transformed NIH-3T3 cells began forming palpable tumors in situ 28 days after injection, whereas tumors in mice injected with SOS1 or triple knockdown cells were first detected two to four weeks later and grew much more slowly (Fig. 3b). Tumors in the two latter groups were much smaller at all time points compared to the group injected with control CrkI-transformed cells (Table 1). 70 days after injection, 40% of the mice (n=10) injected with SOS1 knockdown cells formed very small tumors and the other 60% had no palpable tumors, demonstrating that the tumorigenicity of CrkI-transformed cells was almost totally abolished by reduced SOS1 expression. On the contrary, tumors in mice injected with Abl and Arg knockdown cells began forming earlier, beginning 16 days after injection, and were larger than those in mice injected with control CrkI-transformed cells (Table 1 and Fig. 3a). Mice injected with C3G or DOCK180 knockdown cells showed no significant differences in overall tumor growth rate and tumor size compared to the mice injected with control CrkI-transformed cells (Fig. 3c & 3d). No obvious tumor metastasis was found in any of the mice after necropsy. These results demonstrate that knockdown of SOS1 effectively suppresses CrkI-induced tumorigenicity, whereas knockdown of Abl family proteins enhances it.
Fig. 3.
In vivo tumor formation in athymic nude mice. Knockdown cell lines expressing CrkI were prepared as in Fig. 2, and injected subcutaneously into nude mice. Tumor size was monitored every two days. Each point is the mean ± S.E.M. of 10 mice. Mice injected with control cell line (not overexpressing CrkI) did not form tumors under these conditions. P < 0.05 between wtCrkI-GFPi group and wtCrkI-Abli, wtCrkI-SOSi, or wtCrkI-Triplei groups by ANOVA. No significant difference was found between wtCrkI-GFPi group and wtCrkI-C3Gi or wtCrkI-DOCKi groups.
Table 1.
Tumor volumes in nude mice injected with different knockdown cell lines. Average tumor volume (mm3) formed in nude mice at different times after injection (n = 10). Days = days after injection of different knockdown cell lines into nude mice.
| Days | wtCrk GFPi |
wtCrk Abli |
wtCrk SOSi |
wtCrk Triplei |
wtCrk DOCKi |
wtCrk C3Gi |
|---|---|---|---|---|---|---|
| 16 | 0 | 18.25 | 0 | 0 | 0 | 0 |
| 28 | 3.95 | 73.28 | 0 | 0 | 32.02 | 0 |
| 40 | 120.8 | 241.3 | 0 | 1.76 | 137.9 | 104.3 |
| 50 | 262.9 | 501.1 | 0 | 37.7 | 339.2 | 268.9 |
Growth and apoptosis rates of different knockdown cell lines
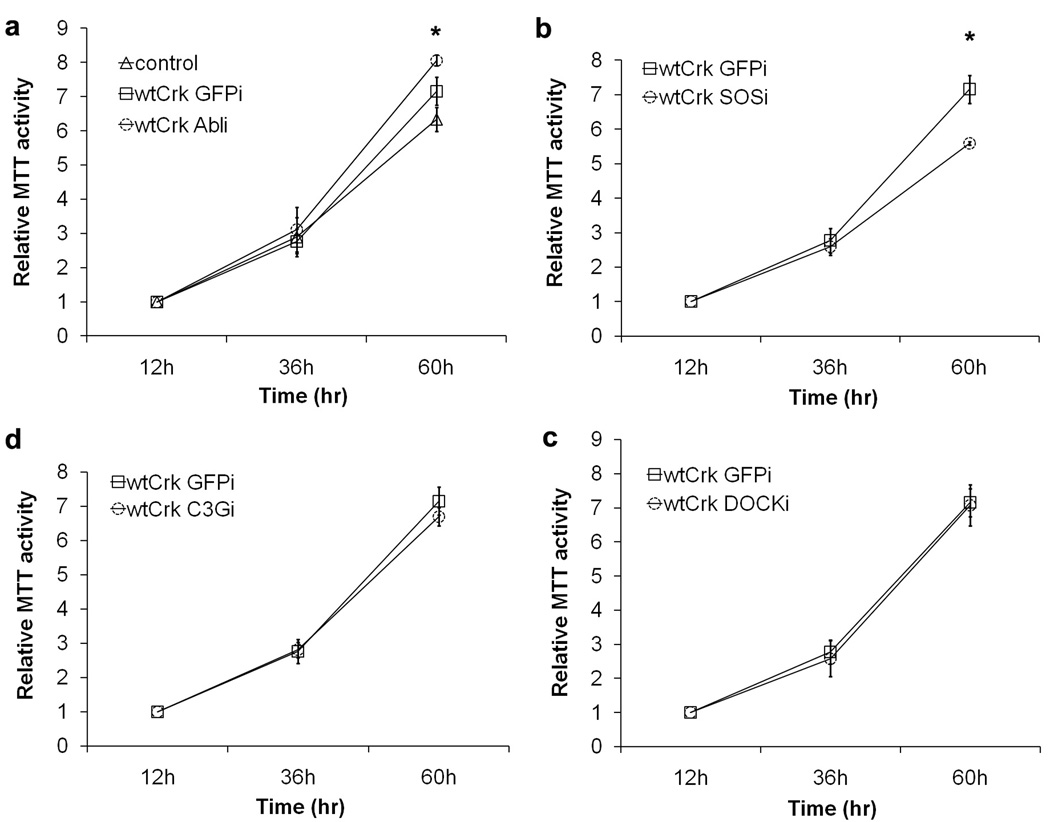
To gain further insight into the underlying causes for differences in tumorigenicity, we investigated the effect of CrkI effector knockdown on the rates of proliferation and apoptosis in CrkI-transformed cells. The rate of cell proliferation was determined using the MTT cell viability assay for cells cultured on tissue culture plastic in complete medium. As expected, the growth rate of CrkI-transformed cells was slightly higher than that of normal NIH-3T3 cells (Fig. 4a). The growth of CrkI-transformed cells was significantly suppressed by knocking down SOS1 and significantly accelerated by knocking down Abl family proteins (P < 0.05 at 60 h) (Fig. 4a & 4b), while the knockdown of DOCK180 or C3G had no significant effect (Fig. 4c & 4d).
Fig. 4.
In vitro proliferation. Knockdown cell lines expressing CrkI were prepared as in Fig. 2, and plated in 96 well plates. Normal NIH-3T3 cells were used as negative control. Cell growth in complete medium was determined via MTT assay over time (hrs). Average relative MTT activity ± standard deviation is shown. *P < 0.05 between wtCrk-GFPi and wtCrk-Abli or wtCrk-SOSi cell lines at 60 h using ANOVA test. No significant difference was found between wtCrk-GFPi and wtCrk-C3Gi or wtCrk-DOCKi cell lines.
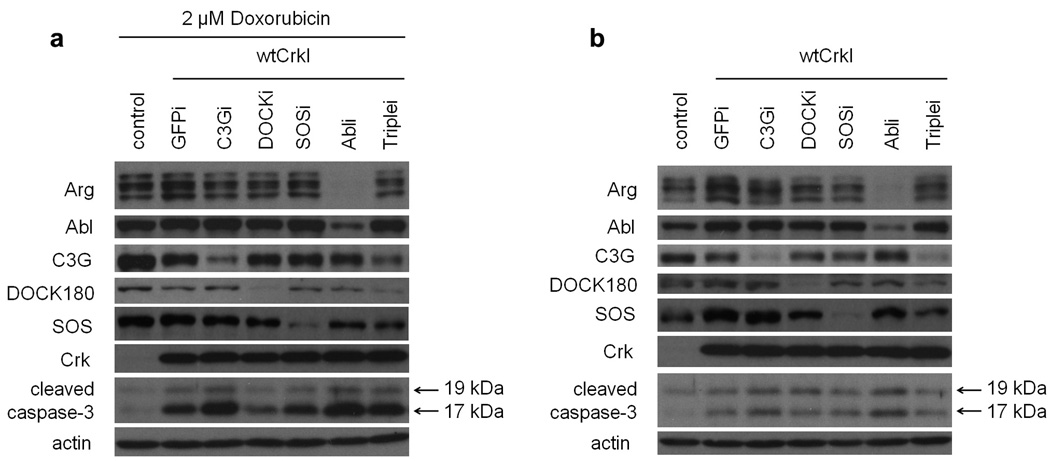
We also tested whether altered sensitivity to apoptosis might contribute to the observed differences in growth rates. Apoptosis was assayed in cells with or without pre-treatment with the DNA-intercalating anthracyclin doxorubicin, a commonly used cancer chemotherapeutic that promotes apoptosis. The levels of cleaved caspase-3, a biochemical marker of apoptosis, were increased in CrkI-expressing cells compared to parental NIH-3T3 cells, both with and without doxorubicin treatment (Fig. 5). Levels of cleaved caspase-3 in the knockdown cells, including the SOS1 and the Abl and Arg knockdowns, were similar to parental CrkI-transformed cells under both conditions; if anything, caspase-3 levels were higher in the Abl knockdown cells. This result was confirmed using a second inducer of apoptosis, UV irradiation (Supplemental Fig. S1). We also performed TUNEL staining of sections of tumors induced in nude mice by knockdown cell lines, and found more apoptotic cells in the more rapidly growing Abl knockdown tumors than in control CrkI tumors (Supplemental Fig. S1). Thus we found no apparent correlation between increased growth and decreased apoptosis. Taken together, the results of biological assays indicate that SOS1 knockdown suppresses the growth of CrkI-transformed cells, whereas Abl knockdown enhances their growth; these effects are most likely to be due to differences in the rate of proliferation, not to differences in the rate of apoptosis.
Fig. 5.
Apoptosis in knockdown cell lines. Knockdown cell lines expressing CrkI were prepared as in Fig. 2. Immunoblotting analysis was performed using cleaved caspase-3 (Asp175) antibody on extracts from different knockdown cell lines either treated with doxorubicin (16 h, 2 µM) (a) or left untreated (b). Cleaved caspase-3, a biochemical apoptotic marker, was detected as a 17–19 kDa doublet. Representative results of two independent experiments are shown.
Analysis of downstream signaling pathways in different knockdown cell lines
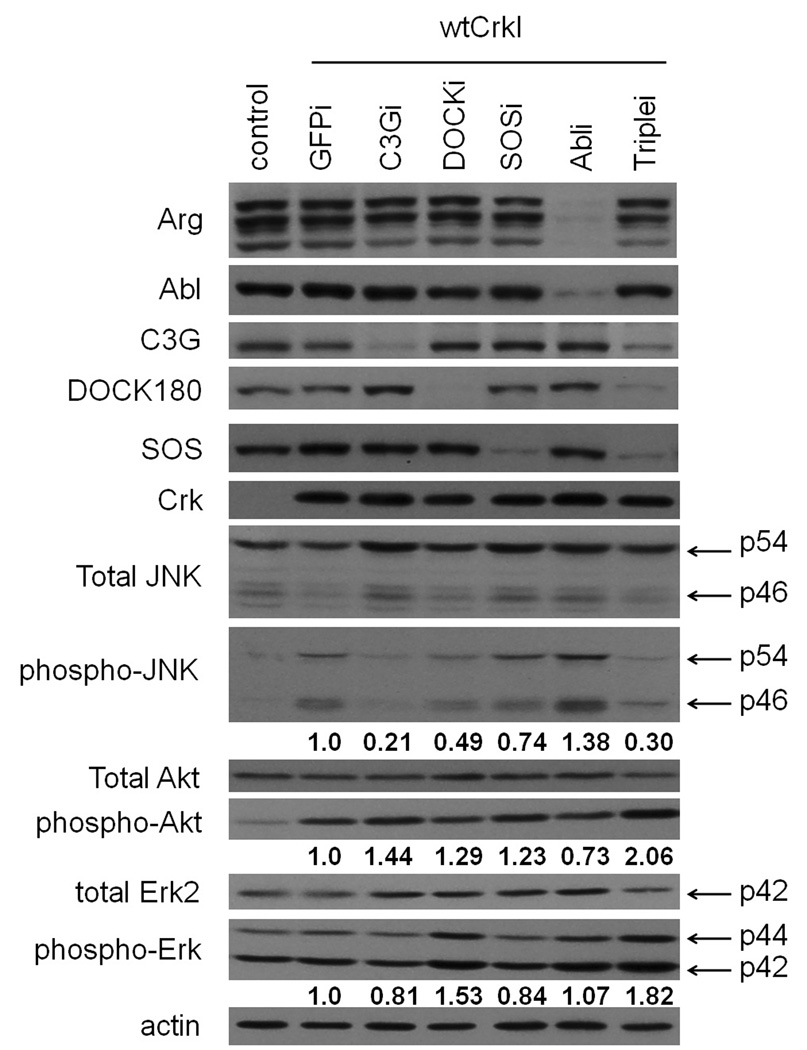
We next examined the effects of Crk SH3 binding protein knockdown on signaling downstream of CrkI. Both JNK and Akt have been reported to be activated in Crk-transformed cells (Akagi et al., 2002; Akagi et al., 2000; Mochizuki et al., 2000; Tanaka et al., 1997), so we assayed pathway activity by immunoblotting with phosphospecific antibodies for the activated kinases. As expected from previous results, both JNK and Akt were activated in CrkI-transformed cells compared with normal NIH-3T3 cells (Fig. 6). The activation of JNK was strongly suppressed in C3G or triple knockdown cells, and modestly enhanced in Abl and Arg knockdown cells. In contrast, the strong Akt activation induced by CrkI overexpression was not suppressed by knockdown of any Crk SH3 binding proteins tested (Fig. 6). Our data are consistent with previous studies indicating that C3G is required for JNK activation in Crk-transformed cells (Mochizuki et al., 2000; Tanaka et al., 1997), and also suggest that Abl family proteins act as negative regulators for this downstream pathway in Crk transformation. Importantly, knockdown of SOS1, which potently suppressed the transforming activity of Crk, had no obvious effect on the activation of either JNK or Akt by CrkI.
Fig. 6.
Effect of knockdown on downstream signaling pathways. Knockdown cell lines expressing CrkI were prepared as in Fig. 2, and total cell lysates subjected to immunoblot analysis with phosphospecific antibodies for JNK (phospho-JNK), Erk1/2 (phospho-Erk) and Akt (phospho-Akt). The same blots were then re-probed with anti-JNK, anti-Erk or anti-Akt antibodies. Arrows show p54/p46 JNK and p44/p42 Erk proteins. Numbers below phospho-JNK, phospho-Erk and phospho-Akt blots are relative values normalized to total JNK, Erk and Akt levels, and represent the average of 3 independent experiments.
We also assessed Erk pathway activity in CrkI transformed knockout cells. The MAP kinases Erk1 and Erk2 are major downstream effectors of Ras, and thus good candidates for regulation by Sos1 in CrkI transformation. Erk activity was assessed by immunoblotting with phosphospecific antibodies for activated Erk. Surprisingly, we found that Erk was not activated in CrkI-transformed cells compared with normal NIH-3T3 cells, and that knockdown of SOS1 did not significantly suppress Erk activity under these conditions (in compete medium with serum) (Fig. 6). Thus we conclude that the dependence of Crk transformation on SOS1 is unlikely to be due to effects on the Erk pathway.
Enhancement of CrkI-induced cell transformation by a small-molecule Abl inhibitor
The results from soft-agar colony formation, proliferation, and tumorigenicity assays all suggested a surprising negative role for Abl family kinases in CrkI-induced cell transformation. To address whether Abl kinase activity was required for suppression of Crk transformation, we tested the effect of imatinib (Gleevec®), a small-molecule tyrosine kinase inhibitor specific for Abl family proteins (and to a lesser extent for PDGF and Kit family receptors) (Druker, 2009).
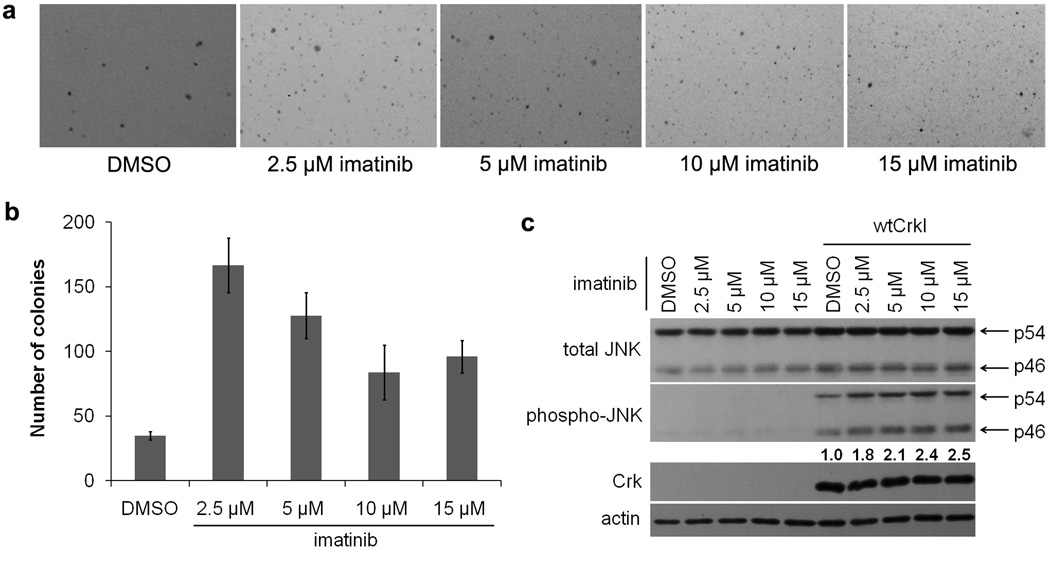
When anchorage-independent growth of CrkI-transformed cells was assayed by soft-agar colony formation, we found that imatinib increased the number of colonies more than four-fold compared to the untreated control (Fig. 7a,b). 2.5 µM imatinib was most potent, consistent with published IC50 values (Druker et al., 1996). Higher concentrations were somewhat less effective, likely due to nonspecific toxicity (Deininger et al., 1997). Inhibition of Abl family proteins by imatinib stimulated the anchorage-independent growth of CrkI-transformed cells more efficiently than knockdown of Abl and Arg (Fig. 2), most likely because the drug can effectively inhibit kinase activity in all cells, whereas shRNA cannot totally eliminate the proteins.
Fig. 7.
Enhancement of CrkI-induced transformation by Abl kinase inhibitor. (a,b) Anchorage-independent growth of CrkI-transformed NIH-3T3 cells in the presence or absence of imatinib or vehicle (DMSO). (a) 4 weeks after plating, photographs of the colonies were taken. Representatives of three independent experiments are shown. (b) Number of colonies was quantified. Bars represent average of three independent experiments. Error bars represent standard deviations. (c) Increased JNK activity in CrkI-transformed NIH-3T3 cells in the presence of Abl kinase inhibitor. Cells were cultured with imatinib at indicated concentrations or vehicle (DMSO) for 3 days. Total cell lysates were subjected to immunoblot analysis with phosphospecific antibody for JNK (phospho-JNK) and then re-probed with anti-JNK antibody. Arrows show p54 and p46 JNK proteins. Numbers are relative phospho-JNK values normalized to total JNK, representing the average of 3 independent experiments.
We also assayed JNK pathway activation in CrkI-transformed cells in the presence or absence of imatinib by immunoblotting with phospho-specific JNK antibody. We found that imatinib treatment of CrkI-transformed cells led to further elevation of JNK activity, without any obvious effect in normal NIH-3T3 cells (Fig. 7c). Thus chemical inhibition of Abl family kinases enhances both JNK activation and the anchorage-independent growth of CrkI-transformed cells, consistent with the negative role for Abl family kinases suggested by gene silencing experiments.
Dependence on SOS1 and Abl activity is specific for Crk transformation
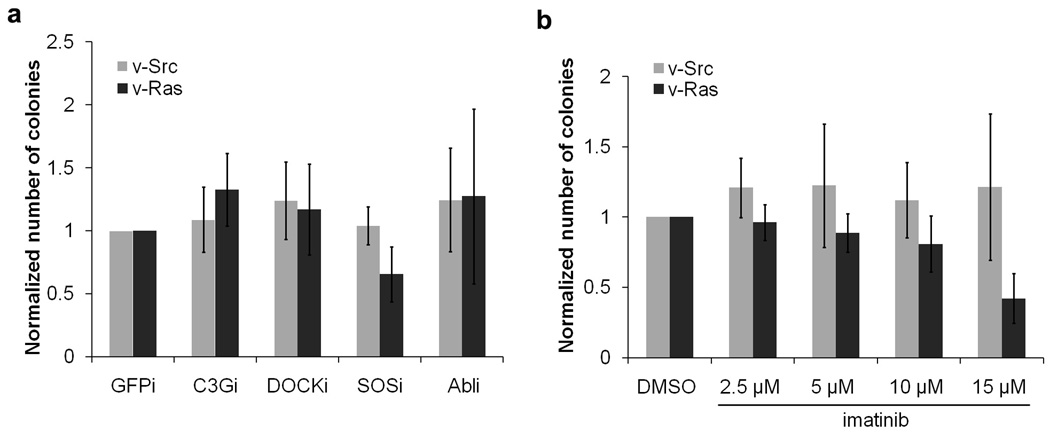
Next we examined the effects of knockdown of Crk effectors on cell transformation induced by other oncogenes, to see if those effects are specific to Crk transformation. NIH-3T3 cells were infected by retroviruses carrying v-Src (Schmidt-Ruppin A strain) or v-Ras (Ha-RasVal12, a constitutively active Ras mutant), and infected cells were subjected to drug selection. These Src- or Ras-transformed cells were then used to generate different Crk SH3 binding protein knockdown cell lines, using the same retroviral vector-based shRNAs used for CrkI-transformed cells. Knockdown of targeted proteins was confirmed by immunoblotting (data not shown). We found that, in contrast to CrkI-transformed cells, knockdown of Crk SH3 binding proteins had little effect on the anchorage-independent growth of Src- or Ras-transformed cells (Fig. 8a--compare to Fig. 2d). These results show that Crk transformation is particularly sensitive to levels SOS1, and to a lesser extent C3G, and that this sensitivity is not a general property of all transformed cells. In order to further test the role of Abl family proteins on transformation induced by v-Src or v-Ras, soft-agar colony formation assays were done in the presence of imatinib. Inhibition of Abl family kinases did not enhance anchorage-independent growth of v-Src or v-Ras transformed cells, in contrast to CrkI-transformed cells (Fig. 8b--compare to Fig. 7b). Thus inhibition of Abl family kinases has no obvious effect on v-Src- or v-Ras-induced cell transformation. We conclude that both the dependence of transformation on SOS1, and its enhancement by Abl activity, are specific to CrkI-induced transformation.
Fig. 8.
Effect of SH3 binding protein knockdown or Abl inhibition on anchorage-independent growth of v-Ras- or v-Src-transformed cells. NIH-3T3 cells were infected with retrovirus expressing v-Ras or v-Src and subjected to puromycin drug selection. Colonies were counted 2 weeks after plating. Bars represent the averages of colony numbers (normalized to control cell line) from three independent experiments. Error bars represent standard deviations.
(a) Cells expressing v-Ras or v-Src were infected with retrovirus stocks producing shRNAs targeting different Crk SH3 binding proteins. shRNA targeting GFP was used as control. Infected cells were subjected to hygromycin drug selection, and then used in the soft-agar colony formation assay. (b) v-Ras- or v-Src-transformed cells were plated in soft agar with imatinib or vehicle (DMSO).
Discussion
The overall goal of this study was to identify, out of several candidate Crk SH3 binding proteins, those that are critical for transformation by the Crk oncogene. Because the only known biochemical activity of Crk is protein binding, and since the binding activity of its SH2 and SH3 domains is essential for transformation, the association of Crk with one or more endogenous binding partners must be directly responsible for transformation. However, because the Crk SH2 and SH3 domains bind to many cellular proteins, teasing out which interactions are important for a particular biological activity of Crk is a difficult problem. Here we have taken the approach of knocking down the expression of candidate nSH3 binding partners and assaying the effects on various parameters of Crk transformation. We found that Crk transformation was highly dependent on expression of SOS1, while on the other hand decreased abundance or activity of Abl family kinases actually enhanced Crk transformation. Both of these effects were specific for Crk transformation, as they were not seen in cells transformed by the Src or Ras oncogenes.
SOS1 is important for CrkI-induced cell transformation
Our study shows that SOS1 is critical for CrkI-induced cell transformation. Knockdown of SOS1 suppresses the anchorage-independent growth of CrkI-transformed NIH-3T3 cells and their tumorigenicity in nude mice. Previous studies suggested that Ras is required for Crk transformation (Akagi et al., 2002; Greulich and Hanafusa, 1996). As a GEF for Ras, SOS1 is a plausible candidate for linking Ras activation to Crk, and our study provides the first direct evidence that SOS1 is required for CrkI-induced cell transformation. Somewhat surprisingly, the canonical pathway downstream of Ras (Raf/MAP kinase) is not stimulated strongly by CrkI transformation, as we saw no increase in Erk activity in cells overexpressing CrkI (Fig. 6). Thus activation of the Raf/MAPK pathway is unlikely to explain the requirement for Sos1 in Crk transformation. Akagi et al. found that Akt was activated in v-Crk transformed chicken fibroblasts, and that overexpression of H-Ras or SOS1 further enhanced Akt activation in these cells (Akagi et al., 2002), suggesting a role for SOS1 in Akt activation. However, we found that knockdown of SOS1 did not suppress activation of Akt in CrkI-transformed NIH-3T3 cells (Fig. 6). In mouse cells, there are two closely related homologs of Drosophila Sos, SOS1 and SOS2. In our study, shRNA was used to target SOS1, and it is possible that SOS2 can compensate for activating Akt in Crk transformation. In any case, it is clear that the dependence of Crk transformation on SOS1 cannot be fully explained by its role in Akt activation, as we find Akt is unaffected in SOS1 knockout cells that are severely defective in growth rate, anchorage independence, and tumorigenicity. Because SOS1 can act as a GEF for both Ras and Rac (Innocenti et al., 2002), the critical downstream targets of SOS1 in Crk transformation may include effectors for both of these families of small GTPases.
We also tested the effect of SOS1 knockdown on v-Ras- and v-Src-induced transformation. Previous studies showed that transformation of rodent fibroblasts by v-Src is dependent on SOS1 (Qian et al., 2000). Qian et al. generated a sos1−/− cell line by targeted disruption of both alleles of mouse sos1. The transforming activity of v-Src was abolished in sos1−/− cells, but only slightly decreased in sos1+/− cells. The transforming activity of v-Ras was intact in both sos1−/− and sos1+/− cell lines, confirming that transformation by constitutively activated Ras was not dependent on the Ras activator SOS1. In our system, SOS1 knockdown did not significantly affect anchorage-independent growth of v-Ras or v-Src transformed NIH-3T3 cells. It is possible that the decrease in endogenous SOS1 levels achieved by shRNA is insufficient to suppress v-Src transformation. Regardless, our results demonstrate that Crk transformation is more highly dependent on SOS1 than is transformation by other oncogenes tested.
Negative role of Abl family kinases in CrkI-mediated transformation
We found that knockdown of Abl and Arg not only enhanced the anchorage-independent growth of CrkI-transformed cells, but also accelerated the growth of those cells in tissue culture and in a nude mouse tumorigenicity assay. Inhibition of Abl family kinases by imatinib also stimulated colony formation by Crk-transformed cells in soft agar. All these results strongly suggest a surprising negative role for Abl family kinases in suppressing CrkI-induced cell transformation. This is particularly unexpected given the well-known transforming activity of activated Abl variants (Pendergast, 2002), along with reports that Crk can trans-activate Abl in vivo (Hemmeryckx et al., 2001; Shishido et al., 2001).
It has long been appreciated that Abl family kinases can negatively regulate the longer CrkII isoform by phosphorylating Tyr221, located in the linker region between the two SH3 domains of CrkII. Intramolecular Crk SH2-pTyr221 interaction prevents binding of the SH2 and nSH3 domains to other proteins (Feller et al., 1994; Kobashigawa et al., 2007). A number of studies have shown that this mechanism is indeed important for regulating activities such as cell migration and apoptosis (Cipres et al., 2007; Kain and Klemke, 2001), and decreased CrkII phosphorylation was seen in highly invasive carcinoma cells (Kain et al., 2003). Recently, Noren et al. found that in human breast cancer cells, Abl negatively regulates CrkII upon activation of EphB4 receptor tyrosine kinases, leading to induction of apoptosis and inhibition of cell migration and invasion in those cells. Knockdown of Abl by siRNA abolished the tumor-suppressive effects of EphB4 in those breast cancer cells (Noren et al., 2006). These findings received considerable attention because Abl family kinase inhibitors now used to treat cancer have the potential to deregulate endogenous Crk activity.
In contrast to CrkII, however, the CrkI protein used in our studies lacks the linker region containing the regulatory Tyr221 (although the nSH3 domain that binds to Abl kinases is present—see Fig. 1). Thus the ability of Abl to suppress Crk transformation cannot be explained by phosphorylation of the regulatory site. So how do Abl family kinases negatively regulate CrkI-induced cell transformation? One potential explanation would be that Abl induces apoptosis upon binding to CrkI. Previous studies demonstrated that nuclear c-Abl tyrosine kinase is a regulator of apoptosis, and c-Abl-deficient cells show defects in apoptosis (Wang, 2000). However, we found that knockdown of Abl family kinases in CrkI-transformed cells actually stimulated apoptosis (Fig. 4, Fig. S1), making it highly unlikely that apoptosis underlies the transformation-suppressing activity of Abl kinases.
Another possible explanation could be that endogenous CrkII works together with CrkI in cells transformed by overexpression of CrkI. Certainly the two proteins are likely to co-localize, because their SH2 and nSH3 domains are identical and thus can bind the same partners. CrkI overexpression leads to increased steady-state levels of tyrosine-phosphorylated Crk SH2 binding sites (Matsuda et al., 1992), which are likely to recruit and bind endogenous CrkII. We have recently shown that CrkII and CrkI differ in their ability to activate downstream small GTPases: specifically, CrkII is more effective in activating Rac1, whereas CrkI is more effective in activating Rap1 (Antoku and Mayer, 2009). Thus CrkII might cooperate with CrkI in transformation by recruiting and/or activating a distinct set of downstream effectors. Because CrkII can be inhibited effectively by Abl-mediated phosphorylation, such cooperative interactions between CrkI and CrkII would be promoted by Abl downregulation.
The ability of Abl kinases to suppress Crk transformation has important implications for clinical oncology, because small molecule inhibitors targeting Abl family kinases are now being used to treat cancer patients, and others are under development (Druker, 2009). To date the most prominent example is the success of imatinib (Gleevec®) in treating chronic myelogenous leukemia, a disease caused by the activation of Abl via chromosomal translocation. Imatinib has also been used to treat gastrointestinal stromal tumors (GIST). Dasatinib (Sprycel®), a dual inhibitor targeting both Abl and Src family kinases, is used to treat certain types of leukemia, and is currently in clinical trials for solid tumors. Since elevated expression of CrkI has been found in different types of human cancers, and our study showed that knockdown or inhibition of Abl family kinases enhances CrkI-induced cell transformation, the potential for undesirable consequences when using Abl inhibitors to treat human cancers should not be ignored.
C3G is important for some aspects of CrkI-induced cell transformation
Our results show that C3G is important for some aspects of CrkI-induced cell transformation in tissue culture. Knockdown of C3G suppressed the anchorage-independent growth of CrkI-transformed NIH-3T3 cells, although not to the extent seen with knockdown of SOS1 (Fig. 2). We also showed that activation of the downstream JNK pathway in CrkI-transformed cells was dependent on C3G. Finally, we found that C3G knockdown caused the morphological reversion of CrkI-transformed cells, which was not observed for knockdown of other Crk SH3 binding proteins. These results are consistent with previous studies, in which dominant-negative mutants of C3G or other downstream effectors were employed (Mochizuki et al., 2000; Tanaka et al., 1997). However, we found that knockdown of C3G had no apparent effect on the proliferation rate of CrkI-transformed cells in monolayer culture, or on the rate of tumor growth in nude mice, in contrast to the dramatic effects seen upon knockdown of SOS1. Thus we conclude that C3G plays a supporting but limited role in CrkI-mediated transformation.
C3G is a GEF for both Rap1 and R-Ras (Gotoh et al., 1995; Gotoh et al., 1997). Mochizuki et al. found that expression of dominant-negative R-Ras also caused the morphological reversion of v-Crk-transformed cells, leading them to propose that R-Ras is the critical C3G effector for Crk transformation (Mochizuki et al., 2000). However, the limited effects of C3G knockdown on anchorage-independent growth and tumorigenicity seen in our studies are consistent with the notion that cell morphology in monolayer culture correlates imperfectly at best with these more biologically relevant transformation parameters. Furthermore, our results demonstrate that JNK activation is not essential for anchorage independence or tumorigenicity induced by CrkI.
Conclusions
It has long been known that the modular protein binding domains of Crk can bind to many cellular targets, making it difficult to ascribe a particular biological activity (such as transformation) to a specific interaction partner. In principle, a single binding partner could be solely responsible for an activity, or alternatively a biological output might depend on the concerted action of many different binding partners. Our results indicate that for cell transformation by CrkI, the reality lies somewhere between these two extremes. SOS1 is clearly required for most aspects of cell transformation, but other binding partners contribute to the transformed phenotype—for example, C3G is uniquely required for morphological changes. Indeed some binding partners appear to oppose the activity of others: the activity of Abl family kinases suppresses the transforming activity of CrkI, whereas SOS1 and to a lesser extent C3G are required for transformation. It is important to note that our results reveal which binding partners are necessary, but do not address which are sufficient for transformation. Preliminary experiments in which the Crk SH2 domain was forced to interact uniquely with SOS1 in vivo, using the Functional Interaction Trap system (Sharma et al., 2004), suggested that CrkI interaction with SOS1 was not sufficient to induce transformation (data not shown). Furthermore, nSH3 binding proteins not addressed in this study could play important roles in Crk transformation. Thus a variety of experimental approaches will be required to unravel the contributions of each binding partner to the many biological activities of Crk.
Materials and Methods
Antibodies and reagents
Antibody for Abl (8E9) was purchased from BD Pharmingen. Antibody for CrkI/II (clone 22) was purchased from BD Transduction Laboratories. Antibodies for DOCK180 (C19), C3G (C19), SOS1/2 (D21), Abl (K-12), HA (Y11), and actin (I-19) were purchased from Santa Cruz Biotechnology. Antibodies for SAPK/JNK, phospho-SAPK/JNK (Thr183/Tyr185), Akt, phospho-Akt (Ser473), Erk2, phospho-Erk1/2 (Thr202/Tyr204), and cleaved caspase-3 (Asp175) were purchased from Cell Signaling Technology. Antibody for FLAG (M2) was purchased from Sigma-Aldrich. Monoclonal antibody to Arg (Dr. Peter Davies, Albert Einstein College of Medicine) was a generous gift. Imatinib was provided by Novartis Pharmaceuticals and purchased from LC Laboratories. Doxorubicin-HCl (GR-319) was purchased from BIOMOL.
Plasmids
The cDNA encoding human CrkI (Michiyuki Matsuda, Kyoto University) was a generous gift. Human CrkI cDNA was inserted into pMSCV-puro (Clontech). Short hairpin RNAs (shRNA) targeting mouse C3G, DOCK180, SOS1, Abl and Arg were inserted into pSUPER.retro vector (Oligoengine). shRNA targeting GFP was used as control and also inserted into pSUPER.retro vector. The sequences for shRNA knockdown constructs targeting mouse C3G (Wang et al., 2006), DOCK180 (generous gift from Michiyuki Matsuda), SOS1, Abl, Arg, and GFP (Ui-Tei et al., 2004) were: 5'- GGACTTTGATGTTGAATGT-3' (C3G); 5'-GAAGCCATTGTTGAAGGAA-3' (DOCK180); 5'-AGATCGGACCTCTATATCA-3' (SOS1); 5’-GAGTACTTGGAGAAGAAGA-3’(Abl/Arg); 5’-GGAGCCAAATTTCCTATTA-3’ (Arg) and 5’-GCCACAACGTCTATATCAT-3’ (GFP) respectively.
Cell culture, viral infection and soft agar colony formation assay of cells
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Mediatech) containing 10% (v/v) fetal bovine serum (Gemini Bio-products) and 1x penicillin-streptomycin solution (Mediatech). NIH-3T3 cells were maintained in DMEM containing 10% (v/v) bovine serum (Gemini Bio-products) and 1x penicillin-streptomycin solution. For viral production, HEK293T cells were transfected with retroviral vector plus packaging plasmids pMD.env and pMD.gag.pol by the calcium phosphate coprecipitation method with 25 mM chloroquine as described previously (Tanaka et al., 1995), and medium containing virus was harvested at 24 hours post-transfection. NIH-3T3 cells were infected with virus in the presence of 2 µg/ml polybrene (Millipore). NIH-3T3 cells infected with pMSCV-puro derived virus were drug selected with 5.0 µg/ml puromycin (Sigma) for 4 days, recovered for 2 days without drug and kept for further experiments. CrkI-transformed NIH-3T3 cells infected with pSUPER vector-derived viruses were re-plated 72 hours post infection for later experiments. For the soft agar colony formation assay, 1×105 CrkI-transformed NIH-3T3 cells were plated per 6 cm culture dish as a suspension in 4 ml of Iscove’s modified Dulbecco’s medium (Invitrogen) containing 10% fetal bovine serum and 0.4% agar (Becton, Dickinson) on a layer of 4 ml of Iscove’s medium containing 10% calf serum and 0.6% agar. 3 ml of Iscove’s medium containing 10% fetal bovine serum and 0.4% agar was replenished on the top agar layer once a week. Plates were incubated at 37 °C for 4 weeks until colonies formed. For v-Ras or v-Src transformed NIH-3T3 cells, 1×104 cells were plated in soft agar and colonies were counted after 2 weeks. Colonies larger than 0.3 mm in diameter were counted under a microscope. For imatinib treatment, different concentrations (2.5, 5, 10, 15 µM) of imatinib were continuously maintained during the soft agar assay (0.5 ml of medium containing the same concentration of imatinib was added to the top agar layer every 4 days) or in the medium until colonies were counted or the cells were harvested.
In vivo tumor formation assay in nude mice
Different knockdown cell lines were prepared as suspended cells in DMEM without calf serum (1×107 cells/ml). 0.4 ml (4 × 106 cells) of different knockdown cell lines were injected subcutaneously into right flanks of 6-week-old male athymic nude mice (10 mice per cell line). Mice were monitored every other day, measuring weight and tumor size. Tumor volume (TV) was calculated by the formula TV (mm3) = (d2 × D)/2, where d and D are the shortest and longest diameters of the tumor, respectively, measured in millimeters.
Immunoblotting
Cells were lysed with kinase lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% (v/v) Triton X-100, 10% (v/v) glycerol, 1 mM Na3VO4, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 1 mM phenylmethylsufonyl fluoride and 1 µg/ml aprotinin). Cell lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotted onto nitrocellulose membranes (Whatman, Schleicher & Schuell). The membrane was blocked with 5% milk and then incubated with primary antibodies followed by peroxidase-conjugated secondary antibodies. Positive signals were detected by enhanced chemiluminescence reagent (Amersham, GE Healthcare). Band quantification was performed with NIH ImageJ (http://rsbweb.nih.gov/ij/) with phosphoproteins normalized against their respective total proteins.
Cell growth assay
Cell growth was determined using methylthiazolyldiphenyl-tetrazolium bromide (MTT) (Sigma) following the manufacturer’s protocol. 3000 cells in 100 µl media were plated per well in multiple wells (n=12) in 96-well plates and cultured with DMEM containing 10% (v/v) bovine serum and 1x penicillin-streptomycin solution. MTT was added to the cell culture media at 1/10th volume and incubated for 3 h at 37 °C. Isopropanol was added and spectrophotometric absorbance was measured at 570 nm and background at 690 nm using SmartSpec Plus spectrophotometer (Biorad). Experiments were performed at least three times and results are reported as mean ± SD.
Supplementary Material
Acknowledgements
We thank Dr. Michiyuki Matsuda (Kyoto University, Japan) for providing the shRNAs targeting DOCK180 and the antibody against DOCK180, Peter Davies (Albert Einstein College of Medicine, Bronx NY) for Arg antibody, and Novartis (Basel, Switzerland) for imatinib. We thank Kathryn Phoenix and Frank Vumbaca for the technical help. This work was supported by grants CA82258 (to BJM) and CA064436 (to KPC) from the National Institutes of Health.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Akagi T, Murata K, Shishido T, Hanafusa H. v-Crk activates the phosphoinositide 3-kinase/AKT pathway by utilizing focal adhesion kinase and H-Ras. Mol Cell Biol. 2002;22:7015–7023. doi: 10.1128/MCB.22.20.7015-7023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Shishido T, Murata K, Hanafusa H. v-Crk activates the phosphoinositide 3-kinase/AKT pathway in transformation. Proc. Natl Acad. Sci. USA. 2000;97:7290–7295. doi: 10.1073/pnas.140210297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoku S, Mayer BJ. Distinct roles for Crk adaptor isoforms in actin reorganization induced by extracellular signals. J Cell Sci. 2009;122:4228–4238. doi: 10.1242/jcs.054627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge RB, Fajardo JE, Reichman C, Shoelson SE, Songyang Z, Cantley LC, et al. Identification and characterization of a high-affinity interaction between v-Crk and tyrosine-phosphorylated paxillin in CT10-transformed fobroblasts. Mol. Cell. Biol. 1993;13:4648–4656. doi: 10.1128/mcb.13.8.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipres A, Abassi YA, Vuori K. Abl functions as a negative regulator of Met-induced cell motility via phosphorylation of the adapter protein CrkII. Cell Signal. 2007;19:1662–1670. doi: 10.1016/j.cellsig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, et al. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373:607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121 doi: 10.1242/jcs.018044. 2435-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defilippi P, Di Stefano P, Cabodi S. p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol. 2006;16:257–263. doi: 10.1016/j.tcb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Deininger MW, Goldman JM, Lydon N, Melo JV. The tyrosine kinase inhibitor CGP57148B selectively inhibits the growth of BCR-ABL-positive cells. Blood. 1997;90:3691–3698. [PubMed] [Google Scholar]
- Druker BJ. Perspectives on the development of imatinib and the future of cancer research. Nat Med. 2009;15:1149–1152. doi: 10.1038/nm1009-1149. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- Feller SM, Knudsen B, Hanafusa H. c-Abl kinase regulates the protein binding activity of c-Crk. EMBO J. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh T, Hattori S, Nakamura S, Kitayama H, Noda M, Takai Y, et al. Identification of Rap1 as a target for the Crk SH3 domain-binding guanine nucleotide-releasing factor C3G. Mol. Cell. Biol. 1995;15:6746–6753. doi: 10.1128/mcb.15.12.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh T, Niino Y, Tokuda M, Hatase O, Nakamura S, Matsuda M, et al. Activation of R-Ras by Ras-Guanine Nucleotide-releasing Factor. J Biol Chem. 1997;272:18602–18607. doi: 10.1074/jbc.272.30.18602. [DOI] [PubMed] [Google Scholar]
- Greulich H, Hanafusa H. A role for Ras in v-Crk transformation. Cell Growth Differ. 1996;7:1443–14451. [PubMed] [Google Scholar]
- Hasegawa H, Kiyokawa E, Tanaka S, Nagashima K, Gotoh N, Shibuya M, et al. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol Cell Biol. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmeryckx B, van Wijk A, Reichert A, Kaartinen V, de Jong R, Pattengale PK, et al. Crkl enhances leukemogenesis in BCR/ABL P190 transgenic mice. Cancer Res. 2001;61:1398–1405. [PubMed] [Google Scholar]
- Innocenti M, Tenca P, Frittoli E, Faretta M, Tocchetti A, Di Fiore PP, et al. Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J Cell Biol. 2002;56:125–136. doi: 10.1083/jcb.200108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara T, Akagi T, Fujitsuka Y, Hanafusa H. CrkII regulates focal adhesion kinase activation by making a complex with Crk-associated substrate, p130Cas. Proc Natl Acad Sci U S A. 2004;101:17693–17698. doi: 10.1073/pnas.0408413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara T, Akagi T, Shishido T, Hanafusa H. CrkII induces serum response factor activation and cellular transformation through its function in Rho activation. Oncogene. 2003;22:5946–5957. doi: 10.1038/sj.onc.1206633. [DOI] [PubMed] [Google Scholar]
- Kain K, Klemke R. Inihibition of cell migration by Abl family tyrosine kinases through uncoupling of Crk-CAS complexes. Journal of Biological Chemistry. 2001;276:16185–16192. doi: 10.1074/jbc.M100095200. [DOI] [PubMed] [Google Scholar]
- Kain KH, Gooch S, Klemke RL. Cytoplasmic c-Abl provides a molecular 'Rheostat' controlling carcinoma cell survival and invasion. Oncogene. 2003;22:6071–6080. doi: 10.1038/sj.onc.1206930. [DOI] [PubMed] [Google Scholar]
- Kobashigawa Y, Sakai M, Naito M, Yokochi M, Kumeta H, Makino Y, et al. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat Struct Mol Biol. 2007;14:503–510. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- Linghu H, Tsuda M, Makino Y, Sakai M, Watanabe T, Ichihara S, et al. Involvement of adaptor protein Crk in malignant feature of human ovarian cancer cell line MCAS. Oncogene. 2006;25:3547–3556. doi: 10.1038/sj.onc.1209398. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Hashimoto Y, Muroya K, Hasegawa H, Kurata T, Tanaka S, et al. CRK protein binds to two guanine nucleotide-releasing proteins for the Ras family and modulates nerve growth factor-induced activation of Ras in PC12 cells. Mol Cell Biol. 1994;14:5495–5500. doi: 10.1128/mcb.14.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Ota S, Tanimura R, Nakamura H, Matuoka K, Takenawa T, et al. Interaction between the amino-terminal SH3 domain of CRK and its natural target proteins. J Biol Chem. 1996;271:14468–14472. doi: 10.1074/jbc.271.24.14468. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Tanaka S, Nagata S, Kojima A, Kurata T, Shibuya M. Two species of human CRK cDNA encode proteins with distinct biological activities. Mol. Cell. Biol. 1992;12:3482–3489. doi: 10.1128/mcb.12.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ, Hamaguchi M, Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988;332:272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- Mayer BJ, Hanafusa H. Mutagenic analysis of the v-crk oncogene: requirement for SH2 and SH3 domains and correlation between increased cellular phosphotyrosine and transformation. J. Virol. 1990;64:3581–3589. doi: 10.1128/jvi.64.8.3581-3589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Chen G, Gharib TG, Wang H, Thomas DG, Misek DE, et al. Increased C-CRK proto-oncogene expression is associated with an aggressive phenotype in lung adenocarcinomas. Oncogene. 2003;22:7950–7957. doi: 10.1038/sj.onc.1206529. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Ohba Y, Kobayashi S, Otsuka N, Graybiel AM, Tanaka S, et al. Crk activation of JNK via C3G and R-Ras. J Biol Chem. 2000;275:12667–12671. doi: 10.1074/jbc.275.17.12667. [DOI] [PubMed] [Google Scholar]
- Nievers MG, Birge RB, Greulich H, Verkleij AJ, Hanafusa H, van Bergen en Henegouwen PM. v-Crk-induced cell transformation: changes in focal adhesion composition and signaling. J Cell Sci. 1997;110:389–399. doi: 10.1242/jcs.110.3.389. [DOI] [PubMed] [Google Scholar]
- Noren NK, Foos G, Hauser CA, Pasquale EB. The EphB4 receptor suppresses breast cancer cell tumorigenicity through an Abl-Crk pathway. Nat Cell Biol. 2006;8:815–825. doi: 10.1038/ncb1438. [DOI] [PubMed] [Google Scholar]
- Pendergast AM. The Abl family kinases: mechanisms of regulation and signaling. Adv. Cancer Res. 2002;85:51–100. doi: 10.1016/s0065-230x(02)85003-5. [DOI] [PubMed] [Google Scholar]
- Qian X, Esteban L, Vass WC, Upadhyaya C, Papageorge AG, Yienger K, et al. The Sos1 and Sos2 Ras-specific exchange factors: differences in placental expression and signaling properties. EMBO J. 2000;19:642–654. doi: 10.1093/emboj/19.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichman CT, Mayer BJ, Keshav S, Hanafusa H. The product of the cellular crk gene consists primarily of SH2 and SH3 regions. Cell Growth Diff. 1992;3:451–460. [PubMed] [Google Scholar]
- Ren R, Ye Z-S, Baltimore D. Abl protein-tyrosine kinase selects the Crk adapter as a substrate using SH3-binding sites. Genes & Dev. 1994;8:783–795. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- Riggins RB, DeBerry RM, Toosarvandani MD, Bouton AH. Src-dependent association of Cas and p85 phosphatidylinositol 3'-kinase in v-crk-transformed cells. Mol Cancer Res. 2003;1:428–437. [PubMed] [Google Scholar]
- Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, et al. A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 1994a;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Nishida J, et al. Characterization, partial purification, and peptide sequencing of p130, the main phosphoprotein associated with v-Crk oncoprotein. J Biol Chem. 1994b;269:32740–32746. [PubMed] [Google Scholar]
- Senechal K, Heaney C, Druker B, Sawyers CL. Structural requirement for function of the Crkl adapter protein in firbroblasts and hematopoietic cells. Mol. Cell. Biol. 1998;18:5082–5090. doi: 10.1128/mcb.18.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Antoku S, Mayer BJ. The Functional Interaction Trap: A novel strategy to study specific protein-protein interactions. In: Kamp RM, Calvete J, Choli-Papadopoulou T, editors. Methods in Proteome and Protein Analysis (MPSA 2002) Berlin: Springer-Verlag; 2004. pp. 165–181. [Google Scholar]
- Shishido T, Akagi T, Chalmers A, Maeda M, Terada T, Georgescu MM, et al. Crk family adaptor proteins trans-activate c-Abl kinase. Genes Cells. 2001;6:431–440. doi: 10.1046/j.1365-2443.2001.00431.x. [DOI] [PubMed] [Google Scholar]
- Stam JC, Geerts WJ, Versteeg HH, Verkleij AJ, van Bergen en Henegouwen PM. The v-Crk oncogene enhances cell survival and induces activation of protein kinase B/Akt. J Biol Chem. 2001;276:25176–25183. doi: 10.1074/jbc.M009825200. [DOI] [PubMed] [Google Scholar]
- Takino T, Nakada M, Miyamori H, Yamashita J, Yamada KM, Sato H. CrkI adapter protein modulates cell migration and invasion in glioblastoma. Cancer Res. 2003;63:2335–2337. [PubMed] [Google Scholar]
- Tanaka M, Gupta R, Mayer BJ. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol. Cell. Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, et al. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc. Natl. Acad. Sci. U.S.A. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Ouchi T, Hanafusa H. Downstream of Crk adaptor signaling pathway: Activation of Jun kinase by v-Crk throught the guanine nucleotide exchange protein C3G. Proc. Natl. Acad. Sci. USA. 1997;94:2356–2361. doi: 10.1073/pnas.94.6.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hoeve J, Morris C, Heisterkamp N, Groffen J. Isolation and chromosomal localization of CRKL, a human crk-like gene. Oncogene. 1993;8:2469–2474. [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Mysliwiec T, Feller SM, Knudsen B, Hanafusa H, Kruh GD. Proline-rich sequences mediate the interaction of the Arg protein tyrosine kinase with Crk. Oncogene. 1996;13:1379–1385. [PubMed] [Google Scholar]
- Wang JY. Regulation of cell death by the Abl tyrosine kinase. Oncogene. 2000;19:5643–5650. doi: 10.1038/sj.onc.1203878. [DOI] [PubMed] [Google Scholar]
- Wang L, Tabu K, Kimura T, Tsuda M, Linghu H, Tanino M, et al. Signaling adaptor protein Crk is indispensable for malignant feature of glioblastoma cell line KMG4. Biochem Biophys Res Commun. 2007;362:976–981. doi: 10.1016/j.bbrc.2007.08.106. [DOI] [PubMed] [Google Scholar]
- Wang Z, Dillon TJ, Pokala V, Mishra S, Labudda K, Hunter B, et al. Rap1-mediated activation of extracellular signal-regulated kinases by cyclic AMP is dependent on the mode of Rap1 activation. Mol Cell Biol. 2006;26:2130–2145. doi: 10.1128/MCB.26.6.2130-2145.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.