Abstract
We have recently shown that several carbonylated proteins, including GFAP, β-actin and β-tubulin, accumulate within cerebellar astrocytes during the chronic phase of MOG35–55 peptide-induced EAE in C57BL/6 mice. Since protein carbonyls cannot be repaired and there is less oxidative stress in chronic than in acute EAE, we hypothesized that the accumulation of carbonylated proteins in these animals may be due to a defect in the degradation of the modified proteins. Alternatively, oxidized proteins in chronic EAE mice may be more resistant to proteolysis. Using LPS-stimulated astrocytes and several protease inhibitors we identified the 20S proteasome as the proteolytic system responsible for the elimination of most oxidized proteins. We also discovered that the chymotrysin-like and caspase-like activities of the 20S proteasome are impaired in chronic EAE, while the amount of proteasome was unchanged. Proteasome failure in these animals was confirmed by the build-up of ubiquitinated proteins, mostly within astrocytes. In a cell-free system, carbonylated proteins from EAE mice with acute and chronic disease seem to be equally sensitive to proteasomal degradation. Altogether, the results support the notion that diminished activity of the 20S proteasome is a major contributor to the accumulation of carbonylated proteins in astrocytes of chronic EAE mice.
Keywords: astrocyte, experimental autoimmune encephalomyelitis, oxidative stress, proteasome, protein carbonylation
Carbonylation refers to the non-enzymatic addition of aldehyde or ketone groups to specific amino acid residues and constitutes the major and most common oxidative alteration of proteins (Dalle-Donne et al. 2003; Nystrom 2005). Like in several CNS disorders, including Alzheimer's disease (Aksenov et al. 2001), Parkinson's disease (Floor & Wetzel 1998) and amyotrophic lateral sclerosis (Ferrante et al. 1997), multiple sclerosis (MS) (Bizzozero et al. 2005; Hilgart & Bizzozero 2008) and its animal model experimental autoimmune encephalomyelitis (EAE) are also characterized by the accumulation of carbonylated (oxidized) proteins (Smerjac & Bizzozero 2008; Zheng & Bizzozero, 2010a). Carbonylation leads almost always to loss of protein function and is believed to partake in the etiology of these neurological diseases (for a review, see Bizzozero 2009). In MOG35–55 peptide-induced EAE mice, the amount of the most abundant carbonylated proteins (e.g. β-actin, β-tubulin and GFAP) in cerebellar astrocytes was found to augment as disease advances from the inflammatory (acute) phase to the neurodegenerative (chronic) phase (Zheng & Bizzozero 2010a), suggesting that this deleterious protein modification may play a role in disease progression as well.
It is clear that the amount of carbonylated protein is determined by the rates of generation and degradation of carbonyls. Proteolysis is currently considered the only physiological mechanism for elimination of carbonylated proteins, as there is no evidence for enzymatic reduction of protein-bound carbonyl groups to alcohols (Bizzozero 2009). This and the fact that there is less oxidative stress in chronic than in acute EAE (Zheng & Bizzozero 2010a) suggest that the accumulation of carbonylated cytoskeletal proteins in the cerebellum of chronic EAE mice may be due to impaired degradation. This phenomenon, in turn, could be the result of reduced activity of the degradation system and/or decreased susceptibility of the oxidized proteins to proteolysis. In mammalian cells, the removal of carbonylated proteins is mostly carried out by the 20S proteasome in an ATP- and ubiquitin-independent manner (Shringarpure et al. 2003; Divald & Powell 2006). This multi-enzymatic proteolytic particle selectively recognizes and digests partially unfolded (denatured) oxidized proteins through its chymotrypsin-like activity (Ferrington et al. 2005). However, the calcium-dependent cysteine protease calpain and the lysosomal cathepsins have been also implicated in the proteolytic removal of damaged proteins. For instance, oxidized neurofilaments seem to be preferentially digested by calpain in vitro (Troncoso et al. 1995) while heavily oxidized proteins are taken-up by lysosomes for partial proteolysis (Dunlop et al. 2009).
In this study, we initially assessed the role of these three major degradation systems in the accumulation of carbonylated proteins in LPS-stimulated astrocytes by using well-characterized protease inhibitors. The results clearly show that only the proteasome inhibitor epoxomicin leads to a build-up of carbonylated proteins, while inhibition of lysosomal proteases and calpain do not alter protein carbonylation levels. More important, we discovered that the chymotrypsin-like activity of the 20S proteasome is impaired in the cerebellum of mice with chronic, but not acute, EAE. This observation was also consistent with the accumulation of poly-ubiquitinated proteins within cerebellar astrocytes observed in the animals with the chronic disease. Furthermore, experiments in a cell-free system showed that carbonylated cytoskeletal proteins from acute and chronic EAE are equally sensitive to proteasomal degradation. Overall, this work provides clear evidence supporting the notion that the accumulation of carbonylated proteins in chronic EAE is likely the result of reduced proteasomal activity. To the best of our knowledge, this the first report implicating proteasome dysfunction in the pathophysiology of EAE. A preliminary account of this work has been presented in abstract form (Zheng & Bizzozero 2010b).
Materials and methods
Astrocyte culture
Rat C6 glioblastoma cells (CCL-107) were obtained from American Type Culture Collection (Manassas, VA) and were established as a monolayer culture in Dulbecco's modified Eagle's/F-12 medium supplemented with 15% horse serum, 2.5% fetal bovine serum and an antibiotic/antimycotic mixture (Invitrogen Corp., Carlsbad, CA). Cells were maintained in a humidified incubator at 37°C in an atmosphere of 95% air / 5% CO2. To be differentiated into astrocytes, C6 cells were first serum-starved for 1h and then incubated with 1mM N6-2'-O-dibutyryl cyclic-AMP (Bt2AMP; Sigma, St Louis, MO) and 0.25 mM theophylline (Sigma) for up to 72h. At this point, astrocytes were activated with 1μg/ml lipopolysaccharide (LPS; Sigma) for 24 hours. Nitrite concentration in the cell supernatants was determined spectrofluorometrically using 2,3–diaminonaphthalene (Misko et al. 1993) and non-protein thiol (mostly GSH) levels in the cell homogenates were determined with 5,5'-dithiobis-(2-nitrobenzoic acid) (Bizzozero et al. 2006). To identify the proteolytic system responsible for degradation of carbonylated cytoskeletal proteins, LPS-treated and untreated astrocytes were incubated for 24h in the absence or presence of the proteasome inhibitor epoxomicin (2 μM; Boston Biochemical, Cambridge, MA), the lysosomal inhibitor NH4Cl (2mM, Sigma) or the calpain inhibitor calpeptin (20μM, Sigma). Cells were either fixed with methacarn (methanol: chloroform: acetic acid, 60: 30: 10 by vol) or lysed in PEN buffer (20 mM sodium phosphate, pH 7.5, 1 mM EDTA, and 0.1 mM neocuproine) containing 2 mM 4,5 dihydroxy-1, 3-benzene disulfonic acid and 1 mM dithiothreitol (DTT). Protein homogenates were stored at −20°C until use. Protein concentration was assessed with the Bio-Rad DC™ protein assay (Bio-Rad Laboratories; Hercules, CA) using bovine serum albumin as standard.
Induction of experimental autoimmune encephalomyelitis (EAE)
Housing and handling of the animals as well as the euthanasia procedure were in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals, and were approved by the Institutional Animal Care and Use Committee. Eight-week-old female C57BL/6 mice were purchased from Harlan Bioproducts (Indianapolis, IN) and housed in the UNM-animal resource facility. EAE was induced by active immunization with MOG35–55 peptide (AnaSpec, San Jose, CA) as described in our previous study (Zheng & Bizzozero 2010a). Animals were weighed and examined daily for the presence of neurological signs. At prescribed days post-immunization (DPI), EAE mice and CFA-injected controls were euthanized by decapitation. The cerebellum was removed and either fixed with methacarn (methanol : chloroform : acetic acid, 60 : 30 : 10 by vol) or homogenized in PEN buffer with antioxidants. Protein homogenates were stored and processed as described above.
Protease activity assays
The various proteolytic activities of the proteasome were determined in cerebellar homogenates from control and EAE mice using fluorescence assays (Rodgers et al. 2003). Briefly, 50μg of protein were incubated for 2h at 2°C with 50μM of the 7-aminomethyl-4-coumarin (AMC)-labeled peptide Suc-Leu-Leu-Val-Tyr-AMC (for chymotrypsin-like activity), Boc-Leu-Arg-Arg-AMC (for trypsin-like activity) or z-Leu-Leu-Glu-AMC (for caspase-like activity) in the absence or presence of 10μM clastolactacystin-β-lactone (Enzo Life Sciences, Plymouth Meeting, PA) or 50μM epoxomicin (for the trypsin-like activity). The different activities of the 20S proteasome were calculated as the difference in fluorescence intensity at 460nm between the samples without and with inhibitor using an excitation wavelength of 380nm.
Total calpain activity was determined by a similar procedure using the substrate Suc-Leu-Leu-Val-Tyr-AMC in 100mM KCl, 10mM CaCl2, 25mM Hepes buffer pH 7.5, and carrying out the incubation in the absence or presence of 40μg/ml calpeptin (Hassem et al. 2006). To measure soluble (active) calpain activity, membrane-bound calpain was removed prior to the assay by centrifugation at 10,000 g for 25 min.
Lysosomal proteolytic activity was also measured fluorometrically by incubating cerebellar homogenates with 200μM z-Phe-Arg-AMC in 50 mM sodium acetate (pH 5.5) for 30 min at 37°C (Sitte et al. 2000).
Western blot analysis
Proteins (5 μg) from cells or tissue homogenates were separated by SDS-polyacrylamide gel electrophoresis on 10% gels and blotted to polyvinylidene difluoride (PVDF) membranes. Blots then were incubated overnight at 4°C with monoclonal antibodies against GFAP (1:2,000; Sigma), GAPDH (1:2,000; Chemicon, Temecula, CA) or α-subunit of 20S proteasome (1:2,000; Enzo). Membranes were rinsed three times in phosphate-buffered saline (PBS) containing 0.05% Tween-20 and incubated for 2 h with HRP-conjugated goat anti-mouse antibody (1:2,000; Sigma). Blots were developed by enhanced chemiluminescence (ECL) using the Western Lightning ECL™ kit from Perkin-Elmer (Boston, MA, USA).
Protein carbonyl groups were measured by western blot analysis using the OxyBlot™ protein oxidation detection kit (Intergen Co., Purchase, NY) as described earlier (Smerjac & Bizzozero, 2008). In brief, proteins (5 μg) were incubated with 2,4-dinitrophenyl-hydrazine to form the 2,4-dinitrophenyl (DNP) hydrazone derivatives. Proteins were separated by electrophoresis and blotted to PVDF membranes as above. DNP-containing proteins were detected by ECL using rabbit anti-DNP antiserum (1:500) and goat anti-rabbit IgG conjugated to HRP (1:2000).
Quantification of carbonylation levels in specific proteins
The extent of protein carbonylation was determined using a pull-down/western blot method (Bizzozero et al. 2006). Briefly, protein carbonyls were biotinylated by reaction with biotin hydrazide in the presence of cyanoborohydride. A small aliquot of these protein homogenates was saved for western blotting and the rest was used to isolate the biotinylated proteins using streptavidin-agarose. Proteins were eluted from the beads with SDS-sample buffer and analyzed by western blotting on 10% polyacrylamide gels. Blots were probed with antibodies against individual protein species and developed by ECL as described above. Films were scanned in a Hewlett Packard Scanjet 4890 and the images were quantified using the NIH Image 1.63 imaging analysis program. Band intensities from the total and streptavidin-bound fractions were used to calculate the percentage of protein that is modified by carbonylation.
Immunohistochemical localization of poly-ubiquitinated proteins in cerebellum
Accumulation of poly-ubiquitinated proteins in the cerebellum was assessed by immunohistochemistry. To this end, cerebella from control and EAE animals were fixed overnight in methacarn and then mounted in paraffin. Tissue was sectioned in the sagital plane (6-μm thick) and mounted on Vectabond™-treated slides (Vector Laboratories, Burlingame, CA, USA). Sections were deparafinized with xylenes and a graded alcohol series, and then rinsed with PBS for 10 min. Lesions were detected by staining with hematoxylin and eosin. The adjacent slices were collected, rinsed three times with PBS, blocked with 10% (v/v) normal goat serum and incubated overnight with the mixture of anti-polyubiquitin antibody (1:200, mouse monoclonal; Enzo) and anti-GFAP antibody (1:200, rabbit polyclonal; Dako, Carpinteria, CA). After removing the primary antibodies with PBS containing 0.1% Triton X-100, the sections were incubated for 3 h with fluorescent conjugated secondary antibodies (Alexa Fluor® 488 and Alexa Fluor® 647, Molecular Probes, Eugene, OR, USA). Slide-mounted sections were scanned at a total magnification of 80× and images were imported into Image J software to obtain optical density of polyubiquitin in GFAP immunoreactive cells in cerebella. Briefly, the poly-ubiquitin or GFAP optical density was measured by circling the whole GFAP immunoreactive cell. Three fields per slide and three slides per animal were randomly chosen. In all cases, the background was subtracted from the average density. The final data is presented as average density of polyubiquitin or polyubiquitin divided by GFAP.
Statistical analysis
Results were analyzed for statistical significance with ANOVA utilizing GraphPad Prism® program (GraphPad Software Inc., San Diego, CA).
Results
Differentiation of C6 cells into astrocytes and induction of oxidative stress with LPS
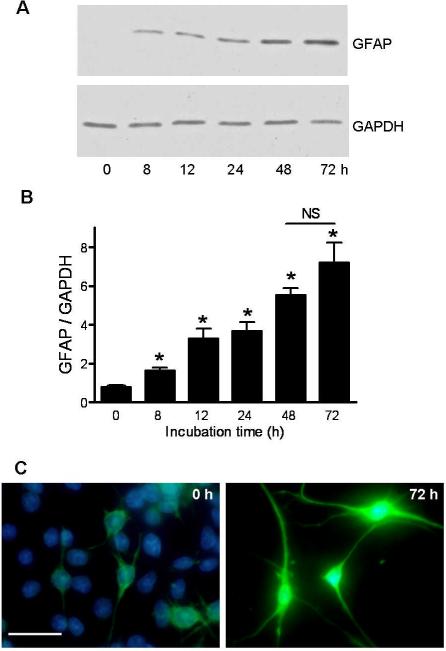
An in vitro study was designed to identify the major proteolytic system(s) responsible for the removal of carbonylated proteins from astrocytes under oxidative stress conditions. To this end, we first established a cell culture system in which C6 glioma cells were differentiated into astrocytes by treatment with a cyclic AMP analogue (Haghighat et al. 2000). As shown in Fig. 1, treatment of C6 cells with Bt2AMP/theophylline elevated GFAP expression already at 8 h of incubation (Fig. 1A–B). GFAP levels in these cells continued to increase until 72 h. At this time cells displayed the morphological characteristics of astrocytes (Fig. 1C).
Fig. 1.
Differentiation of C6 glioma cells into astrocytes. C6 cells were incubated with Bt2AMP/theophylline for up to 72h as described in “Material and Methods”. Levels of GFAP and GAPDH were determined by western blot (panel A). Band intensities were measured by scanning densitometry and were used to calculate the GFAP/GAPDH ratio (panel B). Values represent the mean ± SEM of 3 experiments. Asterisks denote values that are statistically different (p<0.05) from non-stimulated control cells. NS, not significant. Panel C shows a double immunofluorescence picture of untreated and Bt2AMP/theophylline-treated C6 cells. GFAP and nuclear (DAPI) staining are shown in green and blue, respectively. Bar= 50μm. Note that at 72 h, C6 cells are not longer round and flat, but show large amount of GFAP-positive processes that are characteristics of astrocytes.
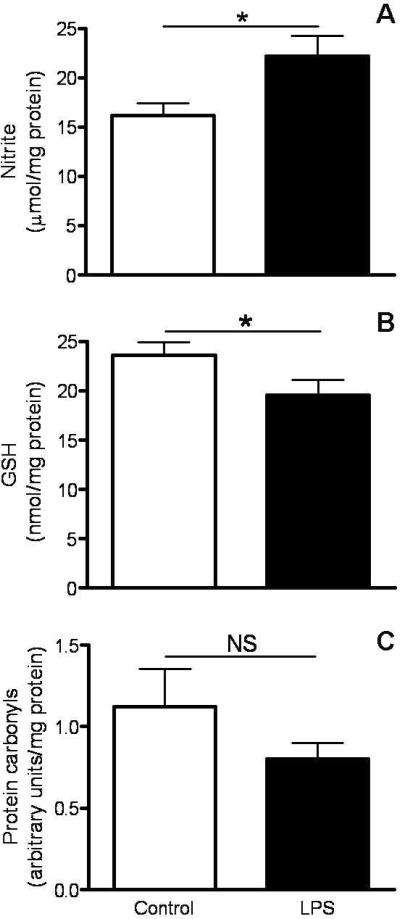
Astrocytes were then incubated with LPS (1μg/ml) for 24 h to induce oxidative stress. LPS is a bacterial endotoxin and a generally accepted inducer of proinflammatory responses (Kalmár et al. 2001). As expected, the concentration of nitrite (a marker of nitric oxide production) in the medium was significantly higher in LPS-treated astrocytes (Fig. 2A). GSH levels in LPS-stimulated astrocytes were reduced to 83% of control values (Fig. 2B). Surprisingly, the amount of protein carbonyls, another marker of oxidative stress, was not increased in LPS-stimulated astrocytes (Fig. 2C). This observation suggests that either the extent of oxidative stress is not enough to induce extensive protein carbonylation or that the proteolytic machinery present in these cells is sufficient to remove oxidized proteins as they are generated.
Fig. 2.
Levels of nitrosative/oxidative stress markers in control and LPS-stimulated astrocytes. Astrocytes were stimulated with 1μg/ml of LPS. After 24h, the levels of nitrite in the medium (panel A), and those of GSH (panel B) and protein carbonyls (panel C) in the cells were determined as described under “Materials and Methods”. Values represent the mean ± SEM of 3 experiments. *p<0.05. NS, not significant.
Proteasome inhibition leads to accumulation of carbonylated proteins in cultured astrocytes
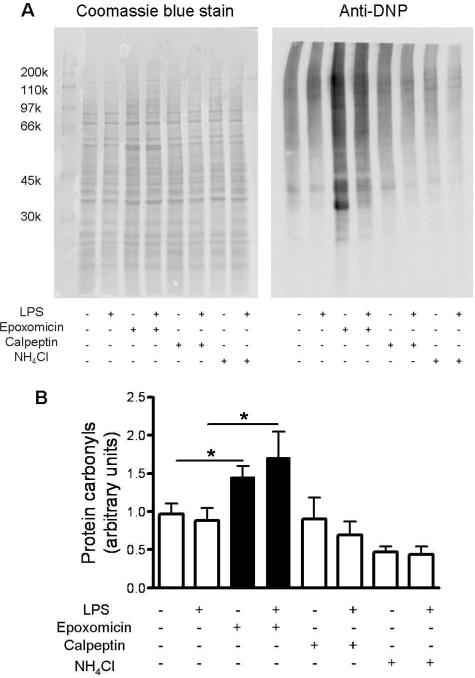
To identify the proteolytic system responsible for the removal of oxidized proteins, control and LPS-stimulated astrocytes were incubated for 24 h in the absence or presence of the proteasome inhibitor epoxomicin (Meng et al. 1999), the lysosomal inhibitor NH4Cl (Brown et al. 1985) or the calpain inhibitor calpeptin (Tsujinaka et al. 1988). To achieve maximal inhibition, these drugs were used at concentrations between 20- and 200-fold higher than their respective IC50. As shown in Fig. 3, only epoxomicin causes the build-up of carbonylated proteins in both control and LPS-stimulated cells. These findings suggest not only that the proteasome is responsible for removal of oxidized proteins in vitro, but also that proteasomal proteolytic activity is more important than the redox environment in determining the carbonylation status of proteins.
Fig. 3.
Only the proteasome inhibitor epoxomicin causes a build-up of carbonylated proteins in cultured astrocytes. Astrocytes were incubated in the presence or absence of LPS with/without the proteasome inhibitor epoxomicin (2μM), the lysosomal inhibitor NH4Cl (2mM) or the calpain inhibitor calpeptin (20μM) as described in “Materials and Methods”. After 24 h, protein carbonyl levels were determined by oxyblot analysis (Panel A). The intensity of each lane of the oxyblot was determined by scanning densitometry and divided by that of the coomassie blue stained membrane to correct for differences in gel loading (panel B). Values represent the mean ± SEM of 3 experiments. *p<0.05.
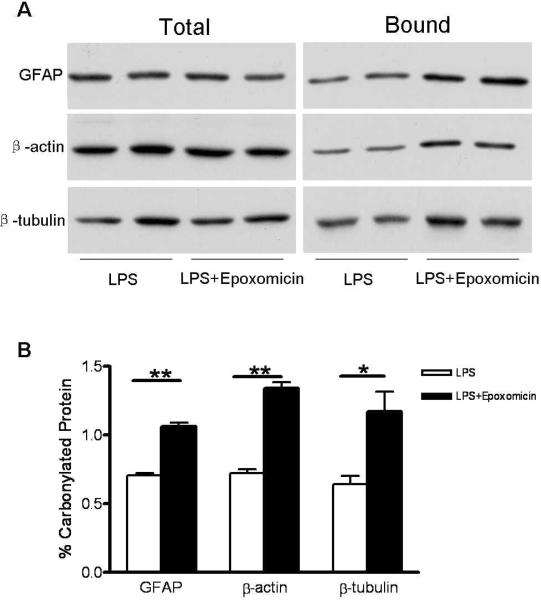
We next investigated whether the major oxidized proteins that accumulate in the cerebellum of mice with chronic EAE (i.e. β-actin, β-tubulin and GFAP) are also degraded by the proteasome in cultured astrocytes. Quantification of the extent of carbonylation of individual proteins was performed using a pull-down/western blot procedure. To this end, protein carbonyl moieties from LPS-treated astrocytes that had been incubated with or without epoxomicin for 24 h were first converted into biotinylated residues by reaction with biotin-hydrazide. Biotin-containing proteins were then isolated with streptavidin-agarose and analyzed by western blotting employing antibodies against each of the three polypeptides. As shown in Fig. 4, the proportion of β-actin, β-tubulin and GFAP present in the streptavidin-bound fraction increased significantly in the epoxomicin-treated cells, indicating that these proteins are also degraded by the proteasome. It is noteworthy that the molecular weight of oxidized proteins present in the bound fraction is identical to those in the total fraction, demonstrating that they do not contain attached ubiquitin moieties (8.5 kDa per monomer). Thus, it is fair to conclude that the degradation of the carbonylated forms of β-actin, β-tubulin and GFAP, and likely those of most other proteins, is carried out by the proteasome via an ubiquitin-independent mechanism.
Fig. 4.
The proportion of carbonylated GFAP, β-actin and β-tubulin in LPS-stimulated astrocytes increases upon incubation with the proteasome inhibitor epoxomicin. LPS-treated astrocytes were incubated in the absence or presence of epoxomicin. After 24 hours, carbonylated proteins were converted into biotinylated proteins and were isolated using streptavidin-agarose as described in “Materials and Methods”. Aliquots of the starting material (total) and the streptavidin-purified fraction (bound) were separated on SDS-gels and transferred to PVDF membranes. Blots were probed with antibodies against β-actin, β-tubulin and GFAP, and were developed by ECL (panel A). Densitometric scans were obtained to calculate the proportion of the various carbonylated species in epoxomicin-treated and untreated activated astrocytes (Panel B). Values represent the mean ± SEM of 3 experiments. *p<0.05, **p<0.01.
Proteasomal proteolytic activity is reduced in chronic EAE
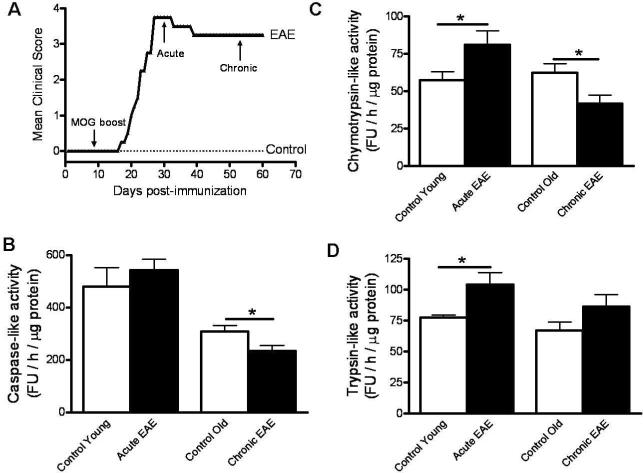
After identifying the 20S proteasome as the proteolytic system responsible for removal of oxidized proteins in cultured astrocytes, we sought to determine whether the proteasome's proteolytic activity is impaired in chronic EAE. To this end, EAE in female C57BL/6 mice was induced by active immunization with MOG35–55 peptide as described under Materials and Methods. Symptoms were graded according to the following scale: 0, no symptoms; 1, tail weakness; 1.5, clumsy gait; 2, hind limb paresis; 2.5, partial hind limb dragging; 3, hind limb paralysis; 3.5, hind limb paralysis with fore limb paresis; 4, complete paralysis; and 5, moribund. In this well-characterized EAE model, neurological symptoms begin at 14 DPI (i.e. 7 days after the boost with MOG peptide) reaching a peak at 30 DPI, and most animals remain ill (score 3.0–3.5) throughout the entire experimental period (60 DPI) (Fig. 5A). Acute disease was defined as having clinical signs of EAE without any signs of improvement for at least three consecutive days while chronic EAE was defined arbitrarily as animals that remain in the stationary phase of the disease for 30 days (60 DPI). A complete morphological description of the cerebellar pathology in this model, including the histological and cellular distribution of carbonyls, has been reported in our previous study (Zheng & Bizzozero 2010a).
Fig. 5.
The chymotrypsin-like and caspase-like activities of the 20S proteasome are significantly reduced in chronic EAE. The disease was induced in C57BL/6 female mice by active immunization with MOG35–55 peptide as described in “Materials and Methods”. Animals were monitored daily for signs of clinical disease and scored as indicated in the text (panel A). Aliquots of cerebellum homogenates from control and EAE mice were used to determine the chymotrypsin-like (panel B), caspase-like (panel C) and trypsin-like (panel D) activity of the 20S proteasome as described in “Materials and Methods”. Values represent the mean ± SEM of 4–10 experiments. *p<0.05; FU, fluorescence units.
Aliquots of cerebellar homogenates from control and EAE mice, both at the peak of the disease and in the chronic phase, were used to determine the various proteolytic activities of the 20S proteasome. The chymotrypsin-like activity, which is believed to be responsible for degrading oxidized proteins (Ferrington et al. 2005), was elevated in acute EAE and greatly reduced (~40%) in chronic EAE relative to the controls (Fig. 5B). This finding agrees with the idea that decreased proteasomal activity is behind the accumulation of protein carbonyls as disease progresses from the inflammatory (acute) to the neurodegenerative (chronic) phase. A similar pattern was found for the proteasome caspase-like activity (Fig. 5C). The trypsin-like activity while increased in acute EAE was unchanged in chronic EAE (Fig. 5D).
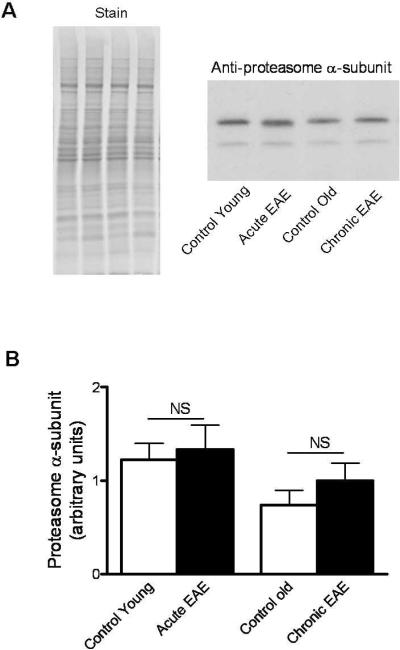
To establish if the changes in proteolytic activity observed during the course of EAE was a consequence of variations in proteasome concentration, we measured the relative levels of the constitutive α-subunit of the 20S proteasome by western blot analysis. As depicted in Fig. 6, the amount of proteasome α-subunit in acute and chronic EAE was the same as those in the controls, suggesting that decrease activity in the chronic phase of the disease may be due to enzyme inactivation.
Fig. 6.
The amount of 20S proteasome is not altered in chronic EAE. Proteins from control and EAE cerebella were separated on SDS-gels and transferred to PVDF membranes. Blots were probed with antibodies against the constitutive α-subunit, and were developed by ECL (panel A). The relative levels of the proteasome were calculated by dividing the α-subunit band intensity on the western blot by that of the commassie blue stained lane. Values are the mean ± SEM of 5–8 experiments. NS, not significant.
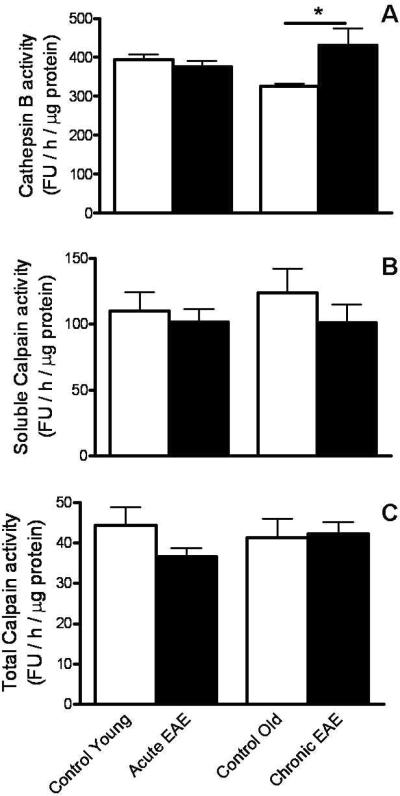
We also looked into the possibility that the proteolytic activity of the lysosome and/or calpain may be decreased in chronic EAE. The activity of cathepsin B, one of the major lysosomal proteases, was assayed with the z-Phe-Arg-AMC peptide at acid pH. Using these conditions, the lysosomal proteolytic activity was increased in chronic EAE as compared to control animals, while no difference was observed between acute EAE and control mice (Fig. 7A). Neither the total (Fig 7C) nor the soluble (active) calpain activity (Fig. 7B) in the cerebellum of affected animals was different from those of controls.
Fig. 7.
Neither cathepsin B nor calpain activity is decreased in chronic EAE. Aliquots of cerebellum homogenates from control and EAE mice were used to determine cathepsin B and total/soluble calpain activity as described in “Material and Methods”. Values are the mean ± SEM of 4–6 experiments. * p<0.05; FU, fluorescence units.
Carbonylated cytoskeletal proteins prepared from acute and chronic EAE tissues are equally sensitive to proteasomal degradation in cell-free system
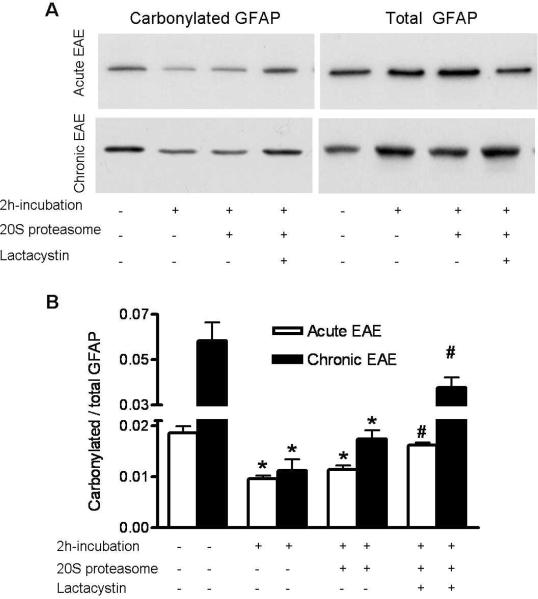
Another possibility that could explain the build-up of oxidized proteins in chronic EAE is that the carbonylated species in the chronic phase are somehow more resistant to proteolysis that those present in the acute phase. To address this issue, we incubated cerebellar homogenates from acute and chronic EAE mice with 20S proteasome in the absence or presence of the proteasome inhibitor clasto-lactacystin-β-lactone. After 2h, the proportion of carbonylated β-actin, β-tubulin and GFAP was determined using the pull-down/western blot procedure described earlier. Since the results were the same for all three proteins, only those corresponding to GFAP are presented herein (Fig. 8). The results clearly show that the proteolytic activity present in the tissue homogenate itself is sufficient to degrade carbonylated GFAP from acute and chronic EAE samples equally well. Addition of 20S proteasome did not produce any significantly increase in the extent of proteolysis. In all cases, degradation of carbonylated GFAP was prevented by addition of clasto-lactacystin-β-lactone, indicating that proteolysis of oxidized proteins in this cell-free system, like in cultured astrocytes, is indeed mediated by the 20S proteasome. The fact that degradation of oxidized GFAP in this system takes place in the absence of added ATP demonstrates once again that the mechanism does not involve ubiquitination. In sum, these data support the notion that oxidized GFAP from acute and chronic EAE are equally sensitive to degradation by the 20S proteasome.
Fig. 8.
Carbonylated GFAP from acute and chronic EAE animals are sensitive to proteasomal degradation in a cell-free system. Cerebellum homogenates from acute and chronic EAE mice (200 μg protein) were incubated with 0.5 μg of 20S proteasome (Sigma) in the absence or presence of 2μg clasto-lactacystin-β-lactone (Enzo Life Sciences). After 2 h, carbonylated proteins were isolated using the pull-down procedure described in “Materials and Methods”. A representative western blot of the total and bound fractions developed with an antibody against GFAP is shown in panel A. Densitometric scans were obtained to calculate the proportion of carbonylated GFAP in the various conditions (Panel B). The 20S chymotrypsin-like activity in the homogenates increased ~12-fold upon addition of 20S proteasome and was reduced by ~90% in the presence of clasto-lactacystin-β-lactone. Values represent the mean ± SEM of 4 experiments. *Significantly different (p<0.05) from non-incubated condition; #Significantly different (p<005) from the 20S proteasome-treated condition.
Ubiquitinated proteins build-up in cerebellar astrocytes in chronic EAE
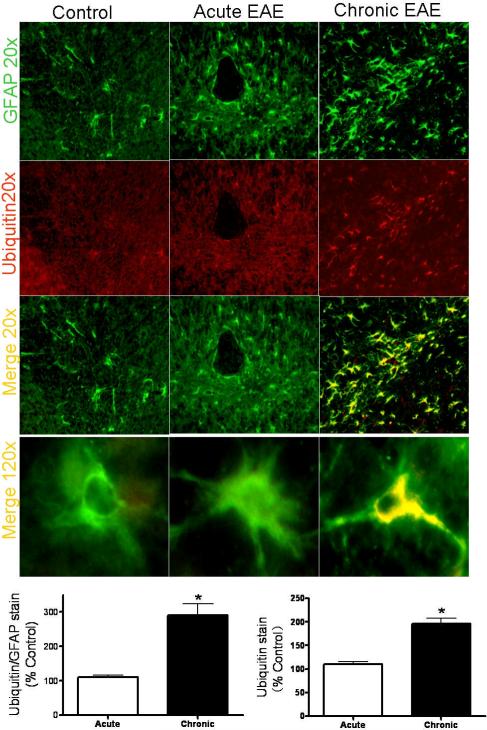
Since a significant proportion of the 20S catalytic particle is part of the 26S ubiquitin-dependent proteasome, we reasoned that the proteolytic activity of the latter, and thus the ability to remove ubiquitinated proteins, might also be compromised in chronic EAE. This possibility was explored by immunohistochemistry using a monoclonal antibody that reacts with mono/poly-ubiquitinated proteins but not with free ubiquitin (Fujimuro et al. 1994). A strong and extensive poly-ubiquitin staining, the majority of which co-localizes with GFAP, was observed in the cerebellum of chronic EAE but not acute EAE mice (Fig. 9). The intensity of poly-ubiquitinated proteins relative to that of GFAP augmented three-fold in chronic EAE compared to the age-matched control while there was no difference in this parameter between acute EAE and its control. The accumulation of ubiquitinated proteins in cerebellar astrocytes of chronic EAE mice demonstrates an impairment of proteasomal activity in this cell type. Furthermore, this finding may also explain why the majority of carbonylated proteins also accumulate within these cells (Zheng & Bizzozero 2010a).
Fig. 9.
Ubiquitinated proteins build-up inside cerebellar astrocytes of mice with chronic EAE. Double immunofluorescence analysis was performed as described in “Methods and Materials”. Green channel is for GFAP while red channel is for mono/poly-ubiquitinated proteins. Immunofluorescence images show extensive poly-ubiquitin staining only in chronic EAE. The majority (~73%) of the cells containing ubiquitinated proteins are also GFAP+. Bar graphs depict the quantification of poly-ubiquitin immunoreactivity and poly-ubiquitin divided by GFAP. *p<0.01
Discussion
We have previously shown that the proportion of carbonylated GFAP, β-tubulin and β-actin is notably higher in chronic EAE mice than in acute EAE animals (Zheng & Bizzozero 2010a). Because oxidative stress is relatively low in the chronic phase, we speculated either that the degradation system responsible for removal of oxidized proteins becomes impaired as disease progresses or that the carbonylated protein species from chronic EAE animals are more resistant to degradation. To investigate these two non-excluding possibilities, we first performed an in vitro study to identify the specific degradation system responsible for the removal of carbonylated proteins. Using LPS-stimulated astrocytes (the cell type where the majority of carbonyls accumulate in EAE) and a number of protease inhibitors, we identified the 20S proteasome as the system involved in the proteolysis of carbonylated proteins. This conclusion agrees with studies carried out in other systems (Shringarpure et al. 2003; Divald & Powell 2006). We then discovered that there is a marked reduction in the 20S proteasome chymotrypsin-like activity in chronic EAE without significant changes in the proteasome level. Moreover, based on studies in a cell-free system (Fig. 8), the possibility that oxidized cerebellar proteins from chronic EAE mice are less susceptible to proteasomal degradation seems unlikely. Thus, we conclude that the accumulation of carbonylated proteins in chronic EAE is probably the result of the direct inhibition of the proteasome's proteolytic activity. The fact that decreased proteasomal activity and increased protein carbonylation take place primarily in the same cell type (i.e. astrocytes) further support such relationship.
Mammalian cells contain several proteolytic systems including the lysosomal cathepsins, calcium-activated proteases (calpains) and the 20S/26S proteasome (Grune et al. 2001). It has been shown that degradation of carbonylated proteins is carried out in an ATP- and ubiquitin-independent manner by the 20S proteasome, which selectively recognizes and digests partially unfolded (denatured) oxidized proteins (Rivett 1985; Pacifici et al. 1993; Grune et al. 1995). However, the calcium-dependent cysteine protease calpain has been also shown to preferentially degrade oxidized neurofilament over non-oxidized protein in cell-free systems (Troncoso et al. 1995). Furthermore, there is some evidence that moderately or heavily oxidized proteins are taken up by lysosomes, where in some cases are incompletely degraded and accumulate in the form of lipofuscin-like, autofluorescent aggregates (Dunlop et al. 2009). Our study clearly shows that degradation of carbonylated proteins in LPS-treated and untreated astrocytes is carried out by 20S proteasome as neither calpain not lysosome inhibition led to an accumulation of the oxidized proteins in the 24h-incubation period. Furthermore, calpain and cathepsin B activities were not impaired in chronic EAE, where there is a build up of carbonylated species. The increased lysosomal cathepsin B activity in the cerebellum of chronic EAE mice is noteworthy. As suggested by Pandley et al. (2007), an increase in the lysosomal degradation machinery when the proteasome system is not functioning may represent a compensatory mechanism for intracellular protein degradation.
The 20S proteasome particle is composed by two outer (alpha) and two inner (beta) rings. Three of the β subunits carry the proteolytic activity, classified as caspase-like (β1), trypsin-like (β2), and chymotrypsin-like (β5), which cleaves after acidic, basic and hydrophobic amino acids, respectively (Coux et al. 1996). Of these, the β5 subunit is believed to be responsible for the degradation of oxidized (carbonylated) proteins (Ferrington et al. 2005). Thus, the decrease in 20S proteasome chymotrypsin-like activity in chronic EAE mice is consistent with the accumulation of carbonylated proteins previously reported in the same animals (Zheng & Bizzozero 2010a). Interestingly, the decrease in chymotrypsin-like activity is not due to a reduction in the number of 20S particles since α-subunit expression was unaltered in the disease. Furthermore, the various activities were not equally affected throughout the course of the disease. For instance, only chymotrypsin-like and caspase-like activities were reduced in chronic EAE while only chymotrypsin-like and trypsin-like activities were augmented in acute EAE. Based on these findings, it is fair to speculate that, as disease progresses, there is a change in the proportion of individual β-subunits within the catalytic core particle and/or that the enzyme activities are specifically inhibited. The increase in the activity associated with β2 and β5 subunits during the inflammatory phase of disease might be due to the γ-interferon-triggered replacement of these catalytic subunits by inducible subunits iβ1, iβ2 and iβ5 to form the so-called immunoproteasome, which exhibits higher chymotrypsin-like and trypsin-like activities and lower caspase-like activity (Gaczynska et al. 1993). Yet this mechanism cannot explain the reduction in caspase-like and chymotrypsin-like activity observed in the chronic animals. Most likely, the occurrence of inhibitory posttranslational modifications such as oxidation or the presence of endogenous proteasomal inhibitors such as cross-linked proteins is responsible for this phenomenon. For example, oxidative injury to the heart during ischemia-reperfusion injury has been found to induce selective rather than global inhibition of proteasomal activity (Bulteau et al. 2001a). Furthermore, specific subunits of the 20S proteasome are targeted for modification by the lipid peroxidation product 4-hydroxy-2-nonenal (4-HNE) (Farout et al. 2006), the glycoxidation product glyoxal (Bulteau et al. 2001b) and γ-ketoaldehydes (isoketals) (Davies et al. 2002). Whether similar types of modifications also take place in EAE is not known. However, using antibodies against the major reactive carbonyl species, we have not been able to detect HNE, MDA or acrolein-adducts in EAE tissue (Zheng & Bizzozero 2010a). A more likely modification of β-subunits could be the direct carbonylation of its amino acid residues as shown for those in proteasomes of hepatocellular carcinoma HepG2 cells under oxidative stress conditions (Kessova & Cederbaum 2005). Beside direct inactivation of the proteasome by oxidation/carbonylation, 4-HNE cross-linked proteins (Friguet et al. 1994) and lipofuscin/ceroid fluorescent pigments (Sitte et al. 2000) have been implicated in the inhibition of proteasome activity. An additional and still unexplored possibility might be the inactivation of proteasome activity by specific autoantibodies. Interestingly, antibodies against several proteasomal subunits have been detected in serum and CSF from MS patients (Mayo et al. 2002). It will be important to know if proteasome autoantibodies occur also in animals with chronic EAE and whether they are indeed capable of causing enzyme inhibition.
While impaired proteasomal activity has been reported in several neurodegenerative diseases, such as Alzheimer's disease (Keller et al. 2000) Parkinson's disease (McNaught et al. 2003) and Huntington's disease (Seo et al. 2004), this is the first study to demonstrate proteasome dysfunction in a chronic demyelinating disorder. Furthermore, preliminary studies in our laboratory suggest that proteasome failure also occur in chronic MS, where accumulation of both carbonylated proteins (Bizzozero et al. 2005) and ubiquitin-conjugates (Giordana et al. 2002) has been already described. The pathophysiological consequences of decreased proteasomal activity in chronic EAE as well as in classical neurodegenerative disorders are unknown. The functional impact resulting from the accumulation of a number of ubiquitinated, misfolded, aggregated and oxidized proteins, along with reduced degradation of various signaling and pro-apoptotic molecules, is likely widespread and difficult to predict. Yet decreased proteasomal activity is likely pathogenic and a contributor to neurodegeneration in chronic EAE. This notion is based mostly on experiments linking proteasomal inhibition to axonal damage (Korhonen 2004), the development of pro-inflammatory responses via up-regulation of cyclooxygenase-2 and prostaglandin E2 (Rockwell et al. 2000) and apoptosis of neurons and oligodendrocytes via mitochondrial dysfunction (Goldbaum et al. 2006).
Finally, it is puzzling why proteasomal inhibition and accumulation of both oxidized and ubiquitinated proteins occur mostly in astrocytes. One possibility is that reactive oxygen species and other molecules produced upon inflammatory activation of astrocytes (Keller et al. 1999) are capable of damaging the 20S proteasome. Since astrocytes are more resistant than neurons and oligodendrocytes to the cytotoxic consequences of proteasomal inhibition (Tsuji et al. 2005; Goldbaum et al. 2006), it is also possible that as disease progresses damaged neurons and oligodendrocytes die and are removed from the tissue leaving behind astrocytes loaded with undigested proteins. Studies in our laboratory are underway to test these possibilities.
Acknowledgements
This work was supported by PHHS grant NS057755 from the National Institutes of Health.
Abbreviations
- CFA
complete Freund's adjuvant
- DPI
days post-immunization
- EAE
experimental autoimmune encephalomyelitis
- DTT
dithiothreitol
- ECL
enhanced chemiluminescence
- GFAP
glial fibrillary acidic protein
- 4-HNE
4-hydroxy-2-nonenal
- LPS
lipopolysaccharide
- MOG
myelin-oligodendrocyte glycoprotein
- MS
multiple sclerosis
- PBS
phosphate-buffered saline
- PVDF
polyvinylidene difluoride
- SDS
sodium dodecyl sulfate
Footnotes
Disclosure/conflict of interest The authors have no conflict of interest.
References
- Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer's disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- Bizzozero OA. Protein carbonylation in neurodegenerative and demyelinating CNS diseases. In: Lajtha A, Banik N, Ray S, editors. Handbook of Neurochemistry and Molecular Neurobiology. Springer; 2009. pp. 543–562. [Google Scholar]
- Bizzozero OA, DeJesus G, Callahan K, Pastuszyn A. Elevated protein carbonylation in the brain white matter and gray matter of patients with multiple sclerosis. J. Neurosci. Res. 2005;81:687–695. doi: 10.1002/jnr.20587. [DOI] [PubMed] [Google Scholar]
- Bizzozero OA, Ziegler JL, De Jesus G, Bolognani F. Acute depletion of reduced glutathione causes extensive carbonylation of rat brain proteins. J. Neurosci. Res. 2006;83:656–667. doi: 10.1002/jnr.20771. [DOI] [PubMed] [Google Scholar]
- Brown JA, Novak EK, Swank RT. Effects of ammonia on processing and secretion of precursor and mature lysosomal enzyme from macrophages of normal and pale ear mice: evidence for two distinct pathways. J. Cell Biol. 1985;100:1894–1904. doi: 10.1083/jcb.100.6.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J. Biol. Chem. 2001a;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- Bulteau AL, Verbeke P, Petropoulos I, Chaffotte AF, Friguet B. Proteasome inhibition in glyoxal-treated fibroblasts and resistance of glycated glucose-6-phosphate dehydrogenase to 20S proteasome degradation in vitro. J. Biol. Chem. 2001b;276:45662–45668. doi: 10.1074/jbc.M105374200. [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Giustarini D, Colombo R, Rossi R, Milzani A. Protein carbonylation in human diseases. Trends Mol. Med. 2003;9:169–176. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- Davies SS, Amarnath V, Montine KS, Bernoud-Hubac N, Boutaud O, Montine TJ, Roberts LJ. Effects of reactive γ-ketoaldehydes formed by the isoprostane pathway (isoketals) and cyclooxygenase pathway (levuglandins) on proteasome function. FASEB J. 2002;16:715–717. doi: 10.1096/fj.01-0696fje. [DOI] [PubMed] [Google Scholar]
- Divald A, Powell SR. Proteasome mediates removal of proteins oxidized during myocardial ischemia. Free Radic. Biol. Med. 2006;40:156–164. doi: 10.1016/j.freeradbiomed.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Dunlop RA, Brunk UT, Rodgers KJ. Oxidized proteins: mechanisms of removal and consequences of accumulation. IUBMB Life. 2009;61:522–527. doi: 10.1002/iub.189. [DOI] [PubMed] [Google Scholar]
- Farout L, Mary J, Vinh J, Szweda LI, Friguet B. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependent on 20S proteasome subtypes. Arch. Biochem. Biophys. 2006;453:135–142. doi: 10.1016/j.abb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Browne SE, Shinobu LA, Bowling AC, Baik MJ, MacGarvey U, Kowall NW, Brown RH, Beal MF. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J. Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- Ferrington DA, Husom AD, Thompson LV. Altered proteasome structure, function and oxidation in aged muscle. FASEB J. 2005;19:644–646. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- Floor E, Wetzel MG. Increased protein oxidation in human substantia nigra pars compacta in comparison with basal ganglia and prefrontal cortex measured with an improved dinitrophenylhydrazine assay. J. Neurochem. 1998;70:268–275. doi: 10.1046/j.1471-4159.1998.70010268.x. [DOI] [PubMed] [Google Scholar]
- Friguet B, Stadman ER, Szweda LI. Modification of glucose-6-phosphate dehydrogenase by 4-hydroxy-2-nonenal: Formation of cross-linked protein that inhibits the multicatalytic protease. J. Biol. Chem. 1994;269:21639–21643. [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- Gaczynska M, Rock KL, Goldberg AL. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- Giordana MT, Richiardi P, Trevisan E, Boghi A, Palmucci L. Abnormal ubiquitination of axons in normally myelinated white matter in multiple sclerosis brain. Neuropathol. Appl. Neurobiol. 2002;28:35–41. doi: 10.1046/j.1365-2990.2002.00372.x. [DOI] [PubMed] [Google Scholar]
- Goldbaum O, Vollmer G, Richter-Landsberg C. Proteasome inhibition by MG-132 induces apoptotic cell death and mitochondrial dysfunction in cultured rat brain oligodendrocytes but not in astrocytes. Glia. 2006;53:891–901. doi: 10.1002/glia.20348. [DOI] [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Joshi M, Davies KJ. Proteolysis in cultured liver epithelial cells during oxidative stress. Role of the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1995;270:2344–2351. doi: 10.1074/jbc.270.5.2344. [DOI] [PubMed] [Google Scholar]
- Grune T, Shringarpure R, Sitte N, Davies KJ. Age-related changes in protein oxidation and proteolysis in mammalian cells. J. Gerontol. A Biol. Sci Med. Sci. 2001;56:459–467. doi: 10.1093/gerona/56.11.b459. [DOI] [PubMed] [Google Scholar]
- Haghighat N, McCandless DW, Geraminegad P. Responses in primary astrocytes and C6-glioma cells to ammonium chloride and dibutyryl cyclic-AMP. Neurochem. Res. 2000;25:277–284. doi: 10.1023/a:1007535922977. [DOI] [PubMed] [Google Scholar]
- Hassen GW, Feliberti J, Kesner L, Stracher A, Mokhtarian F. A novel calpain inhibitor for the treatment of acute experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2006;180:135–146. doi: 10.1016/j.jneuroim.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hilgart AA, Bizzozero OA. J. Neurochem. Vol. 104. Suppl.1: 2008. Carbonylation of major cytoskeletal proteins in multiple sclerosis; pp. PTW06–03. [Google Scholar]
- Kalmár B, Kittel Á, Lemmens R, Környei Z, Madarász E. Cultured aastrocytes react to LPS with increased cyclooxygenase activity and phagocytosis. Neurochem. Int. 2001;38:453–461. doi: 10.1016/s0197-0186(00)00090-5. [DOI] [PubMed] [Google Scholar]
- Kessova IG, Cederbaum AI. The effect of CYP2E1-dependent oxidant stress on activity of proteasomes in HepG2 cells. J. Pharmacol. Exp. Ther. 2005;315:304–312. doi: 10.1124/jpet.105.088047. [DOI] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Gabbita SP, Friebe V, Mattson MP, Kindy MS. Oxidized lipoproteins increase reactive oxygen species formation in microglia and astrocyte cell lines. Brain Res. 1999;830:10–15. doi: 10.1016/s0006-8993(99)01272-x. [DOI] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Marksberry WR. Impaired proteasome function in Alzheimer's disease. J. Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- Korhonen L, Lindholm D. The ubiquitin proteasome system in synaptic and axonal degeneration. J. Cell Biol. 2004;165:27–30. doi: 10.1083/jcb.200311091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaught KS, Belizaire R, Isacson O, Jenner P, Olanow CW. Altered proteasomal function in sporadic Parkinson's disease. Exp. Neurol. 2003;179:38–46. doi: 10.1006/exnr.2002.8050. [DOI] [PubMed] [Google Scholar]
- Mayo I, Arribas J, Villoslada P, Alvarez DoForno R, Rodríguez-Vilariño S, Montalban X, De Sagarra MR, Castaño JG. The proteasome is a major autoantigen in multiple sclerosis. Brain. 2002;125:2658–2667. doi: 10.1093/brain/awf274. [DOI] [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo anti-inflammatory activity. Proc. Natl. Acad. Sci. USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misko TP, Schilling RJ, Salvemini D, Moore WM, Currie MG. A fluorometric assay for the measurement of nitrite in biological samples. Anal. Biochem. 1993;214:11–16. doi: 10.1006/abio.1993.1449. [DOI] [PubMed] [Google Scholar]
- Nystrom T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24:1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici RE, Kono Y, Davies KJ. Hydrophobicity as the signal for selective degradation of hydroxyl radical-modified hemoglobin by the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1993;268:15405–15411. [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray B, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero N, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao T, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:860–864. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Rivett AJ. Preferential degradation of the oxidatively modified form of glutamine synthetase by intracellular mammalian proteases. J. Biol. Chem. 1985;260:300–305. [PubMed] [Google Scholar]
- Rockwell P, Yuan H, Magnusson R, Figueiredo-Pereira ME. Proteasome inhibition in neuronal cells induces a proinflammatory response manifested by upregulation of cyclooxygenase-2, its accumulation as ubiquitin conjugates, and production of the prostaglandin PGE(2) Arch. Biochem. Biophys. 2000;374:325–333. doi: 10.1006/abbi.1999.1646. [DOI] [PubMed] [Google Scholar]
- Rodgers KJ, Dean RT. Assessment of proteasome activity in cell lysates and tissue homogenates using peptide substrates. Int. J. Biochem. Cell Biol. 2003;35:716–727. doi: 10.1016/s1357-2725(02)00391-6. [DOI] [PubMed] [Google Scholar]
- Seo H, Sonntag KC, Isacson O. Generalized brain and skin proteasome inhibition in Huntington's disease. Ann. Neurol. 2004;56:319–328. doi: 10.1002/ana.20207. [DOI] [PubMed] [Google Scholar]
- Shringarpure R, Grune T, Mehlhase J, Davies KJ. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J. Biol. Chem. 2003;278:311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- Sitte N, Huber M, Grune T, Ladhoff A, Doecke W, Von Zglinicki T, Davies JA. Proteasome inhibition by lipofuscin/ceroid during postmitotic aging of fibroblasts. FASEB J. 2000;14:1490–1498. doi: 10.1096/fj.14.11.1490. [DOI] [PubMed] [Google Scholar]
- Smerjac SM, Bizzozero OA. Cytoskeletal protein carbonylation and degradation in experimental autoimmune encephalomyelitis. J. Neurochem. 2008;105:763–772. doi: 10.1111/j.1471-4159.2007.05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso JC, Costello AC, Kim JH, Johnson GV. Metal-catalyzed oxidation of bovine neurofilaments in vitro. Free Radic. Biol. Med. 1995;18:891–899. doi: 10.1016/0891-5849(94)00224-8. [DOI] [PubMed] [Google Scholar]
- Tsuji S, Kikuchi S, Shinpo K, Tashiro J, Kishimoto R, Yabe I, Yamagishi S, Takeuchi M, Sasaki H. Proteasome inhibition induces selective motor neuron death in organotypic slice cultures. J. Neurosci. Res. 2005;82:443–451. doi: 10.1002/jnr.20665. [DOI] [PubMed] [Google Scholar]
- Tsujinaka T, Kajiwara Y, Kambayashi J, Sakon M, Higuchi N, Tanaka T, Mori T. Synthesis of a new cell penetrating calpain inhibitor (calpeptin) Biochem. Biophys. Res. Commun. 1988;153:1201–1208. doi: 10.1016/s0006-291x(88)81355-x. [DOI] [PubMed] [Google Scholar]
- Zheng J, Bizzozero OA. Accumulation of protein carbonyls within cerebellar astrocytes in murine experimental autoimmune encephalomyelitis. J. Neurosci. Res. 2010a doi: 10.1002/jnr.22488. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Bizzozero OA. Reduced proteasomal activity contributes to accumulation of carbonylated proteins within cerebellar astrocytes in chronic EAE. Trans. Am. Soc. Neurochem. 2010b:PTW07–07. doi: 10.1111/j.1471-4159.2010.07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]