Abstract
Recognition by scavenger receptor cysteine rich domains on membrane proteins regulates innate and adaptive immune responses. Two receptors expressed primarily on T cells, CD5 and CD6 are linked genetically and are structurally similar, both containing three scavenger receptor cysteine rich domains in their extracellular regions. A specific cell surface interaction for CD5 has been difficult to define at the molecular level because of the susceptibility of CD5 protein to denaturation. By using soluble CD5 purified at neutral pH to preserve biological activity, we show that CD5 mediates species specific homophilic interactions. CD5 domain 1 only is involved in the interaction. CD5 mAbs which have functional effects in human, rat and mouse block homophilic binding. Antigen specific responses by mouse T cells in vitro were increased when engagement of human CD5 domain 1 was inhibited by mutation or by IgG or Fab fragment from a CD5 mAb. This showed that homophilic binding results in productive engagement. Enhancement of polyclonal immune responses of rat lymph node cells by an Fab fragment from a CD5 mAb shown to block homophilic interactions provided evidence that the extracellular region of CD5 regulates inhibition in normal cells. These biochemical and in vitro functional assays provide evidence that the extracellular region of CD5 regulates immunity through species specific homophilic interactions.
Introduction
CD5 and CD6 are two unusual T cell surface receptors in that their extracellular regions are each comprised of three scavenger receptor cysteine rich (SRCR) domains instead of the more common immunoglobulin superfamily (IgSF) domains (1). SRCR domains are a primitive recognition domain stretching back to invertebrates and have been retained right through evolution in various tissues and the mammalian innate and adaptive immune systems (2).
Expression of CD5 (3-5), like CD6 (6) has an inhibitory effect on the cell which is not necessarily dependent on the extracellular region (3). High levels of CD5 are associated with resistance to activation (7-9). However, there is evidence of positive effects of CD5, like CD6 (6) that depend on extracellular engagement of CD5 and by logical extrapolation that there is an endogenous ligand. A soluble CD5-Ig fusion protein produced in vivo was inhibitory in a mouse experimental allergic encephalomyelitis (EAE) model (10). The effect was species specific, mouse but not human CD5-Ig being inhibitory (10). Positive effects of CD5 were revealed in vivo in the mouse EAE model in which a cytoplasmic mutation in CD5 decreased the severity of disease (11). An overall role for costimulation by CD5 was shown by testing the effect of an immune challenge in the absence of CD5. Mice lacking CD5 were resistant to EAE (10). CD5, like CD6 is required for an optimal immune response (6, 12). The key to understanding the function of CD5 in the immune response is to define when and how the extracellular region is engaged and how this influences the signalling pathway regulated by CD5.
The heterophilic interaction between the membrane proximal SRCR domain of CD6 and its IgSF ligand, CD166 (1) is not a general paradigm for the type of domain recognised by SRCR domains. Other types of recognition by SRCR domains include the interaction between another immune system receptor, CD163 and haemoglobin-haptoglobin complexes (13), and the recognition of pathogens (2, 14). The interaction between CD6 and CD166 (1, 12), is the only example which is understood in terms of a well defined molecular interaction at the cell surface, of regulation of adaptive immune responses by an SRCR domain (6). Engagement of CD6 by CD166 in an antigen specific immune response results in phosphorylation of CD6, direct recruitment of the adaptor protein, SLP-76 by the C-terminal tyrosine motif and a net positive outcome (6).
Cell binding to immune cells has been observed with soluble forms of the extracellular region of CD5 (15-18) and with CD5 purified from cells (19). Consistent with it having a ligand binding function, cell binding was effected by domain 1 alone (17). In none of these experiments was it possible to determine whether binding was mediated by biologically active material. CD5 mAbs did not block binding (17) consistently (19, 20). The cell surface binding partner for CD5 purified from cells was identified as CD72 (19). An interaction between CD72 and CD5 has not been reproduced. Soluble recombinant CD5 did not bind cell surface CD72 (15-17) and there was no evidence for productive engagement of cell surface expressed CD5 and CD72 (16). Attempts to define the molecular identities of alternative cell surface binding partners for recombinant soluble CD5 have not been definitive (15-18) probably due to its unusual lability which may be related to a high degree of conformational flexibility as shown by NMR spectroscopy of CD5 domain 1 (21). We show that preservation of three dimensional structure is key to molecular characterisation of the ligand binding properties of CD5 and the extensive functional data can be interpreted by homophilic interactions of CD5.
Materials and Methods
Recombinant protein production
Chimaeric proteins of the extracellular region of human (h; accession number X04391) and mutants L26K, M124Q, V88D/V97K, E72K, L96P and K69D (21), rat (r; accession number NX78985: bp59A→T, bp258 T→C) and mouse (m; accession number D10728) CD5 were made. Xba I sites were introduced upstream of the initiation ATG (tctAGAAGGCCA) and Sal I sites at the end of the extracellular region (join: gtg gca tcg tcg acc → VAS ST). A BamH I site (bp2035) in hCD5 was removed. CD5, hCD6 and control CD4 leader constructs in frame with rat CD4 domain 3 and 4 and a C-terminal peptide which can be biotinylated enzymatically (rCD4d3+3bio) in the pEFBOS-XB vector were expressed in 293T cells, supernatatants biotinylated as described and tested for binding to a streptividin coated CM5 BIAcore chip (see below) (22). As the biotin tag reduced expression levels, the original rCD4d3+4 tag was used to construct hCD5CD4 and rCD5CD4 pEE14 vectors for production of stable lines (23). hCD5-His and rCD5-His pEE14 vectors were constructed by shuttling fragments via pEFBOS-XB containing a Sal I site and His tag (STHHHHHH). CD4hCD72 chimera was made as a type II chimera with the C-terminal region of human CD72 containing the C-type lectin domain fused to the C-terminus of the leader and domain 3 and 4 of rat CD4. The amino acid sequence at the join was KGLNQQTNRV(CD4 in italics).
Stable lines were produced by transfection using Fugene™ into CHO K1 cells and clones identified by either inhibition ELISA or binding to mAb at 25°C immobilised via anti-mouse Fc on a CM5 BIAcore chip. Supernatant from typically one or two cell factories (Nunc) per selected clone was either purified by affinity chromatography on OX68 (rCD4d3+4 mAb)-Sepharose™ (Pharmacia) with elution in 0.1M Gly-HCl pH 2.5 or nickel coupled to fast flow Sepharose with elution in 250 mM imidazole pH 8. Eluted material was immediately dialysed into HEPES buffered saline or PBS. Eluate was then concentrated with a Viviflow™ 200 (10,000 MW cutoff; Vivisciences, Germany) and subjected to gel filtration using Superdex™ 200 or 75 and fast flow liquid chromatography (Pharmacia). Monomeric and dimeric fractions were collected and analysed by SDS-PAGE (nuPAGE; Invitrogen) under nonreducing and reducing conditions. Yields of purified protein were in the order ~ 15 mg/l for His tagged proteins and CD4 chimaeras. Theoretical molecular weights and molar extinction coefficients are: hCD5CD4, 59,932 kDa, 75390 M−1 cm−1; rCD5CD4, 60,137 kDa, 64020 M−1 cm−1; hCD5-His, 40445 kDa, 55,760 M−1 cm−1; rCD5-His, 40650 kDa, 43670 M−1 cm−1; mCD5-His, 39762 kDa, 43670 M−1 cm−1.
mAbs
Monoclonal antibodies used as tissue culture supernatant or purified IgG were specific for: human CD5 domain 1, LEU1 (mIgG2a) (ATCC) and UCHT2 (mIgG1) (gift from Doreen Cantrell); human CD5 domain 3, CD5-48 (mIgG1; produced against soluble purified hCD5CD4 by Giovanna Roncador and Jackie Cordell, Oxford); rat CD5 domain 1, OX19 (mIgG1) (24); mouse CD5 domain 1, 53.7.131 (rIgG2a) (ATCC), rat TCR mAb, R73 (24), human CD72, BU40; human CD6, T12 (ATCC); rat CD4 domains 3 and 4, OX68 (23); control OX21 (IgG1). Recognition by CD5-48 mAb was mapped using hCD5CD4, hCD5 domain 1 CD4, rCD5CD4 and definitively with CD5 domain 3 made in E. coli by Acely Garcia-Garza (21). Fab fragments were prepared by papain digestion and gel filtration.
BIAcore Analyses
For analysis of homophilic interactions, CM5 chips were coated with streptavidin at 25°C and then the temperature was raised to 37°C for immobilisation of biotinylated CD4d3+4 chimaeric proteins (5 μl/min) at equivalent levels, in the range of 1,500-3,000 Response Units (RU) and subsequent injection (5 μl) of increasing concentrations of soluble protein (20 μl/min) to measure equilibrium binding (12, 22, 25). The rack containing proteins to be injected was cooled to 5-10°C to minimise aggregation and denaturation during the time scale of the experiment. Specific equilibrium binding was determined by subtracting responses in a control flowcell. mAbs were injected as tissue culture supernatant or purified (~1-10 μg/ml) at (5 μl/min) over chimaeric proteins or polyclonal rabbit anti-mouse IgG (Pharmacia.). CD5-His (~1 μM) was injected (5 μl/min) over immobilised mAb. For analysis of mAb blocking flowcells, double volumes of CD5-His were prepared and injected before and after saturation of immobilised CD5 with mAb. CD5 domain 1 mutants were immobilised on the chip from tissue culture supernatant via the rat CD4d3+4 mAb, OX68 for screening their effect on homophilic binding by hCD5-His or hrCD5-His at 25°C.
Functional assays
Antigen specific IL-2 production of the mouse 2B4 hybridoma cells (105) in response to moth cytochrome c peptide (mcc) presented by Chinese hamster ovary cells expressing IEK (CHO IEK; 5 ×104 −105) were carried out as described previously in round bottomed wells of 96 well plates (26). IL-2 production after 18-24h was measured by ELISA. mAbs and Fab fragments were used at a final concentration of 5 μg/ml. Stimulation of lymph node cells from female Lewis rats was carried out as described (24). Following red cell lysis lymph node cells were plated out at 2.5×105/well in round bottomed wells of 96 well plates and preincubated for 30 min at 0°C with antibody or Fab fragments at a final concentration, 5 μg/ml. Concanavalin A (5 μg/ml) and TCR mAb (R73) (5 μg/ml) were added and incubated along with the cells for 3 days at 37°C. 18 hours prior to harvesting, 0.5 μl, [methyl-3H] thymidine (185 MBq/5 ml; Amersham) were added per well and cells and incorporation into cells determined by scintillation counting. Student T-test was carried out on samples in triplicate and significance determined by p<0.05.
Results
Species specific homophilic interactions of CD5
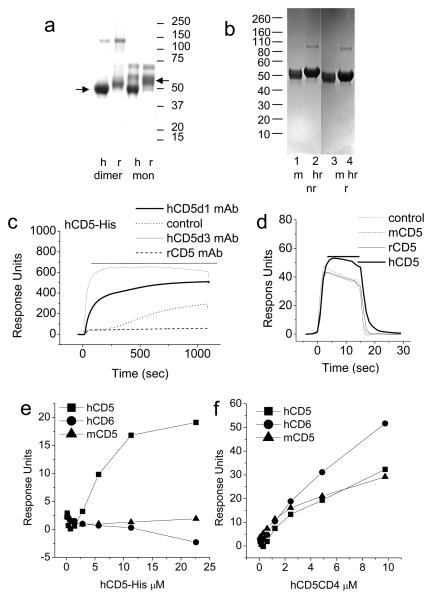
Similarities in the composition of the extracellular regions of CD5 and CD6 suggested ligand binding properties in common. However, during screens using surface plasmon resonance (SPR) for possible interactions of CD5 with cell surface proteins including CD166 and the related protein named BCAM or Lutheran there was no binding (data not shown). Instead, we noticed a weak self association of the purified recombinant human CD5CD4, a fusion protein comprised of the extracellular region of human CD5 and domains 3 and 4 of rat CD4. We examined the potential for CD5 for self association and to eliminate the risk of denaturation due to purification at pH 2.5 using affinity chromatography, we produced His tagged versions of human and rat CD5 which were purified at neutral pH. On gel filtration, hCD5-His and rCD5-His separated as two peaks (not shown), with elution volumes consistent with asymmetric proteins forming dimers. The majority of material was monomeric. Both peaks migrated as monomers on SDS PAGE under nonreducing (Fig. 1a, b) and reducing conditions consistent with a noncovalent association. Species specific saturable binding by purified hCD5-His to immobilised hCD5 domain 1 (LEU1 and UCHT2, not shown) and domain 3 mAbs was observed indicating that the material was antigenically active (Fig. 1c). Rat CD5-His bound the rCD5 mAb, OX19 (not shown). The hCD5-His gave some binding to the high levels of immobilised rabbit IgG (Fig. 1c). Such a slow on rate indicates this is probably due to traces of aggregated material.
Figure 1. Species specific homophilic binding of CD5.
(a, b) hCD5-His, rCD5-His, mCD5-His and hrCD5-His migrated as monomers in SDS-PAGE analysis under nonreducing or reducing (not shown) consistent with noncovalent dimer formation in gel filtration (not shown). (c) Monomeric hCD5-His (~0.3 μM) and rCD5-His (not shown) bound immobilised CD5 domain 1 (LEU1) and 3 (CD5-48) mAbs. Over time slow binding of presumably aggregated material to the rabbit anti- mouse Fc was observed. (d) BIAcore sensorgram traces of dimeric hCD5-His (2 μM) over hCD5CD4 (1474 RU), rCD5CD4 (1941 RU), mCD5CD4 (2790 RU) and control IgSF protein rCD4d3+4 (1582 RU) immobilised on streptavidin coated flowcells. The horizontal line indicates injection period. (e, f) Equilibrium binding data at 37°C from injections of increasing concentrations of monomeric hCD5-His or hCD5CD4 over hCD5CD4 (1434 RU), hCD6CD4 (1931 RU), mCD5CD4 (1477 RU) and control IgSF protein hTREM1CD4 (1439 RU) immobilised on streptavidin coated flowcells. At the end of the experiment, species specific CD5 mAb binding were injected over all flowcells to confirm the identity of immobilised proteins.
We tested soluble CD5-His for self association by SPR and to mimic physiological conditions all experiments were carried out at 37°C. Biotinylated CD5CD4 fusion proteins which had not been subjected to denaturing purification conditions were immobilised on a streptavidin coated chip and serial dilutions of human CD5-His injected (Fig. 1d). Specific homophilic interactions of hCD5-His were detected (Fig. 1d). The interaction was weak as indicated by a rapid dissociation whether the dimeric or monomeric fraction was used (Fig. 1d and data not shown). Plotting the equilibrium binding data for binding a series of CD5 concentrations revealed concentration dependent species specific binding of hCD5-His to hCD5CD4 (Fig. 1e).
In the complementary experiment, rCD5-His bound specifically to immobilised rat and mouse CD5CD4 equally well (see Fig. 2b and data not shown) and vice versa mCD5-His to rCD5CD4 and mCD5CD4 (see Fig. 2d and f). Thus CD5 binds CD5 reproducibly in human, mouse and rat. Human CD5 reacts only with human CD5 whereas mouse and rat CD5 can crossreact.
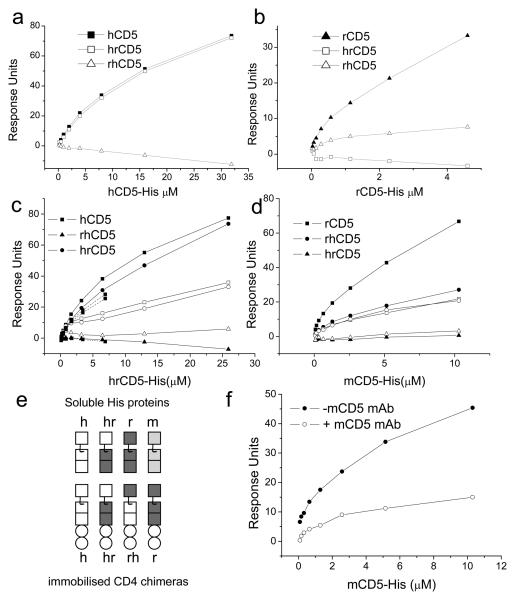
Figure 2. Homophilic interactions between CD5 domain 1.
(a-d) Chimaeric proteins contain rCD4 domains 3 and 4 (circles) hrCD5CD4, rhCD5CD4, hCD5CD4 and rCD5CD4 were immobilised (~2,000-3,000 RU) on streptavidin coated flowcells. Increasing concentrations of (a) hCD5-His, (b) rCD5-His, (c) hrCD5-His or (d) mCD5-His were injected over all flowcells at 37°C. Equilibrium binding data, relative to responses over rCD5CD4 (a, c) or hCD5CD4 (b, d) are plotted. The identity and equivalent levels of immobilised proteins were confirmed with mAb specific for hCD5 domain 1, (LEU1), rCD5 domain 1, (OX19) and hCD5 domain 3 (not shown). (c) Binding before (closed symbols) and after (open symbols) saturation of immobilised proteins with LEU1 (hCD5CD4, hrCD5CD4) or OX19 (rCD5CD4 and rhCD5CD4) is shown. (e) Recombinant proteins are shown schematically. SRCR domains (squares) of human, rat and mouse CD5 are depicted, domain 1 uppermost. (e) mCD5CD4 was saturated with 53.7 or control rCD4 mAb, OX68 mAb and mCD5-His reinjected.
CD5 is susceptible to denaturation
The behaviour of soluble human CD5CD4 purified at pH 2.5 was analysed in the same way as hCD5-His in the same experiment using the same chip (Fig. 1e and f). Soluble hCD5CD4 purified at pH 2.5 gave binding to both human and mouse CD5 but also the control protein, hCD6CD4 and showed none of the discrimination seen with hCD5-His.
Domain 1 is necessary for homophilic interactions of CD5
The N-terminal domain of CD5 is topologically suited to mediate ligand binding (2). To determine if species specific homophilic binding depends on the N-terminal membrane distal domain of CD5, we constructed chimaeric CD5CD4 proteins containing human domain 1 and rat domains 2 and 3 (hrCD5CD4; see Fig 2e) and the complementary protein containing rat domain 1 and human domains 2 and 3 (rhCD5CD4) (Fig. 2). Soluble hCD5-His or rCD5-His was injected over equivalent levels of CD5CD4 or the chimeras (Fig. 2). Species specific binding was calculated by subtracting responses in the rCD5CD4 flowcell for hCD5-His and vice versa for rCD5-His. Human CD5-His bound equally well to hCD5CD4 and hrCD5CD4 showing that human CD5 domain 1 was necessary for species specific homophilic binding (Fig. 2a). Rat CD5-His bound in a species specific manner to rCD5CD4 but poorly to rhCD5CD4 (Fig. 2b). Nevertheless, the weak binding indicates that rat CD5 domain 1 mediates species specific homophilic binding. The identity and levels of immobilisation of all four proteins were checked using mAbs specific for human CD5 domains 1 (LEU1) and 3 (CD5-48) and rat CD5 domain 1 (OX19) (data not shown). Rat CD5 mAb, OX19 was shown to recognise domain 1 using a chimaeric CD4 protein containing rat domain 1 and mouse domains 2 and 3 (data not shown).
Homophilic interactions between CD5 domain 1
Binding between hCD5-His and hrCD5CD4 shows that CD5 domain 1 is necessary for homophilic interactions. There are several possible topologies of homophilic interactions of CD5 that comply with this result. Theoretically, any one of the domains in hCD5-His could bind domain 1 of the immobilised material. To ask whether only CD5 domain 1 is involved in the species specific homophilic interactions, we produced soluble hrCD5-His (Fig. 1b) and tested it for binding to hrCD5CD4. Human rat CD5-His bound equally well to hCD5CD4 and hrCD5CD4 but not to rCD5CD4 or rhCD5CD4 (Fig. 2c). These data show that human CD5 domain 1 binding to itself is the dominant interaction in the species specific homophilic interactions. In the complementary experiment with the same chip (Fig. 2d), mCD5-His bound the chimeras containing rat CD5 domain 1 confirming the identity and activity of the chimeras on the chip and the cross reactivity between mouse and rat CD5 homophilic interactions. The validity of CD5 homophilic binding is emphasized by it being demonstrated in three species and with two types of recombinant proteins, His tagged and CD4d3+4 fusion proteins.
CD5 domain 1 mAbs inhibit homophilic interactions of CD5
An understanding of the potential of CD5 domain 1 mAbs to block ligand binding by CD5 aids in mapping the ligand binding site on CD5 domain 1 and is important for interpretation of their effects in functional assays. Two hCD5 domain 1 mAbs, LEU1 and UCHT2 recognise epitopes which do not overlap (21). We analysed the effect of saturating immobilised hCD5CD4 with LEU1 on homophilic binding by hCD5-His and hrCD5-His. LEU1 specifically blocked binding of hCD5-His to hCD5CD4 (data not shown and Fig. 2c). Blocking of homophilic binding by UCHT2 was not observed (see Fig. 3b).
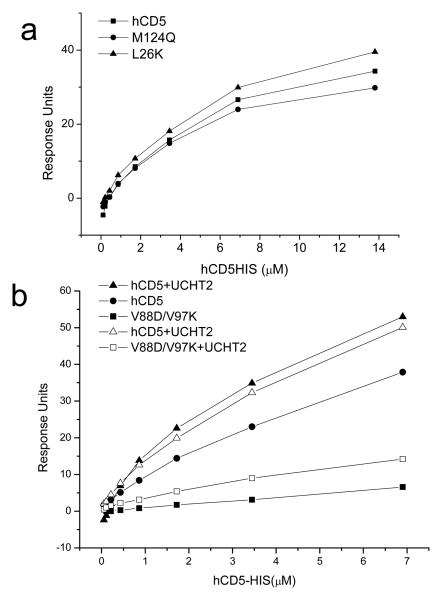
Figure 3. The V88D/V97K CD5 domain 1 mutant does not bind homophilically.
(a, b) hCD5CD4, L26KCD4, M124QCD4, V88D/V97KCD4 and rCD5CD4 (~3,000 RU) were immobilised on streptavidin coated flowcells. (b) Increasing concentrations of hCD5-His were injected over all flowcells at 37°C (closed symbols). (b) hCD5-His was injected over hCD5CD4 saturated with UCHT2 mAb, hCD5CD4 and V88D/V97KCD4 (closed symbols). V88D/V97KCD4 was then saturated with UCHT2 mAb and hCD5-His reinjected (open symbols). Binding to hCD5CD4 saturated with UCHT2 was the same in the two sets of injections. Equilibrium binding data, relative to responses over rCD5CD4 are plotted.
We tested the rat CD5 domain 1 mAb (OX19) for its effect on CD5 ligand binding. OX19 blocked rat CD5 homophilic binding (Fig. 2d). Similarly, saturation of mCD5CD4 with a mAb (53.7) specific for domain 1 (27) inhibited mouse CD5 homophilic binding (Fig. 2f). Cross species blocking by the rat (OX19) (Fig. 2d) and mouse (53.7) CD5 mAbs (not shown) showed that soluble CD5-His did not compete for immobilised mAb.
The CD5 domain 1 V88D/V97K mutant does not bind homophilically
A clear separation between the epitopes of LEU1 and UCHT2 was demonstrated by a single mutation, L26K in hCD5CD4 which specifically abolished recognition by UCHT2 mapping its epitope to the N-terminus of CD5 domain (21). The L26K hCD5CD4 mutant, which is not recognised by UCHT2, bound CD5-His as well as wild type CD5 (Fig. 3a) consistent with the N-terminus of CD5 domain 1 not being essential for homophilic interactions. The mutant gave a clearer result than testing for blocking with UCHT2 which appeared to crosslink immobilised and soluble CD5 whether it be the wild type or a nonbinding mutant (Fig. 3b). Mutation of a residue, M124Q in the region defined as critical for the interaction between CD6 and CD166 did not have a drastic effect on homophilic binding (Fig. 3a). We analysed a panel of human CD5 domain 1 mutants for their effect on homophilic binding (Table 1). Two mutants showed reduced binding, E72K and V88D/V97K (Table 1 and Fig. 3b). Expression and LEU1 mAb binding of E72K was reduced relative to wild type CD5 which may indicate structural instability. V88D/V97K identified as nonbinding is the mutant from which the NMR structure (21) was derived thus maintains structural integrity. In the screen of the panel of mutants, immobilised recombinant human CD72 was used as a negative control and its identity checked with the CD72 mAb, BU40.
Table 1. Identification of residues critical for homophilic binding of human CD5 domain 1.
Soluble hCD5-His or hrCD5-His was passed over CD4 chimeras immobilised directly from tissue culture supernatant onto a BIAcore chip coated with the CD4d3+4 mAb, OX68 at 25°C and binding at equilibrium measured.
| protein | hCD5 binding | LEU1 binding |
|---|---|---|
| CD4hCD72A | − | − |
| E72K | − | 75% wt |
| L96P | + | + |
| V88D/V97K | − | 80-100% wt |
| K69D | + | + |
| M124Q | + | + |
| L26K | + | + |
Homophilic interactions of CD5 regulate an immune response
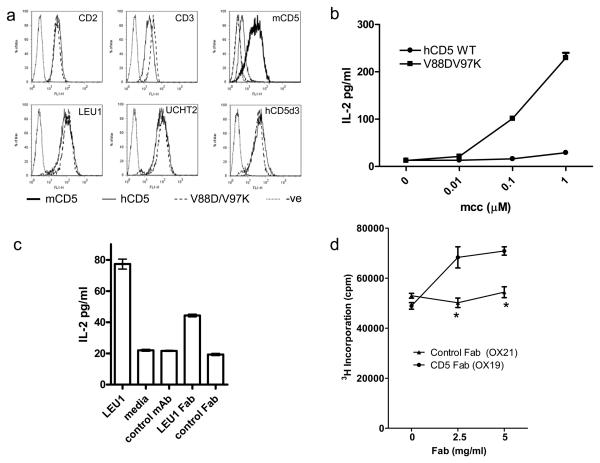
We developed a well controlled in vitro assay in which we could detect functional effects of engagement of CD5 domain 1. Wild type and mutant (V88D/V97K) CD5 were expressed in 2B4 mouse T hybridoma cells. The human CD5 variants were highly expressed as monitored by CD5 domain 1 and 3 mAbs (Fig. 4a). Consistently, CD3 levels were higher on cells expressing mutant CD5 (Fig. 4a). Endogenous mouse CD5 was downregulated on cells expressing human CD5 while transfected mouse CD2 remained unchanged (Fig. 4a). The cells transfected with mouse CD2 were used as the parental line to boost levels of antigen specific IL-2 in the stimulation assays. Cells were stimulated with peptide presented by Chinese hamster ovary cells expressing the MHC Class II, IEK (CHO IEK cells) or CHO IEK cells expressing mouse CD48 (28). A similar pattern of responses was observed in response to both types of antigen presenting cell with higher IL-2 production for the ones expressing mouse CD48 (not shown).
Figure 4. Homophilic interactions of CD5 regulate an immune response.
(a) Mouse 2B4 T hybridoma cells were analysed by flow cytometry for expression of transfected mCD2, mCD3, mCD5 and transduced hCD5 with hCD5 domain 1 (LEU1 and UCHT2) and domain 3 (hCD5d3) mAbs. (b-c) Antigen specific IL-2 production by 2B4 hybridoma cells in response to (b) CHO IEK or CHO IEK expressing mCD48 (c) and (b) different concentrations or (c) 1 μM mcc peptide were enhanced by expression of V88D/V97K compared with wild type hCD5 (b) or blocking with a hCD5 domain 1 (LEU1) or Fab (LEU1) and not with a control mAb (OX21) or Fab (T12) (c). The difference between LEU1 and control is significant for both IgG and Fab (p<0.001) (d) Proliferation of polyclonally activated rat lymph node cells was enhanced by Fab fragments of rat CD5 mAb (OX19) and not control Fab (OX21) (* p<0.01). The data shown are representative of ≥ 3 independent experiments.
Antigen specific IL-2 production by cells expressing mutant CD5 was increased relative to wild type CD5 (Fig 4b). To ensure this was related to engagement of human CD5 domain 1 and not just the higher CD3 levels on the mutant cells, the effect of a soluble blocking CD5 mAb on responses by cells expressing wild type human CD5 was analysed. The human CD5 mAb which blocked species specific homophilic interactions enhanced antigen specific IL-2 production (Fig. 4c). Confirmation that this was due to blocking was obtained by showing a Fab fragment also enhanced IL-2 production (Fig. 4c). Functional effects of human CD5 domain 1 in mouse cells are consistent with species specific homophilic interactions resulting in productive engagement.
Dissection in the hybridoma model revealed the potential of the extracellular region of CD5 to regulate inhibitory effects of CD5. To assess its physiological relevance, we examined the potential of the extracellular region of CD5 to regulate inhibition of immune responses by a normal polyclonal cell population. We analysed the mode of action of the rat CD5 mAb, OX19 which has been shown to have costimulatory effects on primary cells (24). The rat CD5 mAb, OX19 blocks homophilic interactions (Fig. 2c). Comparison of the effects of freshly gel filtrated Fab fragments with Fab'2 or IgG (not shown, (24) ) showed that all three reagents enhanced polyclonal proliferation of rat lymph node cells (Fig. 4d). CD5 was not modulated off the surface by OX19 IgG or Fab in vitro (not shown). The data are consistent with homophilic interactions of CD5 regulating activation of normal cells.
Discussion
Molecular characterisation of homophilic binding by CD5 has resolved two major sources of confusion over immune regulation by this receptor. Firstly, specific and nonspecific binding by the extracellular region of CD5 were distinguished. CD5 interacted with itself in a species specific manner. Secondly, an understanding that the extracellular region of CD5 can engage in homophilic binding provides a molecular mechanism for functional effects of perturbing engagement of the extracellular region of CD5.
The biochemical and functional data indicated that homophilic CD5 interactions were strong enough to mediate productive interactions to regulate immune responses. The micromolar concentrations of soluble protein which yielded measurable binding to immobilised material were in the range of those observed for other weak but physiological relevant interactions (12, 25). The kinetics of CD5 binding revealed rapid on and off rates consistent with transient interactions typical for cell surface proteins (12, 22). Low stoichiometry precluded good mathematical analysis as we have done for other homophilic interactions (12, 25). Low stoichiometry of binding measured by SPR could be due to the presence of inactive protein or a consequence of self association by the soluble material competing with immobilised.
The presence of a region of particular flexibility in the NMR solution structure of human CD5 domain 1 (21) provides a molecular basis for the difficulty in identifying specific ligand binding. Interactions of CD5 across species (15, 16, 19) or on cells which do not express CD5 (17); Alan Johnstone, unpublished) may be explained by the capacity of denatured CD5 in CD5 preparations to give non specific binding. Promiscuous binding by denatured CD5 may reflect a general propensity of SRCR domains including CD5, to multerimise (14, 29, 30).
Homophilic binding by CD5 domain 1 may provide more of a general paradigm for recognition by SRCR domains than the interaction between an IgSF domain of CD166 and the membrane proximal SRCR domain of CD6. Domain 1 of CD6 does not participate in the interaction with CD166 (1, 12) but by analogy with CD5, its membrane distal position indicates a ligand binding function. There are data to indicate a significant role for CD6 domain 1 in regulation by CD6 in immune responses. Inhibitory effects of two CD6 mAbs characterised as specific for different epitopes on the membrane distal N-terminal domain have been described (31-34). As they were effective in soluble form, it is likely they are blocking an interaction. Importantly, the T1 mAb has shown therapeutic immunosuppressive effects in patients (33, 34). This could involve a homophilic interaction of CD6 domain 1. A ligand binding function of CD6 domain 1 is also suggested by the existence of a form of CD6 which lacks the membrane proximal CD166 binding domain on activated T cells (35).
Homophilic binding by CD5 domain 1 has the potential to mediate interactions between cells, in trans or on the same cell, in cis. The lack of involvement or of a subsidiary role for domains 2 and 3 in CD5 homophilic interactions favours trans interactions as might occur between T cells or T cells and B cells (16). Alternatively, the absence of CD5 on dendritic cells at the initiation of an immune response favours cis interactions. The two topologies for CD5 homophilic binding are not mutually exclusive. There is good precedent for competition between cis and trans interactions in the Siglec family (36) and interactions between Ly49 and MHC Class I (37). The highly conserved flexible stalk between CD5 domains 1 and 2 is likely to be important in regulating potential trans versus cis CD5 interactions as has been recently shown for Ly49 (37). The exclusive role of CD5 domain 1 in species specific homophilic interactions differs from the involvement of all three SRCR domains of CD5 in an interaction with fungal wall components (38). The involvement of domains 2 and 3 of CD5 in other interactions (39-41) is not excluded by the homophilic binding data.
Correlation of activation and inhibition by CD5 with a requirement for extracellular engagement is not absolute. Dissection of engagement of CD5 in the cell line model show the potential of the extracellular region of CD5 to regulate its inhibitory effects. Caution must be exercised in dismissing the extracellular region in regulating inhibition by CD5 based on experiments with genetically manipulated mice (3). To demonstrate a role for the extracellular region of CD5, it is important to distinguish between engagement of the extracellular and intracellular region. Simply observing differences between CD5+ and CD5− cells does not make this distinction. An inhibitory role for the cytoplasmic region of CD5 was shown in mice expressing CD5 lacking an extracellular region (3). To test whether interactions of the extracellular region contribute to regulation by CD5, we used a CD5 mAb which blocks CD5 homophilic interactions. In retrospective analysis of the literature with the knowledge that a particular CD5 mAb blocks homophilic binding, rigorous analysis of the mode of action is required before functional effects of the mAb for example, OX19 (24) can be interpreted. To ensure the mode of action of OX19 was blocking, we used an Fab fragment. Enhancement of proliferation by rat lymph node cells by Fab fragments from OX19 provides good evidence that the extracellular region of CD5 is important for controlling activation in normal immune responses. A molecular understanding of the effect of ligand binding by the extracellular region of CD5 allows the relative contributions of extracellular and intracellular engagement of CD5 to be more clearly delineated and provides a molecular basis for interpreting functional data.
Homophilic binding by CD5 provides a molecular basis for species specific inhibition of EAE in mouse by soluble CD5-Ig (10). The mouse CD5 mAb (53.7) which blocks homophilic interactions has also been reported to have functional effects, inhibiting B cell activation (16) which, depending on the mode of action of the mAb in these experiments, would be evidence for ligand dependent positive effects of CD5 in T-B cell collaboration. The difference between the overall effect of CD5 engagement being activating or inhibitory may depend on the number and activation status of responding cells in a polyclonal population. The evidence is that the extracellular region of CD5 is important for regulating both activation and inhibition by CD5 and the molecular mechanism involves species specific homophilic interactions of CD5 domain 1.
Acknowledgements
We thank the following for their support in this project, Steve Simmonds, Andrew McGrath, Alan Johnstone, Nicholas Clarkson, members of the lab and Neil Barclay.
The Medical Research Council, UK (G0400808) provided financial support.
Non standard abbreviations
- CD5CD4
extracellular region of CD5 fused to rat CD4 domains 3 and 4
- CD5-His
extracellular region of CD5 fused to a His tag
- h
human
- r
rat
- m
mouse
- SPR
surface plasmon resonance
References
- 1.Aruffo A, Bowen MA, Patel DD, Haynes BF, Starling GC, Gebe JA, Bajorath J. CD6-ligand interactions: a paradigm for SRCR domain function? Immunol. Today. 1997;18:498–504. doi: 10.1016/s0167-5699(97)01130-4. [DOI] [PubMed] [Google Scholar]
- 2.Sarrias MR, Gronlund J, Padilla O, Madsen J, Holmskov U, Lozano F. The Scavenger Receptor Cysteine-Rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit. Rev. Immunol. 2004;24:1–37. doi: 10.1615/critrevimmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- 3.Bhandoola A, Bosselut R, Yu Q, Cowan ML, Feigenbaum L, Love PE, Singer A. CD5-mediated inhibition of TCR signaling during intrathymic selection and development does not require the CD5 extracellular domain. Eur. J. Immunol. 2002;32:1811–1817. doi: 10.1002/1521-4141(200206)32:6<1811::AID-IMMU1811>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 4.Pena-Rossi C, Zuckerman LA, Strong J, Kwan J, Ferris W, Chan S, Tarakhovsky A, Beyers AD, Killeen N. Negative regulation of CD4 lineage development and responses by CD5. J. Immunol. 1999;163:6494–6501. [PubMed] [Google Scholar]
- 5.Tarakhovsky A, Kanner SB, Hombach J, Ledbetter JA, Muller W, Killeen N, Rajewsky K. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 1995;269:535–537. doi: 10.1126/science.7542801. [DOI] [PubMed] [Google Scholar]
- 6.Hassan NJ, Simmonds SJ, Clarkson NG, Hanrahan S, Puklavec MJ, Bomb M, Barclay AN, Brown MH. CD6 Regulates T-Cell Responses through Activation-Dependent Recruitment of the Positive Regulator SLP-76. Mol. Cell Biol. 2006;26:6727–6738. doi: 10.1128/MCB.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawiger D, Masilamani RF, Bettelli E, Kuchroo VK, Nussenzweig MC. Immunological unresponsiveness characterized by increased expression of CD5 on peripheral T cells induced by dendritic cells in vivo. Immunity. 2004;20:695–705. doi: 10.1016/j.immuni.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Ryan KR, McCue D, Anderton SM. Fas-mediated death and sensory adaptation limit the pathogenic potential of autoreactive T cells after strong antigenic stimulation. J. Leukoc. Biol. 2005;78:43–50. doi: 10.1189/jlb.0205059. [DOI] [PubMed] [Google Scholar]
- 10.Axtell RC, Webb MS, Barnum SR, Raman C. Cutting edge: critical role for CD5 in experimental autoimmune encephalomyelitis: inhibition of engagement reverses disease in mice. J. Immunol. 2004;173:2928–2932. doi: 10.4049/jimmunol.173.5.2928. [DOI] [PubMed] [Google Scholar]
- 11.Axtell RC, Xu L, Barnum SR, Raman C. CD5-CK2 binding/activation-deficient mice are resistant to experimental autoimmune encephalomyelitis: protection is associated with diminished populations of IL-17-expressing T cells in the central nervous system. J. Immunol. 2006;177:8542–8549. doi: 10.4049/jimmunol.177.12.8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassan NJ, Barclay AN, Brown MH. Frontline: Optimal T cell activation requires the engagement of CD6 and CD166. Eur. J. Immunol. 2004;34:930–940. doi: 10.1002/eji.200424856. [DOI] [PubMed] [Google Scholar]
- 13.Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, Moestrup SK. Identification of the haemoglobin scavenger receptor. Nature. 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 14.Ojala JR, Pikkarainen T, Tuuttila A, Sandalova T, Tryggvason K. Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J. Biol. Chem. 2007;282:16654–16666. doi: 10.1074/jbc.M701750200. [DOI] [PubMed] [Google Scholar]
- 15.Biancone L, Bowen MA, Lim A, Aruffo A, Andres G, Stamenkovic I. Identification of a novel inducible cell-surface ligand of CD5 on activated lymphocytes. J. Exp. Med. 1996;184:811–819. doi: 10.1084/jem.184.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bikah G, Lynd FM, Aruffo AA, Ledbetter JA, Bondada S. A role for CD5 in cognate interactions between T cells and B cells, and identification of a novel ligand for CD5. Int. Immunol. 1998;10:1185–1196. doi: 10.1093/intimm/10.8.1185. [DOI] [PubMed] [Google Scholar]
- 17.Calvo J, Places L, Padilla O, Vila JM, Vives J, Bowen MA, Lozano F. Interaction of recombinant and natural soluble CD5 forms with an alternative cell surface ligand. Eur. J. Immunol. 1999;29:2119–2129. doi: 10.1002/(SICI)1521-4141(199907)29:07<2119::AID-IMMU2119>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 18.Haas KM, Estes DM. The identification and characterization of a ligand for bovine CD5. J. Immunol. 2001;166:3158–3166. doi: 10.4049/jimmunol.166.5.3158. [DOI] [PubMed] [Google Scholar]
- 19.Van de Velde H, von Hoegen I, Luo W, Parnes JR, Thielemans K. The B-cell surface protein CD72/Lyb-2 is the ligand for CD5. Nature. 1991;351:662–665. doi: 10.1038/351662a0. [DOI] [PubMed] [Google Scholar]
- 20.Van de Velde H, Thielemans K. Native soluble CD5 delivers a costimulatory signal to resting human B lymphocytes. Cell. Immunol. 1996;172:84–91. doi: 10.1006/cimm.1996.0218. [DOI] [PubMed] [Google Scholar]
- 21.Garza-Garcia A, Esposito D, Rieping W, Harris R, Briggs C, Brown MH, Driscoll PC. Three-dimensional solution structure and conformational plasticity of the N-terminal scavenger receptor cysteine-rich domain of human CD5. J. Mol. Biol. 2008;378:129–144. doi: 10.1016/j.jmb.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Brown MH, Boles K, van der Merwe PA, Kumar V, Mathew PA, Barclay AN. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J. Exp. Med. 1998;188:2083–2090. doi: 10.1084/jem.188.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown MH, Barclay AN. Expression of immunoglobulin and scavenger receptor superfamily domains as chimeric proteins with domains 3 and 4 of CD4 for ligand analysis. Prot. Eng. 1994;7:515–521. doi: 10.1093/protein/7.4.515. [DOI] [PubMed] [Google Scholar]
- 24.Dallman MJ, Thomas ML, Green JR. MRC OX-19: a monoclonal antibody that labels rat T lymphocytes and augments in vitro proliferative responses. Eur. J. Immunol. 1984;14:260–267. doi: 10.1002/eji.1830140311. [DOI] [PubMed] [Google Scholar]
- 25.Mavaddat N, Mason DW, Atkinson PD, Evans EJ, Gilbert RJ, Stuart DI, Fennelly JA, Barclay AN, Davis SJ, Brown MH. Signaling lymphocytic activation molecule (CDw150) is homophilic but self-associates with very low affinity. J. Biol. Chem. 2000;275:28100–28109. doi: 10.1074/jbc.M004117200. [DOI] [PubMed] [Google Scholar]
- 26.Clarkson NG, Simmonds SJ, Puklavec MJ, Brown MH. Direct and Indirect Interactions of the Cytoplasmic Region of CD244 (2B4) in Mice and Humans with FYN Kinase. J. Biol. Chem. 2007;282:25385–25394. doi: 10.1074/jbc.M704483200. [DOI] [PubMed] [Google Scholar]
- 27.Starling GC, Llewellyn MB, Whitney GS, Aruffo A. The Ly-1.1 and Ly-1.2 epitopes of murine CD5 map to the membrane distal scavenger receptor cysteine-rich domain. Tissue Antigens. 1997;49:1–6. doi: 10.1111/j.1399-0039.1997.tb02702.x. [DOI] [PubMed] [Google Scholar]
- 28.Clarkson NG, Brown MH. Inhibition and activation by CD244 depends on CD2 and phospholipase C-gamma1. J. Biol. Chem. 2009;284:24725–24734. doi: 10.1074/jbc.M109.028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAlister MS, Brown MH, Willis AC, Rudd PM, Harvey DJ, Aplin R, Shotton DM, Dwek RA, Barclay AN, Driscoll PC. Structural analysis of the CD5 antigen--expression, disulphide bond analysis and physical characterisation of CD5 scavenger receptor superfamily domain 1. Eur. J. Biochem. 1998;257:131–141. doi: 10.1046/j.1432-1327.1998.2570131.x. [DOI] [PubMed] [Google Scholar]
- 30.McAlister MS, Davis B, Pfuhl M, Driscoll PC. NMR analysis of the N-terminal SRCR domain of human CD5: engineering of a glycoprotein for superior characteristics in NMR experiments. Prot. Eng. 1998;11:847–853. doi: 10.1093/protein/11.10.847. [DOI] [PubMed] [Google Scholar]
- 31.Singer NG, Richardson BC, Powers D, Hooper F, Lialios F, Endres J, Bott CM, Fox DA. Role of the CD6 glycoprotein in antigen-specific and autoreactive responses of cloned human T lymphocytes. Immunology. 1996;88:537–543. [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmerman AW, Joosten B, Torensma R, Parnes JR, van Leeuwen FN, Figdor CG. Long-term engagement of CD6 and ALCAM is essential for T-cell proliferation induced by dendritic cells. Blood. 2006;107:3212–3220. doi: 10.1182/blood-2005-09-3881. [DOI] [PubMed] [Google Scholar]
- 33.Alonso R, Huerta V, de Leon J, Piedra P, Puchades Y, Guirola O, Chinea G, Montero E. Towards the definition of a chimpanzee and human conserved CD6 domain 1 epitope recognized by T1 monoclonal antibody. Hybridoma. 2008;27:291–301. doi: 10.1089/hyb.2008.0007. [DOI] [PubMed] [Google Scholar]
- 34.Montero E, Falcon L, Morera Y, Delgado J, Amador JF, Perez R. CD6 molecule may be important in the pathological mechanisms of lymphocytes adhesion to human skin in psoriasis and ior t1 MAb a possible new approach to treat this disease. Autoimmunity. 1999;29:155–156. doi: 10.3109/08916939908995386. [DOI] [PubMed] [Google Scholar]
- 35.Castro MA, Oliveira MI, Nunes RJ, Fabre S, Barbosa R, Peixoto A, Brown MH, Parnes JR, Bismuth G, Moreira A, Rocha B, Carmo AM. Extracellular isoforms of CD6 generated by alternative splicing regulate targeting of CD6 to the immunological synapse. J. Immunol. 2007;178:4351–4361. doi: 10.4049/jimmunol.178.7.4351. [DOI] [PubMed] [Google Scholar]
- 36.Nitschke L. CD22 and Siglec-G: B-cell inhibitory receptors with distinct functions. Immunol. Rev. 2009;230:128–143. doi: 10.1111/j.1600-065X.2009.00801.x. [DOI] [PubMed] [Google Scholar]
- 37.Back J, Malchiodi EL, Cho S, Scarpellino L, Schneider P, Kerzic MC, Mariuzza RA, Held W. Distinct conformations of Ly49 natural killer cell receptors mediate MHC class I recognition in trans and cis. Immunity. 2009;31:598–608. doi: 10.1016/j.immuni.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vera J, Fenutria R, Canadas O, Figueras M, Mota R, Sarrias MR, Williams DL, Casals C, Yelamos J, Lozano F. The CD5 ectodomain interacts with conserved fungal cell wall components and protects from zymosan-induced septic shock-like syndrome. Proc. Natl .Acad. Sci. U S A. 2009;106:1506–1511. doi: 10.1073/pnas.0805846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castro MA, Tavares PA, Almeida MS, Nunes RJ, Wright MD, Mason D, Moreira A, Carmo AM. CD2 physically associates with CD5 in rat T lymphocytes with the involvement of both extracellular and intracellular domains. Eur. J. Immunol. 2002;32:1509–1518. doi: 10.1002/1521-4141(200205)32:5<1509::AID-IMMU1509>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 40.Pospisil R, Fitts MG, Mage RG. CD5 is a potential selecting ligand for B cell surface immunoglobulin framework region sequences. J. Exp. Med. 1996;184:1279–1284. doi: 10.1084/jem.184.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pospisil R, Silverman GJ, Marti GE, Aruffo A, Bowen MA, Mage RG. CD5 is A potential selecting ligand for B-cell surface immunoglobulin: a possible role in maintenance and selective expansion of normal and malignant B cells. Leuk Lymphoma. 2000;36:353–365. doi: 10.3109/10428190009148857. [DOI] [PubMed] [Google Scholar]