Abstract
Notch receptors participate in a highly conserved signalling pathway that regulates normal development and tissue homeostasis in a context- and dose-dependent manner. Deregulated Notch signalling has been implicated in many diseases, but the clearest example of a pathogenic role is found in T cell lymphoblastic leukaemia/lymphoma (T-LL), in which the majority of human and murine tumours have acquired mutations that lead to aberrant increases in Notch1 signalling. Remarkably, it appears that the selective pressure for Notch mutations is virtually unique among cancers to T-LL, presumably reflecting a special context-dependent role for Notch in normal T cell progenitors. Nevertheless, there are some recent reports suggesting that Notch signalling has subtle yet important roles in other forms of hematologic malignancy as well. Here, we review the role of Notch signalling in various blood cancers, focusing on T-LL with an eye toward targeted therapeutics.
Keywords: Notch signaling, oncogene, T-cell lymphoblastic leukemia/lymphoma, chromosomal rearrangements, targeted therapy
Notch Signalling
Notch receptors are single-pass transmembrane proteins that participate in a highly conserved signalling pathway that regulates many aspects of cellular differentiation and tissue homeostasis in metazoans. Mammals have four Notch receptors (Notch1-4); this review will largely focus on Notch1, the mammalian family member with the greatest homology to Drosophila Notch, the founding member of the family. The best characterized Notch ligands belong to two families, the Delta-like ligands and the Jagged/Serrate-like ligands. Mammals have three Delta-like ligands (DLL1, DLL3, and DLL4), and two Jagged ligands, (Jagged1 and Jagged2), all of which are also single-pass transmembrane proteins.
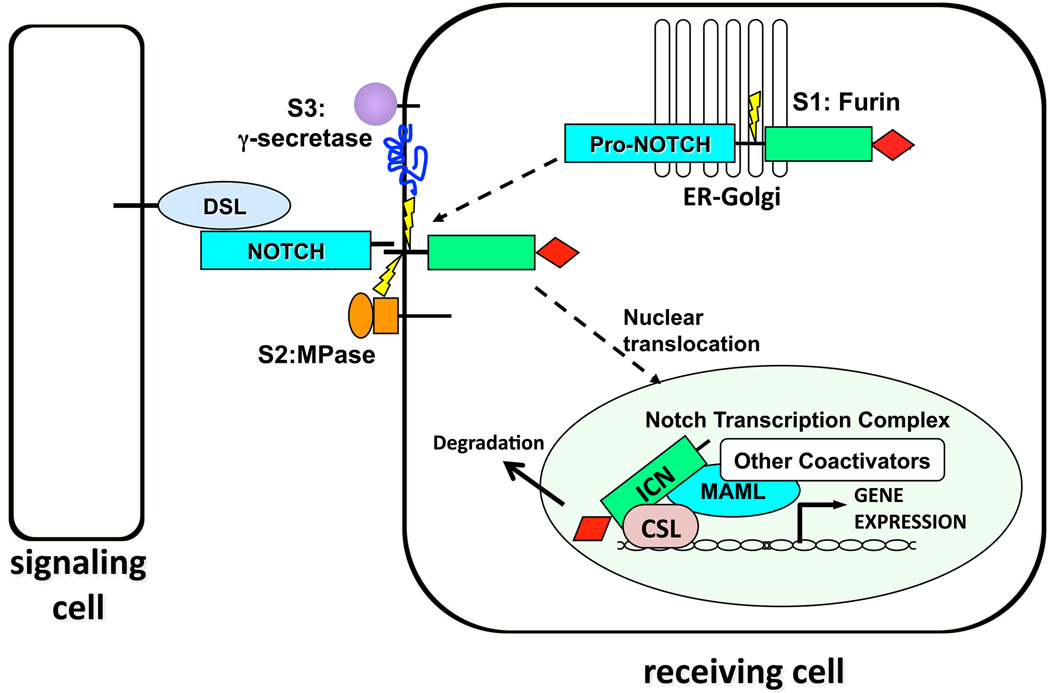
Signal transduction by Notch receptors relies on a series of proteolytic cleavages (Figure 1; for recent review, see ref. [1]). The extracellular domain of Notch1 consists of 36 N-terminal EGF repeats followed by a juxtamembrane negative regulatory region (NRR) consisting of three Notch-Lin12 repeats and a heterodimerization (HD) domain [2, 3]. During trafficking to the cell surface, Notch1 is cleaved by a furin-like protease at a site termed S1 in an unstructured loop within the HD domain [4]. As a result, mature Notch1 receptors are heterodimers made up of non-covalently associated extracellular (NEC) and transmembrane subunits (N™) that are held together by the intrinsic stability of the HD domain [2, 3, 5].
Figure 1.
Canonical Notch Signalling. Notch receptors are processed in the ER/Golgi compartment by furin-like proteases at site S1 during trafficking to the cell surface, producing heterodimeric surface receptors. Engagement of Notch receptors with ligands of the DSL (Delta, Serrate, Lag-2) family expressed on neighbouring cells initiates two successive cleavages by the metalloproteinase ADAM10 at site S2, followed by intramembranous cleavage at site S3 by the mulitsubunit protease γ-secretase. This permits the intracellular domain of Notch (ICN) to translocate to the nucleus and form a Notch transcription complex (NTC) consisting of the DNA-binding protein CSL, ICN, and co-activators of the Mastermind-like (MAML) family. This NTC is normally short-lived due to the presence of a C-terminal PEST domain in ICN (shown as a red diamond).
Signalling is initiated when a Notch receptor on one cell binds to a Notch ligand expressed on a neighbouring cell. This sets in motion events that lead to the cleavage of Notch by two additional proteases. The first cleavage is carried out preferentially by the metalloproteinase ADAM10 at site S2 [6, 7], which lies about 12–14 amino acids external to the transmembrane domain of the N™ subunit. In the resting state, the S2 site is deeply buried within the NRR [2, 8], indicating that a substantial conformational change must precede S2 cleavage. S2 cleavage requires the endocytosis of ligand into the ligand-expressing cell, leading to speculation that mechanical force transmitted to the NRR via endocytosis are responsible for the intramolecular movements that precede S2 cleavage; however, direct evidence for this model is lacking. Cleavage of N™ at S2 in turn creates a short-lived transmembrane intermediate that is cleaved within its transmembrane domain by γ-secretase, a multisubunit protease consisting of presenilin 1 or 2, PEN-2, APH-1, and nicastrin. The ultimate cleavage at site S3 frees the intracellular domain of Notch (ICN) from the membrane, allowing it to translocate to the nucleus. Whether γ-secretase cleavage occurs at the cell surface or following endocytosis within endocytic vesicles remains controversial (as discussed in ref. [9]).
ICN contains several protein-protein interaction domains, including seven iterated ankyrin repeats (ANKs) that are needed for all known Notch functions and a so-called RAM domain. Nuclear ICN associates with the DNA-binding factor CSL (for CBF1, Suppressor of Hairless, Lag-2; also known as RBP-Jκ) through high affinity RAM contacts and lower affinity ANK contacts [10, 11]. Binding of ANK to CSL creates an extended, composite surface groove that recruits scaffold proteins of the Mastermind-like (MAML) family, the third component of the core Notch transcription complex (NTC). The NTC in turn interacts with chromatin modifying factors such as the histone acetyltransferases p300 and pCAF [12–14] as well as components of the mediator complex [15] to transactivate target genes with CSL-binding sites. In the absence of ICN, CSL can associate with multiple proteins that suppress transcription, including multiple complexes with histone deacetylase activity [16–21] and other factors with histone demethylase activity [22]. For this reason, CSL has been likened to a molecular switch capable of actively suppressing or stimulating transcription, depending on the Notch activation status of a cell. Normal Notch transcription complexes are believed to have a half-life on the order of minutes [12, 15], in part due to the presence of a PEST degron domain in the C-terminus of ICN that marks activated Notch for ubiquitinylation and proteasomal degradation.
Several aspects of the canonical Notch signalling pathway warrant emphasis. Because the pathway relies on protein-protein interactions and the NTC is short-lived, a single activated Notch receptor is likely to transactivate only one target gene for a short period of time (on the order of minutes), a “design” that enables very precise temporal and quantitative control. It follows that subtle perturbations in Notch signalling may have significant phenotypic consequences. In line with this idea, haploinsufficiency of Notch pathway signalling components is associated with several human diseases. For example, haploinsufficiency of Notch1 is associated with bicuspid aortic valve deformities [23], and haploinsufficiency of Jagged1 or Notch2 with Alagille syndrome [24–26], which is characterized by developmental abnormalities of a number of organs, particularly the liver and heart.
Notch1 as an oncogene in T-LL
Notch1 was discovered in 1991 through analysis of rare T cell lymphoblastic leukaemias/lymphomas (T-LLs) with balanced (7;9) translocations [27]. The translocation breakpoints were shown to fall within NOTCH1 on chromosome 9 and the T cell receptor β (TCRβ) locus on chromosome 7 and to result in the fusion the 3’ end of NOTCH1 to TCRβ enhancer/promoter elements, which drive the expression of aberrant NOTCH1 transcripts encoding truncated, constitutively nuclear Notch1 polypeptides. Expression of similar forms of Notch1 in murine haematopoietic stem cells (HSCs) induces the rapid appearance of T-LL [28, 29], whereas no tumours are observed when the same polypeptides are expressed in HSCs with genetic defects that abrogate T-cell development [30, 31]. Together, these studies revealed that Notch1 had a special oncotropism for T cell progenitors. Work focused on the role of Notch1 in T cell progenitors elaborated on this relationship by showing that Notch1 is both necessary [32] and sufficient [33] for T cell development from HSCs, indicating that the T cell oncotropicity of Notch1 is likely a reflection of the normal role of Notch1 in T cell progenitors.
The degree of Notch1 involvement in human T-LL became apparent in 2004 when we identified NOTCH1 gain-of-function mutations in roughly 60% of primary human T-LLs [34], findings that have been confirmed repeatedly in subsequent series from other groups [35–41]. Most Notch1 mutations in human T-LL fall into two regions, the HD domain and the C-terminal PEST domain (for review, see [42]). Remarkably, individual tumours may harbour as many as 3 NOTCH1 mutations, typically aligned in cis in a single allele. Subsequent studies of murine T-LLs arising in many different genetic backgrounds also revealed the presence of acquired gain-of-function Notch1 mutations at frequencies varying from 30% to 80%, depending on the genetic model [43–49]. Together, these observations moved Notch1 to the centre of T-LL pathogenesis, and set the stage for more recent work focused on the mechanisms by which Notch1 mutations cause gains in function, the downstream consequences of oncogenic Notch1 signalling, and therapeutic targeting of Notch1 in T-LL.
Of Mice and Men: Oncogenic Notch1 Mutations in T-LL
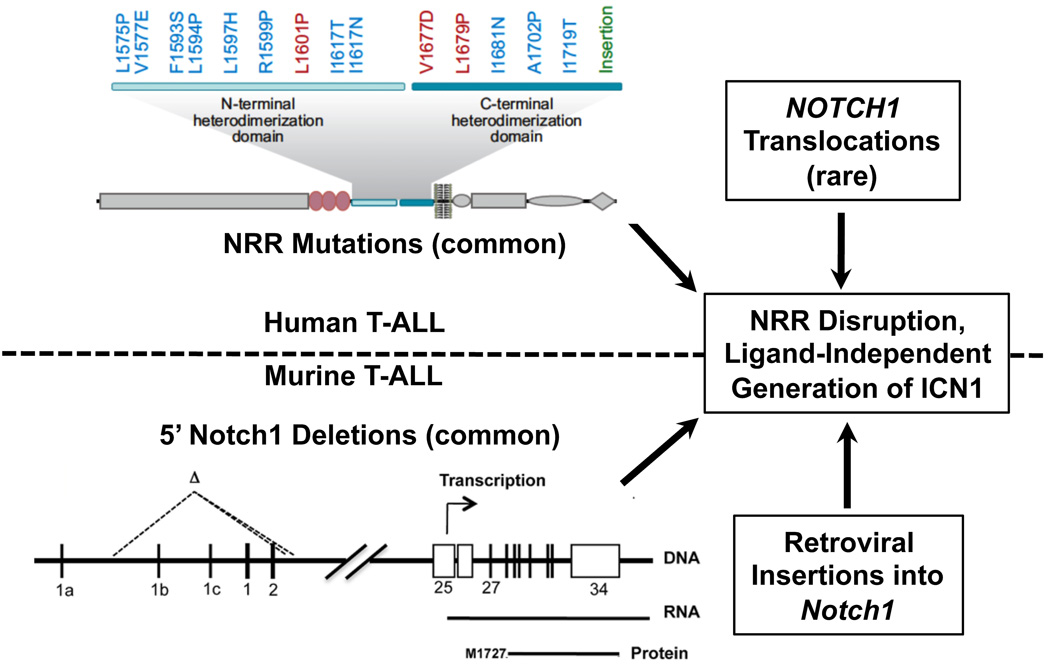
The most common Notch1 mutations in human T-LL consist of point substitutions or small in-frame insertions or deletions that cluster within the hydrophobic core of the HD domain, destabilizing it sufficiently to permit ligand-independent metalloproteinase cleavage at site S2 [50]. The degree of destabilization varies among mutations, such that some cause spontaneous dissociation of Notch1 heterodimers, whereas others do not [50]. An uncommon, second type of mutation consisting of juxtamembrane insertions near the C-terminal end of the HD domain appear to create new, “deprotected” S2 cleavage sites, and also result in ligand-independent proteolysis and activation of Notch1 [50, 51]. Rare mutations in the LNR portion of the NRR have also been described [8]. Given the strong selection for Notch1 gain-of-function in T-LL and the ability of leukemic blasts to proliferate in many tissues where access to ligand is likely limited, it is probable that other mechanisms of ligand-independent Notch1 activation remain to be discovered in human T-LL. This is particularly true of tumours that only have PEST deletions, as such mutations have little or no effect on Notch1 signalling [34, 52] in the absence of some mechanism (e.g., an NRR mutation) that promotes Notch1 proteolysis.
Murine T-LLs initially challenged the idea that lesions in the extracellular domain of Notch1 are required for leukaemogenic increases in Notch1 signalling. In mouse tumours, PEST domain mutations are very common in T-LLs arising in diverse genetic backgrounds, such as loss of E2A [47], p53 [45], or Ikaros [44]; mis-expression of transcription factors such Tal1 [45]; and conditionally active Ras signalling [52, 53]. Yet HD domain mutations are found in only a small minority of murine T-LLs, leaving it unclear how Notch1 activation is initiated in these tumours.
It is now apparent that genetic lesions that disrupt the NRR of Notch1 are the rule rather than the exception in murine T-LL (Figure 2). The first clue to this came from retroviral mutagenesis studies [54–57], which revealed that portions of Notch1 within or just 5’ of the region encoding the NRR are common sites of proviral insertions in murine T-LL. A second important insight came from the work of Tsuji, et al., who noted that T-LLs arising in SCID mice or ATM−/− mice following irradiation had frequent 5’ deletions in Notch1 [43, 58]. In a subset of tumours analyzed by Tsuji, et al., the breakpoints giving rise to the deletions were located immediately adjacent to sequences homologous to RAG recombinase signal sequences (RSSs) and created DNA joins with all of the structural features expected of RAG-mediated rearrangements [59]. The clustering of deletions around these ectopic RSSs permitted Tsuji, et al. to develop a sensitive PCR-based assay that detected identical deletions at low frequencies in normal thymocytes, suggesting that this apparent error in V(D)J recombination occurs at a appreciable background frequency.
Figure 2.
Mechanisms of ligand-independent Notch1 activation in T-LL. In human T-LL, the Notch negative regulatory region (NRR) is often disrupted by point mutations or less frequently by chromosomal translocations, whereas in murine T-LL the NRR is most commonly disrupted are 5’ deletions (most RAG mediated) followed by retroviral insertions. Examples of dissociative (red), destabilizing (blue), and insertional (green) Notch1 NRR mutations found in human T-LL are listed.
More recently, we have “rediscovered” the 5’ deletions in Notch1 in a high fraction of murine tumours arising in diverse T-LL-prone genetic backgrounds [60, 61] (Figure 2). The most common deletions appear to be RAG-mediated and are identical to some of the deletions described by Tsuji, et al. These deletions uniformly remove the proximal promoter elements and exon 1 of Notch1, yet paradoxically activate a cryptic internal promoter that lies within exon 25 that drives the expression of 5’ deleted Notch1 transcripts. Of note, ChIP-Seq analysis has shown that RAG2 associates with the proximal promoter of the Notch1 gene in normal thymocytes [60], possibly through binding to lysine 4-trimethylated histone H3 (H3K4-me3) [62], providing a plausible mechanism for recruitment of RAG recombinase to chromatin near the ectopic RSSs in Notch1. Less often, murine T-LLs bear interstitial Notch1 deletions that remove the region from exon 2 to exon 26-exon28. These deletions appear to be RAG-independent and may stem from random DNA breakage and non-homologous DNA end joining.
Although the structure of the aberrant transcripts expressed by cells with RAG-mediated and RAG-independent deletions is different, the functional consequences of both appear to be identical. Specifically, translation of both types of aberrant transcripts initiates at M1727, a conserved methionine residue that lies within the transmembrane domain of Notch1 [60]. The hydrophobic amino-terminus of the encoded polypeptide is apparently sufficient for membrane insertion, as signalling in cells expressing the aberrant transcripts is strictly γ-secretase dependent, in line with observations showing that murine T-LLs are highly sensitive to γ-secretase inhibitors.
In parallel studies, Jeannet, et al., observed that Cre-mediated deletion of exon 1 of Notch1 paradoxically accelerated development of T-ALL in mice expressing a hypomorphic form of Ikaros [61], a transcriptional repressor and master regulator of lymphoid development that associates with histone deacetylase complexes [63, 64]. These deletions also activate the same cryptic promoter in exon 25 of Notch1 that was responsible for generating aberrant transcripts in tumours with RAG-mediated deletions. In yeast, transcriptional elongation induces epigenetic changes that inhibit internal transcriptional initiation [65]; assuming this principle applies to mammalian cells, deletion of the proximal promoter and exon 1 of Notch1 would be expected to contribute to activation of the cryptic 3’ promoter in exon 25. However, such deletions are insufficient to explain internal promoter activation, as Cre-mediated deletion of the same floxed Notch1 allele in a wild type background produces loss of Notch1 function in several tissues, including the thymus [32]. Given the results of Jeannet, et al., Ikaros may be an additional factor that suppresses transcriptional initiation from the 3’ end of Notch1.
These observations have reinforced the notion that a large proportion of both human and murine T-ALLs have genetic lesions that lead to ligand-independent Notch1 activation, albeit through different mechanisms (Figure 2). Since there is no reason to think that leukaemogenic NRR mutations analogous to those found in human disease could not occur in the mouse, one must posit instead that 5’ Notch1 deletions occur at a much higher frequency in the mouse than in man. This may be because the ectopic RSSs in the murine Notch1 locus are not conserved in human NOTCH1. We speculate that these RSS sites underlie the high frequency of T-LL in the mouse; while T-LL is a rare disease in man (occurring in roughly 600 individuals per year in the USA), it is one of the most common tumours arising in genetically engineered mouse models.
In contrast to the extracellular mutations, intracellular Notch1 mutations in human and murine T-LL are quite similar. Both consist of frameshift or stop codon mutations that remove the C-terminal PEST degron, a conserved negative regulatory domain. In human T-LL, these deletions inevitably occur in cis to NRR mutations when both mutations are present, whereas in the mouse they are usually found in cis to 5’ Notch1 deletions [60]. Several short motifs in this region have been implicated in proteasomal degradation of ICN1, including a short sequence that can be recognized by the E3 ligase Fbw7 [15, 66–69], and the motif WSSSSP, which promotes turnover of ICN1 through uncertain mechanisms [70]. Overall, PEST deletions are more frequent in murine T-LL than human T-LL for reasons that are unclear. One possibility is that mutations in other components of the degradation machinery may be more common in man, which would reduce the selective pressure for Notch1 mutations. Fbw7 loss-of-function mutations, present in roughly 10% to 30% of human T-LLs [35, 37, 38, 71, 74], represent one such potential complementary mutation. Alternatively, a higher fraction of human T-ALL could arise through Notch-independent mechanisms. A full accounting of the true incidence of Notch-associated T-ALL in mouse and man will be necessary before this conclusion can be reached.
Notch1 Targets in T-LL
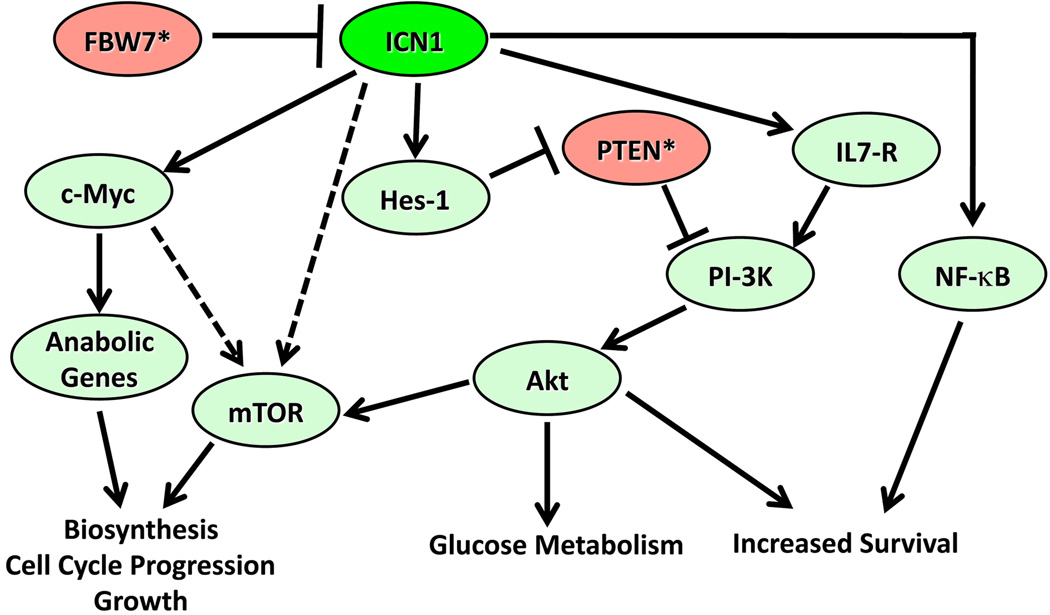
In the context of transformed T cell progenitors, Notch signalling induces and reinforces a program of gene expression that supports cell growth (Figure 3). The most important direct target genes include c-Myc [72–75] and Hes-1 [72, 73, 76, 77], a transcriptional repressor of the bHLH family. Key pathways activated by Notch1 include the PI3-kinase/Akt [78–80] and mTOR [81, 82]. Crosstalk between Notch and these pathways is incompletely understood and probably occurs at several levels. Notch1 signalling via Hes-1, down-regulates PTEN [78], an important negative regulator of PI3-kinase/Akt signalling. Notch1 also upregulates the expression of the IL7 receptor [83], which activates PI3K/Akt signalling in an IL7-dependent fashion. In ex vivo culture systems the growth of primary human T-LL cells is IL7-dependent [84, 85], suggesting that the IL7/IL7-receptor signalling axis is an important mediator of growth. It should be noted that the in vivo significance of both the Notch:Hes:PTEN and the Notch:IL7R axes in T-LL await further confirmation.
Figure 3.
Oncogenic Notch1 Signalling. Signalling pathways downstream of activated Notch1 (ICN1) in T-LL cells are shown. Fbw7 and PTEN are tumour suppressors (*) whose function is often lost, events that increase the consequences of ICN1 signalling. In some tumours, there is evidence that ICN1 stimulates mTOR independently of effects on the PI3K/Akt pathway; this uncharacterized pathway is shown as a dashed line.
The oncotropism of Notch1 for T cell progenitors undoubtedly reflects the normal role of Notch1 in developing T cells, in which Notch1 activates many of the same pathways that contribute to the growth of T-LL cells (for review, see [86]). Notch1 first acts on multipotent progenitors to induce commitment to T cell fate, which within the thymus appears to stem from Notch1 activation by the ligand Delta-like 4 [87]. Later in T cell development, Notch1 is required for progression of cells through β-selection [88–90], a stage when cells that successfully rearrange a TCRβ gene express the pre-T-cell receptor, which generates essential survival signals. Expression of c-Myc closely parallels Notch1 signal strength and peaks during β-selection [72], and constitutively active Akt can rescue, in part, T cell development in the absence of Notch1 [79]; thus, at least some of the pathways implicated in Notch-associated T-LL are also regulated by Notch1 in normal thymocytes.
Notch-related T-LL is also associated with constitutive activation of NF-κB [91, 92] and abnormalities of E2A [47], but the mechanistic bases for these relationships are not fully elucidated. Notch may augment NF-κB activity in T-LL through transactivation of NF-κB pathway genes as well as through direct physical interaction with regulators of the pathway [92]. Other recent data suggest that MAML1 directly co-activates the NF-κB subunit RelA (p65) and promotes the degradation of an inhibitor of NF-κB, IκBα [93], providing an additional possible link between Notch signalling and NF-κB. The relationship between Notch and E2A is even more complex. On the one hand, E2A appears to be important in positive regulator of Notch1 expression in thymocytes and T-LL cells [94], but at the same time Notch1 has been implicated in induction of proteasomal degradation of E2A [95], and Notch1 mutations are common in murine T-LLs arising in E2A deficient backgrounds [47]. One resolution to this apparent discrepancy would be for Notch1 expression in E2A deficient T-LLs to be coming from 5’-deleted Notch1 alleles, which may not require E2A for expression of internally initiated Notch1 transcripts.
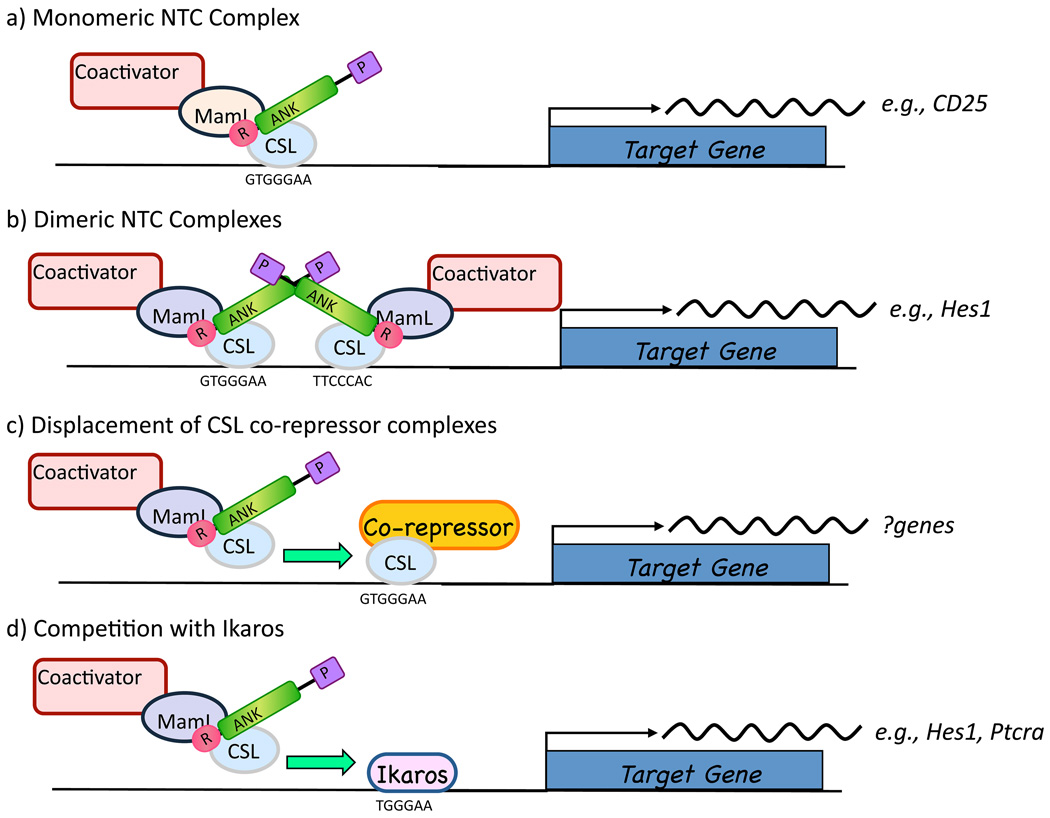
How activated Notch1 triggers transcription of target genes in T-LL cells and other cell types is incompletely understood (Figure 4). Some of the best-characterized direct targets of Notch1 such as Hes-1 have special response elements within their promoters that consist of a pair of head-to-head CSL binding sites separated by spacers of 15–19 base pairs [96]. Activation of promoters containing such elements, termed sequence paired sites or SPSs, requires loading of NTCs onto both CSL sites. Dimeric NTCs associate through weak contacts involving the ANK domain and are stabilized by MAML binding to CSL/ICN binary complexes [97, 98]. Identification of likely paired sites by bioinformatic approaches has been complicated recently by recognition that a high affinity CSL binding site may be flanked by a sequence with no significant homology to the CSL monomer consensus binding sequence, but which nevertheless is competent for NTC binding and dimer formation [99]. HES5 is one long-recognized Notch target gene that has this type of “cryptic” paired site in its proximal promoter element.
Figure 4.
Notch transcription complexes. Evidence suggests that NTC complexes stimulate transcription on various promoters a) as monomers, b) as dimers, c) by competing co-repressor complexes off of CSL, or d) by competing with Ikaros, a zinc-finger protein and transcriptional repressor, for genomic binding sites in Notch responsive genes.
Promoters with paired sites are completely dependent on NTC dimerization for transactivation [98], whereas promoters with other arrangements of CSL binding sites (e.g., multimerized head-to-tail sites or single binding sites) show no such dependency. It is not clear what dictates these two diametrically opposed patterns of response; it is possible that binding of other factors (e.g., E2A) to flanking sites functionally complements monomeric NTCs. It will be of interest to determine the full complement of genes that depend on NTC dimerization, and whether these genes play a role in the induction or maintenance of T-LL.
Another unsettled issue is whether CSL binds stably to DNA and successively interacts with repressive and activating complexes, or whether complete complexes are assembled prior to loading onto DNA. In either case, the factors that control whether CSL associates with repressive or activating complexes on any particular CSL-binding site in the genome are unknown, as are the dynamics of complex exchange. As already mentioned, it has also been suggested that Ikaros, a key regulator of lymphoid development, competes directly with CSL for genomic binding sites [100–103], providing another level of control of Notch target gene expression that may go awry in T-ALL. Ikaros is a frequent target of inactivating mutations in murine T-LL, for example by retroviral insertion [100, 104], but inactivating Ikaros mutations appear to be rare in human T-LL [105], representing yet another apparent distinction between T-LL in mice and men.
Do other Notch receptors contribute to T-LL?
In addition to Notch1, developing thymocytes also express Notch3, which appears to be a downstream target of Notch1 [72, 73]. Transgenic mice expressing ICN3 develop T-LL [31, 91], but the relative importance of Notch3 as compared to Notch1 has been uncertain. Recently, Jeannet, et al. observed that Cre-mediated deletion of Notch3 had no effect on development of T-LL in a hypomorphic Ikaros mutant background [61]. T-LLs arising in this background had acquired Notch1 mutations and were sensitive to γ-secretase inhibition. However, Screpanti and co-workers have shown that ICN3 can promote the appearance of splice variants that encode dominant negative forms of Ikaros [106]; if this is the primary contribution of Notch3 to T-ALL development, it would not be selected for in the hypomorphic Ikaros background used by Jeannet et al. Thus, an ancillary role for Notch3 in T-LLs remains a possibility. In addition, Notch2 is activated by feline leukaemia virus in some T-LLs in the cat [107], but has not been implicated in T-LL in other species.
Is Notch a Useful Biomarker in T-LL?
Multiple clinical series worldwide have confirmed that Notch1 mutations in T-LL are frequent in all genetic and clinical subtypes of human T-LL, leading investigators to ask if mutational status is a useful biomarker. However, associations between mutation status and outcome have been inconsistent (summarized in Table 1). While a few series have suggested that Notch1 mutations are associated with worse outcomes, most have shown no association or a trend towards more favourable responses. Although Notch signalling has been linked to resistance to glucocorticoids in studies of T-LL cell lines [108], no such association has been found in primary tumours; in fact, in some series the trend is towards better responses to glucocorticoids among tumours with Notch1 mutations [36]. Another potential relationship under investigation is that between Notch activation and involvement of the central nervous system. This is based on murine models showing that Notch1-induced murine T-LLs express CCR7, a chemokine receptor that is associated with spread of T-LL to the meninges [109].
Table 1.
Notch1/Fbw7 Mutational Status and Clinical Outcome in T-LL
| Reference | Patient Group | Notch1 Mutations |
Fbw7 Mutations |
Wild Type |
Association with Outcome |
|---|---|---|---|---|---|
| [34] | Children (n = 96) | 56% | N.A. | 44% | None |
| [35] | Adults (n = 141) | 62% | 24% | 28% | Positive (better event free survival and overall survival) |
| [36] | Children (n = 157) | 52% | N.A. | 48% | Positive (better early treatment response and long-term outcome) |
| [37] | Children (n = 47) | 34% | 11% | 62% | None |
| [38] | Adults (n = 88) | 60% | 18% | 34% | None |
| [39] | Children (n = 69) | 33% | 16% | 42% | Favourable (better event free survival and overall survival) |
| [130] | Children (n = 70) | 57% | N.A. | 43% | None |
| [41] | Children and adults (n = 77) | 38% | N.A. | 62% | Unfavourable (higher blast counts in the peripheral blood, worse overall survival) |
| [71] | Children (n = 26) | 31% | 31% | 54% | Favourable (better overall survival) |
Part of the difficulty of linking Notch1 signalling to various clinical correlates is that there remains no straightforward way to quantify Notch1 signal strength. Ideally, levels of ICN1 would be measured directly using immunohistochemistry or flow cytometry, but development of reliable methods to do so has proved daunting. In lieu of such methods, testing for mutations in NOTCH1 and Notch1 regulatory genes such as FBW7 will remain the method of choice, albeit one that is less than ideal in terms of stratification of T-LLs into “Notch-on” and “Notch-off” groups. Beyond these technical limitations, there remain questions about the biological existence of distinct “Notch-on” and “Notch-off” groups. It is difficult, for example, to identify tumours with and without Notch1 mutations using unsupervised clustering of gene expression profiles. Given this, it is perhaps not surprising that associations between Notch1 mutation status and clinical outcomes are subtle at best.
Notch signalling in other hematologic malignancies: Is there a role?
Multiple reports have raised the possibility of involvement of Notch signalling in a wide spectrum of haematological malignancies, but to date a “smoking gun” analogous to the gain-of-function mutations identified in human T-LL is lacking. A small subset of tumours classified as acute myeloid leukemias (AMLs) have Notch1 gain-of-function mutations [110, 111], but many of these tumours express T cell markers and may be better considered very immature forms of T-LL. Another rare form of AML associated with megakaryocytic differentiation and a 1;22 translocation that produces a OTT-MAL (also termed Rbm15-MKL1) fusion protein has also been hypothesized to transform in part through upregulation of CSL-dependent genes via a novel mechanism [112, 113]. Specifically, Mercher, et al., proposed that the OTT-MAL fusion protein competes with OTT for binding to CSL[113]. OTT, a member of the SPEN family of proteins, functions as a co-repressor, whereas the OTT-MAL fusion protein is a co-activator that upregulates transcription from promoters containing CSL binding sites. Of further interest, this same group has shown that exaggerated Notch signalling can skew myeloid development towards megakaryocytic fate [114], suggesting that the selection for increased expression of CSL-regulated genes in AML with megakaryocytic differentiation may reflect a physiologic role for the same set of target genes during megakaryocytic development. However, most of the data supporting a role for Notch in acute myeloid leukaemia with megakaryocytic differentiation has come from studies in mice and a very limited number of human cell lines, and further work is needed to firmly establish that upregulation of CSL target genes is a general phenomenon in human AMLs associated with the t(1;22).
Aberrant Notch signalling has also been proposed to contribute to a number of lymphoid malignancies other than T-LL. It has been suggested that part of the global epigenetic reprogramming that is the hallmark of the Reed-Sternberg cells of Hodgkin lymphoma might be mediated by Notch1 activation [115], but it is not clearly established whether upregulation of Notch1 expression is causative or merely a marker of these changes. Expression of the Notch ligand Jagged1 has been reported to enhance the growth of the malignant plasma cells of multiple myeloma [116], but the in vivo significance of this observation remains uncertain. Based on the requirement for Notch2 signalling for marginal zone B lymphocyte development in the mouse [117], it has been suggested that Notch2 might serve as an oncogene in marginal zone B cell lymphomas in man, but data to support this idea have not been forthcoming. A single report of Notch gain-of-function mutations in a small subset of diffuse large B cell lymphomas in a Japanese cohort of patients [118] has yet to be confirmed by others. Similarly, several reports suggesting that deregulated Notch signalling is commonplace in chronic lymphocytic leukemia/small lymphocytic lymphoma [119–122] remain to be corroborated.
At the end of the day, in may be that the most important roles for Notch signalling in cancer will be non-cell-autonomous. Notch is integrally important in regulating angiogenesis (for review, see [123]) (which is coordinated in large part by the interplay of the Notch and VEGF signalling pathways) and both the adaptive and innate arms of the immune system (for review, see [124]), through which it is likely to have an important impact on tumour immunity.
Therapeutic opportunities
The unique circuitry of the Notch signalling pathway makes it an attractive therapeutic target. When administered chronically, gamma-secretase inhibitors cause dose-limiting goblet cell hyperplasia of the gut, as inhibition of both Notch1 and Notch2 in multipotent proliferating epithelial progenitors skews differentiation towards goblet cell fate and away from enterocyte fate [125]. However, intermittent dosing with gamma-secretase inhibitors largely obviates this toxicity in both murine models [84] and clinical trials conducted in patients with solid tumours and Alzheimer disease. Moreover, recent work from several groups describes the development of antibodies that selectively inhibit individual Notch receptors [126–128], an approach that also avoids gut toxicity [128]. Unfortunately, it appears that the activity of these antibodies against human T-LL cell lines bearing Notch1 mutations is not as great as that of gamma-secretase inhibitors [126], either because the antibodies fail to bind to the mutated receptors with high affinity, or because proteolysis of the mutated receptors occurs in part intracellularly. Another approach that has been pioneered is to directly inhibit formation of active NTCs using stapled peptides that mimic the amino-terminus of MAML1 [129], but whether such lead molecules can be transformed into drugs remains to be determined. Regardless of what type of inhibitor is used, it will be important to design trials that use biomarkers to determine the extent of Notch inhibition in tumour cells. Since direct quantification of activated Notch is difficult, measurement of the transcript levels of Notch target genes will likely remain the biomarker test of choice.
In the short run, T-LL remains the best hope for successful translation of Notch-directed therapeutics into clinical practice. Single drug therapy with a Merck gamma-secretase inhibitor failed to produce objective responses in patients with relapsed/refractory T-LL, possibly because Notch mutations are not initiating events but rather collaborating secondary acquired “hits” [52]. Nevertheless, the genetics of T-LL in man and mouse strongly argue that Notch1 is the best available rational target in this disease. Work from the Ferrando laboratory showing strong synergy between glucocorticoids and gamma-secretase inhibitors have provided the rationale for second line trials of this drug combination [108], which is now being tested in humans. These and other attempts to target Notch over the next few years should reveal if the therapeutic promise of Notch inhibitors in T-LL and other cancers is to be fulfilled.
Acknowledgements
This work was supported by grants from the National Institute of Health to J.C.A. (P01CA119070), S.C.B. (P01CA119070, add dimer grant [AQ; is this grant identified?]), and W.S.P. (R01AI047833, P01CA119070) and a Leukemia and Lymphoma Society SCOR Award to J.C.A., S.C.B. and W.S.P.
Footnotes
Author Contributions
J.C.A., W.S.P., and S.C.B. jointly wrote and edited the manuscript and produced the accompanying figures.
Conflict of Interest.
J.C.A., W.S.P., and S.C.B. have no conflicts to declare.
References
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon WR, Vardar-Ulu D, Histen G, et al. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 3.Gordon WR, Vardar-Ulu D, L'Heureux S, et al. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS ONE. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logeat F, Bessia C, Brou C, et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rand MD, Grimm LM, Artavanis-Tsakonas S, et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Tetering G, van Diest P, Verlaan I, et al. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem. 2009;284:31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol. 2009;29:5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon WR, Roy M, Vardar-Ulu D, et al. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113:4381–4390. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorensen EB, Conner SD. gamma secretase-dependent cleavage initiates Notch signaling from the plasma membrane. Traffic. 2010;11:1234–1235. doi: 10.1111/j.1600-0854.2010.01090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lubman OY, Ilagan MX, Kopan R, et al. Quantitative dissection of the Notch:CSL interaction: insights into the Notch-mediated transcriptional switch. J Mol Biol. 2007;365:577–589. doi: 10.1016/j.jmb.2006.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Bianco C, Aster JC, Blacklow SC. Mutational and energetic studies of Notch 1 transcription complexes. J Mol Biol. 2008;376:131–140. doi: 10.1016/j.jmb.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fryer CJ, Lamar E, Turbachova I, et al. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16:1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oswald F, Tauber B, Dobner T, et al. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallberg AE, Pedersen K, Lendahl U, et al. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh JJ, Zhou S, Chen L, et al. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci U S A. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao HY, Ordentlich P, Koyano-Nakagawa N, et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oswald F, Winkler M, Cao Y, et al. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou S, Hayward SD. Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex. Mol Cell Biol. 2001;21:6222–6232. doi: 10.1128/MCB.21.18.6222-6232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroda K, Han H, Tani S, et al. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- 21.Sakano D, Kato A, Parikh N, et al. BCL6 canalizes Notch-dependent transcription, excluding Mastermind-like1 from selected target genes during left-right patterning. Dev Cell. 2010;18:450–462. doi: 10.1016/j.devcel.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liefke R, Oswald F, Alvarado C, et al. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24:590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garg V, Muth AN, Ransom JF, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Krantz ID, Deng Y, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 25.Oda T, Elkahloun AG, Pike BL, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 26.McDaniell R, Warthen DM, Sanchez-Lara PA, et al. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 28.Aster J, Pear W, Hasserjian R, et al. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harb Symp Quant Biol. 1994;59:125–136. doi: 10.1101/sqb.1994.059.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Pear WS, Aster JC, Scott ML, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allman D, Karnell FG, Punt JA, et al. Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J Exp Med. 2001;194:99–106. doi: 10.1084/jem.194.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellavia D, Campese AF, Checquolo S, et al. Combined expression of pTalpha and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc Natl Acad Sci U S A. 2002;99:3788–3793. doi: 10.1073/pnas.062050599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radtke F, Wilson A, Stark G, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 33.Pui JC, Allman D, Xu L, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 34.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 35.Asnafi V, Buzyn A, Le Noir S, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood. 2009;113:3918–3924. doi: 10.1182/blood-2008-10-184069. [DOI] [PubMed] [Google Scholar]
- 36.Breit S, Stanulla M, Flohr T, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108:1151–1157. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 37.Larson Gedman A, Chen Q, Kugel Desmoulin S, et al. The impact of NOTCH1, FBW7 and PTEN mutations on prognosis and downstream signaling in pediatric T-cell acute lymphoblastic leukemia: a report from the Children's Oncology Group. Leukemia. 2009;23:1417–1425. doi: 10.1038/leu.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansour MR, Sulis ML, Duke V, et al. Prognostic implications of NOTCH1 and FBXW7 mutations in adults with T-cell acute lymphoblastic leukemia treated on the MRC UKALLXII/ECOG E2993 protocol. J Clin Oncol. 2009;27:4352–4356. doi: 10.1200/JCO.2009.22.0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park MJ, Taki T, Oda M, et al. FBXW7 and NOTCH1 mutations in childhood T cell acute lymphoblastic leukaemia and T cell non-Hodgkin lymphoma. Br J Haematol. 2009;145:198–206. doi: 10.1111/j.1365-2141.2009.07607.x. [DOI] [PubMed] [Google Scholar]
- 40.van Grotel M, Meijerink JP, Beverloo HB, et al. The outcome of molecular-cytogenetic subgroups in pediatric T-cell acute lymphoblastic leukemia: a retrospective study of patients treated according to DCOG or COALL protocols. Haematologica. 2006;91:1212–1221. [PubMed] [Google Scholar]
- 41.Zhu YM, Zhao WL, Fu JF, et al. NOTCH1 mutations in T-cell acute lymphoblastic leukemia: prognostic significance and implication in multifactorial leukemogenesis. Clin Cancer Res. 2006;12:3043–3049. doi: 10.1158/1078-0432.CCR-05-2832. [DOI] [PubMed] [Google Scholar]
- 42.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuji H, Ishii-Ohba H, Ukai H, et al. Radiation-induced deletions in the 5' end region of Notch1 lead to the formation of truncated proteins and are involved in the development of mouse thymic lymphomas. Carcinogenesis. 2003;24:1257–1268. doi: 10.1093/carcin/bgg071. [DOI] [PubMed] [Google Scholar]
- 44.Dumortier A, Jeannet R, Kirstetter P, et al. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol. 2006;26:209–220. doi: 10.1128/MCB.26.1.209-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Neil J, Calvo J, McKenna K, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin YW, Nichols RA, Letterio JJ, et al. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 2006;107:2540–2543. doi: 10.1182/blood-2005-07-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reschly EJ, Spaulding C, Vilimas T, et al. Notch1 promotes survival of E2A-deficient T cell lymphomas through pre-T cell receptor-dependent and -independent mechanisms. Blood. 2006;107:4115–4121. doi: 10.1182/blood-2005-09-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mantha S, Ward M, McCafferty J, et al. Activating Notch1 mutations are an early event in T-cell malignancy of Ikaros point mutant Plastic/+ mice. Leuk Res. 2007;31:321–327. doi: 10.1016/j.leukres.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 49.Maser RS, Choudhury B, Campbell PJ, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malecki MJ, Sanchez-Irizarry C, Mitchell JL, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sulis ML, Williams O, Palomero T, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112:733–740. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiang MY, Xu L, Shestova O, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kindler T, Cornejo MG, Scholl C, et al. K-RasG12D-induced T-cell lymphoblastic lymphoma/leukemias harbor Notch1 mutations and are sensitive to gamma-secretase inhibitors. Blood. 2008;112:3373–3382. doi: 10.1182/blood-2008-03-147587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Girard L, Hanna Z, Beaulieu N, et al. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 55.Hoemann CD, Beaulieu N, Girard L, et al. Two distinct Notch1 mutant alleles are involved in the induction of T-cell leukemia in c-myc transgenic mice. Mol Cell Biol. 2000;20:3831–3842. doi: 10.1128/mcb.20.11.3831-3842.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen H, Suzuki T, Munroe DJ, et al. Common sites of retroviral integration in mouse hematopoietic tumors identified by high-throughput, single nucleotide polymorphism-based mapping and bacterial artificial chromosome hybridization. J Virol. 2003;77:1584–1588. doi: 10.1128/JVI.77.2.1584-1588.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Howard G, Eiges R, Gaudet F, et al. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 58.Tsuji H, Ishii-Ohba H, Katsube T, et al. Involvement of illegitimate V(D)J recombination or microhomology-mediated nonhomologous end-joining in the formation of intragenic deletions of the Notch1 gene in mouse thymic lymphomas. Cancer Res. 2004;64:8882–8890. doi: 10.1158/0008-5472.CAN-03-1163. [DOI] [PubMed] [Google Scholar]
- 59.Tsuji H, Ishii-Ohba H, Noda Y, et al. Rag-dependent and Rag-independent mechanisms of Notch1 rearrangement in thymic lymphomas of Atm(−/−) and scid mice. Mutat Res. 2009;660:22–32. doi: 10.1016/j.mrfmmm.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Ashworth T, Pear W, Chiang MY, et al. Key role for a conserved internal translational start site in Notch1 in deletion-based mechanisms of Notch1 activation in T-ALL. Blood. 2010 doi: 10.1182/blood-2010-05-286328. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeannet R, Mastio J, Macias-Garcia A, et al. Oncogenic activation of the Notch1 gene in T-ALL by deletion of its promoter. Blood. 2010 doi: 10.1182/blood-2010-05-286658. DOI 10.1182/blood-2010-05-286658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji Y, Resch W, Corbett E, et al. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sridharan R, Smale ST. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J Biol Chem. 2007;282:30227–30238. doi: 10.1074/jbc.M702541200. [DOI] [PubMed] [Google Scholar]
- 64.Kim J, Sif S, Jones B, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 65.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 66.Tetzlaff MT, Yu W, Li M, et al. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc Natl Acad Sci U S A. 2004;101:3338–3345. doi: 10.1073/pnas.0307875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsunematsu R, Nakayama K, Oike Y, et al. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J Biol Chem. 2004;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- 68.O'Neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson BJ, Buonamici S, Sulis ML, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiang MY, Xu ML, Histen G, et al. Identification of a conserved negative regulatory sequence that influences the leukemogenic activity of NOTCH1. Mol Cell Biol. 2006;26:6261–6271. doi: 10.1128/MCB.02478-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malyukova A, Dohda T, von der Lehr N, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 72.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palomero T, Lim WK, Odom DT, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma VM, Calvo JA, Draheim KM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X, Gounari F, Protopopov A, et al. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med. 2008;205:2851–2861. doi: 10.1084/jem.20081561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jarriault S, Brou C, Logeat F, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 77.Dudley DD, Wang HC, Sun XH. Hes1 potentiates T cell lymphomagenesis by up-regulating a subset of notch target genes. PLoS One. 2009;4:e6678. doi: 10.1371/journal.pone.0006678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palomero T, Sulis ML, Cortina M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 80.Sade H, Krishna S, Sarin A. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J Biol Chem. 2004;279:2937–2944. doi: 10.1074/jbc.M309924200. [DOI] [PubMed] [Google Scholar]
- 81.Cullion K, Draheim KM, Hermance N, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009 doi: 10.1182/blood-2008-02-136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan SM, Weng AP, Tibshirani R, et al. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110:278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gonzalez-Garcia S, Garcia-Peydro M, Martin-Gayo E, et al. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7R{alpha} gene expression in early human thymopoiesis and leukemia. J Exp Med. 2009;206:779–791. doi: 10.1084/jem.20081922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barata JT, Cardoso AA, Boussiotis VA. Interleukin-7 in T-cell acute lymphoblastic leukemia: an extrinsic factor supporting leukemogenesis? Leuk Lymphoma. 2005;46:483–495. doi: 10.1080/10428190400027852. [DOI] [PubMed] [Google Scholar]
- 85.Armstrong F, Brunet de la Grange P, Gerby B, et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 2009;113:1730–1740. doi: 10.1182/blood-2008-02-138172. [DOI] [PubMed] [Google Scholar]
- 86.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 87.Hozumi K, Mailhos C, Negishi N, et al. Delta-like 4 is indispensable in thymic environment specific for T cell development. J Exp Med. 2008;205:2507–2513. doi: 10.1084/jem.20080134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sambandam A, Maillard I, Zediak VP, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 89.Tan JB, Visan I, Yuan JS, et al. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 90.Krueger A, von Boehmer H. Identification of a T lineage-committed progenitor in adult blood. Immunity. 2007;26:105–116. doi: 10.1016/j.immuni.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bellavia D, Campese AF, Alesse E, et al. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19:3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vilimas T, Mascarenhas J, Palomero T, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med. 2007;13:70–77. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]
- 93.Jin B, Shen H, Lin S, et al. The maml1 co-activator regulates constitutive NF-{kappa}B signaling and cell survival. J Biol Chem. 2010;285:14356–14365. doi: 10.1074/jbc.M109.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yashiro-Ohtani Y, He Y, Ohtani T, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nie L, Xu M, Vladimirova A, et al. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 2003;22:5780–5792. doi: 10.1093/emboj/cdg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 97.Nam Y, Sliz P, Song L, et al. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 98.Nam Y, Sliz P, Pear WS, et al. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci U S A. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arnett K, Hass MR, McArthur DG, et al. Structural and mechanistic insights into cooperative assembly of dimeric notch transcription complexes. Nature Structural and Molecular Biology. doi: 10.1038/nsmb.1938. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beverly LJ, Capobianco AJ. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell. 2003;3:551–564. doi: 10.1016/s1535-6108(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 101.Chari S, Winandy S. Ikaros regulates Notch target gene expression in developing thymocytes. J Immunol. 2008;181:6265–6274. doi: 10.4049/jimmunol.181.9.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kathrein KL, Chari S, Winandy S. Ikaros directly represses the notch target gene Hes1 in a leukemia T cell line: implications for CD4 regulation. J Biol Chem. 2008;283:10476–10484. doi: 10.1074/jbc.M709643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kleinmann E, Geimer Le Lay AS, Sellars M, et al. Ikaros represses the transcriptional response to Notch signaling in T-cell development. Mol Cell Biol. 2008;28:7465–7475. doi: 10.1128/MCB.00715-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dail M, Li Q, McDaniel A, et al. Mutant Ikzf1, KrasG12D, and Notch1 cooperate in T lineage leukemogenesis and modulate responses to targeted agents. Proc Natl Acad Sci U S A. 2010;107:5106–5111. doi: 10.1073/pnas.1001064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marcais A, Jeannet R, Hernandez L, et al. Genetic inactivation of Ikaros is a rare event in human T-ALL. Leuk Res. 2010;34:426–429. doi: 10.1016/j.leukres.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 106.Bellavia D, Mecarozzi M, Campese AF, et al. Notch3 and the Notch3-upregulated RNA-binding protein HuD regulate Ikaros alternative splicing. EMBO J. 2007;26:1670–1680. doi: 10.1038/sj.emboj.7601626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rohn JL, Lauring AS, Linenberger ML, et al. Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. J Virol. 1996;70:8071–8080. doi: 10.1128/jvi.70.11.8071-8080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Real PJ, Tosello V, Palomero T, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buonamici S, Trimarchi T, Ruocco MG, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature. 2009;459:1000–1004. doi: 10.1038/nature08020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Palomero T, McKenna K, J ON, et al. Activating mutations in NOTCH1 in acute myeloid leukemia and lineage switch leukemias. Leukemia. 2006;20:1963–1966. doi: 10.1038/sj.leu.2404409. [DOI] [PubMed] [Google Scholar]
- 111.Wouters BJ, Jorda MA, Keeshan K, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110:3706–3714. doi: 10.1182/blood-2007-02-073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ma X, Renda MJ, Wang L, et al. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol. 2007;27:3056–3064. doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mercher T, Raffel GD, Moore SA, et al. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. J Clin Invest. 2009;119:852–864. doi: 10.1172/JCI35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mercher T, Cornejo MG, Sears C, et al. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell. 2008;3:314–326. doi: 10.1016/j.stem.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jundt F, Acikgoz O, Kwon SH, et al. Aberrant expression of Notch1 interferes with the B-lymphoid phenotype of neoplastic B cells in classical Hodgkin lymphoma. Leukemia. 2008;22:1587–1594. doi: 10.1038/leu.2008.101. [DOI] [PubMed] [Google Scholar]
- 116.Jundt F, Probsting KS, Anagnostopoulos I, et al. Jagged1-induced Notch signaling drives proliferation of multiple myeloma cells. Blood. 2004;103:3511–3515. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- 117.Saito T, Chiba S, Ichikawa M, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 118.Lee SY, Kumano K, Nakazaki K, et al. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci. 2009;100:920–926. doi: 10.1111/j.1349-7006.2009.01130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rosati E, Sabatini R, Rampino G, et al. Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood. 2009;113:856–865. doi: 10.1182/blood-2008-02-139725. [DOI] [PubMed] [Google Scholar]
- 120.Schwarzmeier JD, Hubmann R, Duchler M, et al. Regulation of CD23 expression by Notch2 in B-cell chronic lymphocytic leukemia. Leuk Lymphoma. 2005;46:157–165. doi: 10.1080/10428190400010742. [DOI] [PubMed] [Google Scholar]
- 121.Duechler M, Shehata M, Schwarzmeier JD, et al. Induction of apoptosis by proteasome inhibitors in B-CLL cells is associated with downregulation of CD23 and inactivation of Notch2. Leukemia. 2005;19:260–267. doi: 10.1038/sj.leu.2403592. [DOI] [PubMed] [Google Scholar]
- 122.Hubmann R, Schwarzmeier JD, Shehata M, et al. Notch2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood. 2002;99:3742–3747. doi: 10.1182/blood.v99.10.3742. [DOI] [PubMed] [Google Scholar]
- 123.Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by notch. Dev Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 124.Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 125.Riccio O, van Gijn ME, Bezdek AC, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aste-Amezaga M, Zhang N, Lineberger JE, et al. Characterization of notch1 antibodies that inhibit signaling of both normal and mutated notch1 receptors. PLoS One. 2010;5:e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li K, Li Y, Wu W, et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- 128.Wu Y, Cain-Hom C, Choy L, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 129.Moellering RE, Cornejo M, Davis TN, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.van Grotel M, Meijerink JP, van Wering ER, et al. Prognostic significance of molecular-cytogenetic abnormalities in pediatric T-ALL is not explained by immunophenotypic differences. Leukemia. 2008;22:124–131. doi: 10.1038/sj.leu.2404957. [DOI] [PubMed] [Google Scholar]