Abstract
We performed a phylogenetic character mapping on 26 stocks of Trypanosoma cruzi, the parasite responsible for Chagas disease, and 2 stocks of the sister taxon T. cruzi marinkellei to test for possible associations between T. cruzi–subspecific phylogenetic diversity and levels of protein expression, as examined by proteomic analysis and mass spectrometry. We observed a high level of correlation (P < 10−4) between genetic distance, as established by multilocus enzyme electrophoresis, and proteomic dissimilarities estimated by proteomic Euclidian distances. Several proteins were found to be specifically associated to T. cruzi phylogenetic subdivisions (discrete typing units). This study explores the previously uncharacterized links between infraspecific phylogenetic diversity and gene expression in a human pathogen. It opens the way to searching for new vaccine and drug targets and for identification of specific biomarkers at the subspecific level of pathogens.
Keywords: infectious disease, parallel evolution, gene expression, Chagas disease, parasite
Despite control improvement over the last two decades, Chagas disease remains a serious health problem in Latin America. The 2002 World Health Organization report reveals that, on the American continent, 20 million people are infected and 100 million are at risk. Moreover, because of migration, blood transfusion, and organ transplantation transmission, cases have been identified in nonendemic countries in Europe, Canada, and the United States. In the United States, a few cases have been identified that are seemingly transmitted by local vectors (1, 2).
The clinical manifestations of the infection include an acute phase and a chronic phase with different levels of severity: 27% of the cases affect the heart, 6% affect the digestive system, and 3% affect the peripheral nervous system (3). Two main drugs exist for the treatment, both of which are toxic and poorly efficient in the chronic phase. Vaccines are not available.
The causes of the clinical variability of the disease are not clear. It has been long suggested (4) that the genetic variability of the parasite could be partly responsible for it. This genetic diversity is strongly correlated to many relevant biomedical properties of the parasite, such as in vitro and in vivo culture growth, pathogenicity in mice, transmissibility by the insect vector, and susceptibility to antichagasic drugs (5–7).
Because of the potential implications of their biomedical properties, the genetic diversity and population structure of Trypanosoma cruzi have received much attention. Surveys have used various genetic markers, including multilocus enzyme electrophoresis (MLEE) (8), random-primer DNA (9), microsatellites (10), miniexon gene variability (11), and multilocus sequencing (12). Other than Escherichia coli, T. cruzi is one of the microparasites best characterized by population genetics (13).
The parasite undergoes predominant clonal evolution (14), as evidenced by considerable linkage disequilibrium, with occasional bouts of hybridization (15). The clonal evolution model applies to all relevant situations: mitotic propagation, parthenogenesis, self-fertilization in the homozygous state, and extreme homogamy. Offspring genotypes are identical or virtually identical to the parental genotypes, with very little or no genetic recombination (16). T. cruzi natural populations consist of six main genetic subdivisions, or discrete typing units (DTUs) (17), numbered I to VI (18). Because of predominant clonal evolution, these DTUs are extremely stable in space and time. The DTUs are the relevant units of analysis for epidemiological tracking and experimental evolution (13).
The genome sequence of T. cruzi (19) predicts 22,570 codified proteins for the deployed genome. A putative function could be assigned to 50.8% of the predicted genes based on significant homologies with characterized proteins or known functional domains in other organisms. The remaining 49.2% correspond to 11,104 proteins without identified function.
The huge majority of parasite species exhibit a metapopulation structure over their entire geographic range, occupying habitats that are fragmented and heterogeneous in space and/or time. Proteomics, because of the level of integration it promotes, has the potential to resolve relevant issues specific to metapopulation biology and adaptive processes (20–22). To identify relevant biomedical properties, the study of protein expression is more effective than genomic studies, although it is technologically more complex. Ascertaining protein expression may be the best approximation for understanding the biological function of a gene.
Proteomics, which seeks to investigate the translation of genomic information, opens up the possibility of studying the global changes in protein expression between T. cruzi’s different genotypes (23, 24). There are differences between the extent of gene diversity and the complexity of the proteome, which prompts the relevance of proteomic analysis. Proteome complexity may be of consequence for an organism's functioning (25).
The aim of the present work is to use the well-accepted subspecific phylogenetic diversity model of T. cruzi established by genetic markers (MLEE, 22 loci; ref. 8) as a framework to perform a phylogenetic character mapping (26) of the parasite's gene expression evidenced by proteomic analysis [2D fluorescence difference gel electrophoresis (2D-DIGE) coupled with mass spectrometry (MS)]. Contrary to previous preliminary studies (27), the present work relies on a firm phylogenetic framework to explore T. cruzi proteomic diversity.
Two hypotheses are tested: (i) the null hypothesis that no correlation exists between T. cruzi phylogenetic diversity as established by genetic markers and proteomic diversity; (ii) the working hypothesis that some proteomic signals are DTU-specific and could evince certain protein functions that are specifically associated to given DTUs.
We have selected MLEE as a phylogenetic reference tool because data from this marker were available for the set of stocks under study (8). It is worth emphasizing that the T. cruzi phylogenies established from MLEE data have been fully corroborated by other genetic markers, including random-primer DNA (9, 28, 29), random amplified differentially expressed sequences (RADES; ref. 30), microsatellites (10), and multigene sequencing (12). This corroboration demonstrates that MLEE, at least in the case of T. cruzi, is a reliable phylogenetic marker, although it is based on phenotypic characters (enzyme electrophoretic variants).
A first set of experiments included 9 T. cruzi laboratory-cloned stocks representative of the whole phylogenetic diversity of the parasite. A second set of experiments involved 26 stocks, including the 9 stocks of the first set. Results from the second set of experiments that concerned the 9 initial stocks were also treated separately to estimate the reproducibility of our experiments. In each of the two experiments, all stocks were cultured two times. Two stocks of the related subspecies T. cruzi marinkellei, a bat parasite, have been included for comparison purposes, because this subspecies has been consistently evidenced as a reliable outgroup in phylogenies established with genetic markers (8, 28, 29, 31) (Table S1). See Materials and Methods for details.
Results
Proteomics Data.
For each of our experiments, we made a visual cleaning of each spot to eliminate artifacts that could interfere with the significant results. With the analysis of 2D-DIGE gels with the Progenesis SameSpots 3.1 software (Nonlinear Dynamics), we sought to identify those spots that had a significant difference of expression (ANOVA < 0.05) between stocks. By this method, we have identified 261 proteic spots (first experiment with 9 stocks), and 172 proteic spots (second experiment with 26 stocks). The number of spots was higher in the first experiment because we set a higher level of visual cleaning in the second experiment (Materials and Methods). Results concerning the 9 stocks that were common to both sets of experiments were compared and showed excellent reproducibility.
Proteomic Expression and Phylogenetic Diversity of T. cruzi.
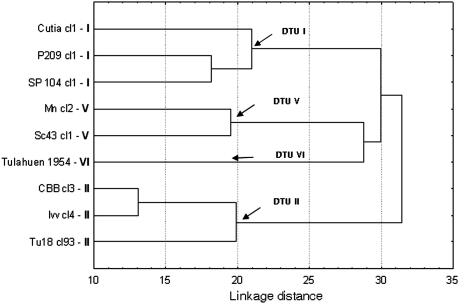
We used hierarchical ascendant classification to determine similarities among stocks and identify groups of stocks according to the normalized volume of the spots obtained from the 261 (experiment 1) and 172 (experiment 2) proteic spots. (Normalization is the process of making the spot volumes comparable between gel images in relation to technical differences in staining, scanning, sample volume, and so on.) The hierarchical ascendant classification of 9 stocks, both in the first and in the second experiment, revealed the existence of four distinct clusters. This clustering (Fig. 1) was roughly congruent with the phylogeny of T. cruzi (Fig. 2) as established by other markers in earlier studies (DTU I, DTU II, DTU V, and DTU VI).
Fig. 1.
Hierarchical cluster analysis (Ward's grouping method, Euclidean distances) of proteomic variability in nine T. cruzi stocks.
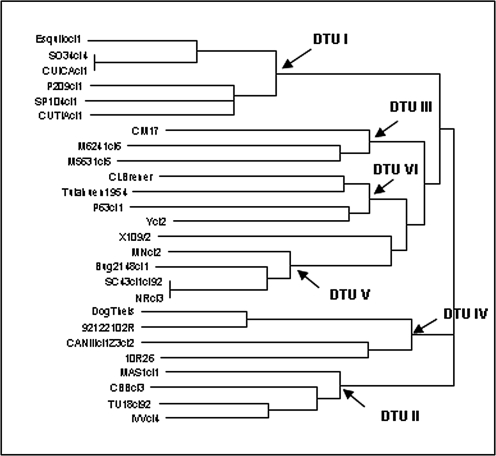
Fig. 2.
Phylogenetic relationships among 26 T. cruzi stocks based on 22 isoenzyme loci.
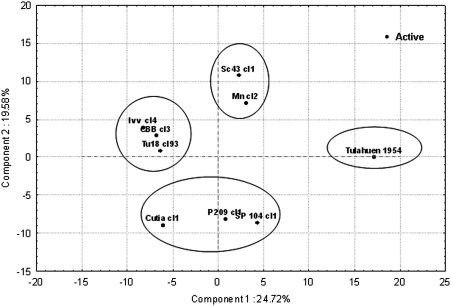
We used principal component analysis (PCA) to determine the proteomic similarities among the different stocks, by comparison with the phylogeny previously established with genetic markers. Eight principal components account for most of the initial variation. These eight principal components each represent 3–25% of the total variation of the data set. The projection of individuals (i.e., stocks) obtained through the PCA analysis clearly shows the four separate DTUs, which indicates that there is a definite tendency for each DTU to exhibit a specific protein expression profile (Fig. 3).
Fig. 3.
Proteomic variability among nine T. cruzi stocks based on the first and second principal components.
To ascertain the strength of the association between previously established phylogenies and proteomic diversity, we performed a correlation between MLEE genetic distances and the proteomic Euclidian distances established by the PCA analysis. We used the two matrices of distances obtained for the nine T. cruzi stocks, and we obtained a highly significant correlation (P < 10−4), based on 104 iterations with the nonparametric Mantel test (32).
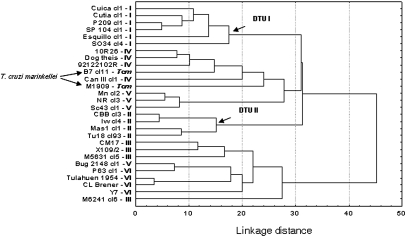
When we used the second set of parasite stocks (i.e., the second experiment), including 26 T. cruzi and 2 T. cruzi marinkellei stocks, the analysis reveals that the DTU I group of stocks remains well-defined and distinct from the DTU II–VI stocks. However, the clustering of the DTU II–VI stocks does not display the same topology as the one evidenced by genetic markers (8, 14, 28–30) (Figs. 2 and 4). Among these DTUs, the DTU II stocks appear more clearly defined than other DTUs (Fig. 4). Nevertheless, the correlation between MLEE and proteomic distances remains highly significant (Mantel test: P < 10−4) for the sample of the 26 T. cruzi stocks (excluding the 2 T. cruzi marinkellei stocks).
Fig. 4.
Hierarchical cluster analysis (Ward's grouping method, Euclidean distances) of proteomic variability among 26 T. cruzi stocks and 2 T. cruzi marinkellei stocks.
The two T. cruzi marinkellei stocks yield an unexpected, although interesting, result: T. cruzi marinkellei is clearly an outgroup from a phylogenetic point of view (8, 28, 29), yet these two stocks are not clearly distinct from the T. cruzi stocks (Fig. 4). The T. cruzi marinkellei stocks appear to be very close to the DTU IV stocks.
Unfortunately, MLEE data that would include both T. cruzi and T. cruzi marinkellei stocks are limited (six T. cruzi stocks only). In any case, the Mantel test performed on these six T. cruzi stocks and the two T. cruzi marinkellei stocks again shows a significant correlation between MLEE and proteomic distances (P < 0.05).
Identification of DTU-Specific Proteins.
We established subclasses, each constituted by the stocks of each DTU (Table S1). ANOVA was performed on each DTU and each proteic spot by using the Newman–Keuls method. We thus identified 29 significant proteic spots that appeared to be specific to given DTUs (P = 10−3). Fig. 5 shows the proteic expression profiles of one discriminating protein spot. This procedure enabled us to evidence the expression profile of stocks belonging to a particular DTU (i.e., DTU II), whereas significant subexpression is maintained in all other DTUs. With these criteria, we identified 29 DTU-specific spots, although, because of technical difficulties, we were only able to unambiguously identify nine specific spots (in a preliminary study; P < 10−3) by using mass spectrometry with MALDI-TOF analysis. Table 1 shows the spots identified.
Fig. 5.
Protein expression profile of one representative proteic spot. (A) Position of the spot in the master gel. (B) Visual display for each DTU. (C) Normalized volume for individual stocks. (D) Curve of the average values for a given spot.
Table 1.
Identification of nine proteic spots by mass spectrometry
| Spot no. | DTU attribution | Protein identity | SwissProt/ TrEMBL accession no. | Mr, kDa | Isoelectric point | Score* | Coverage, % |
| 91 | I | β-Tubulin | TBB_TRYCR | 49.54 | 4.69 | 69 | 21 |
| 276 | IV | α-Tubulin | Q26973_TRYCR | 46.93 | 5.49 | 62 | 17 |
| 90 | II | α-Tubulin | TBA_TRYCR | 49.7 | 4.9 | 92 | 33 |
| 48 | III | β-Tubulin | TBB_TRYCR | 49.54 | 4.69 | 89 | 17 |
| 346 | III | β-Tubulin | TBB_TRYCR | 49.54 | 4.69 | 88 | 25 |
| 171 | V | Heat shock-like 85-kDa protein | HSP85_TRYCR | 80.71 | 5.07 | 118 | 21 |
| 239 | VI | Elongation factor 2 (putative) | Q4D3T1_TRYCR, Q4D5X0_TRYCR, Q6IWF6_TRYCR | 94.13 | 5.73 | 113 | 17 |
| 134 | III/T. c. marinkellei | Heat shock-like 85-kDa protein | HSP85_TRYCR | 80.71 | 5.07 | 40 | 11 |
| 36 | T. c. marinkellei | Iron superoxide dismutase (putative) | Q4DI29_TRYCR | 21.89 | 6.64 | 107 | 43 |
*MASCOT scores higher than 51 indicate significant identity or extensive homology (P < 0.05).
Discussion
The aim of this study was to perform a phylogenetic character mapping (26) of proteomic characters as a first step to test the null hypothesis that no links exist between T. cruzi phylogenetic diversity as established by genetic markers (8) and proteomic diversity. This null hypothesis has been clearly rejected, because correlations between genetic and proteomic distances as measured by the Mantel test (32) were consistently highly significant (P < 10−4).
When only nine T. cruzi stocks were considered, we found a good correspondence between the phylogenetic tree based on genetic markers (MLEE) and the dendrogram established with proteomic hierarchical ascendant classification Euclidean distances (Fig. 1). The fact that the results were identical for the nine stocks that were common to the two proteomic experiments shows that our protocols are quite reproducible. It is worth noting that we based spot characterization on the average of spots obtained from two independent culturing procedures for each stock under analysis.
The correspondence between genetic (MLEE) and proteomic data became clouded when a larger number of stocks was considered (Fig. 4). The dendrogram constructed for 26 stocks corroborated the individualization of the DTU I and DTU II clusters but showed no definite individualization for stocks from other DTUs. This lack of individualization is likely because of two facts: (i) when the number of stocks considered becomes large, it becomes difficult to evidence any clear hierarchization among them, and (ii) genes coding for expressed functional proteins are subject to selective pressure and, hence, often are poor phylogenetic markers. An extreme case of it is the phylogenetic patterns of HLA genes, which sometimes show human HLA alleles that are more closely related to alleles from macaques and other primates than to other human alleles (33). Typically, genes not subject to selective pressure (neutral markers) provide a much more reliable phylogenetic signal than those subject to selection do. Expressed proteins revealed by proteomic analysis should, for that reason, be considered as less reliable phylogenetic markers than neutral traits.
Although the phylogenetic tree and the proteomic dendrogram are not precisely coincident with the 26 stocks, the correlation between genetic and proteomic distances remains highly significant (P < 10−4), which clearly indicates that the evolution of genetic and proteomic characters are far from being independent.
The two T. cruzi marinkellei stocks, which are phylogenetically distinct from all T. cruzi stocks by means of genetic markers (8, 28, 29, 31), exhibited no proteic expression specificity and were not distinct from T. cruzi stocks. The pathogenicity of T. cruzi marinkellei has been explored at length (34). It is morphologically undistinguishable from T. cruzi. The present results corroborate that T. cruzi marinkellei and T. cruzi, although phylogenetically distinct (but closely related), are phenotypically similar. This phenotypic similarity could impact pathogenic properties as well.
The phylogenetic character mapping sought, in a second step, to test the working hypothesis that some protein profiles could be strongly linked to phylogenetic diversity and might prove to be DTU-specific. Such specific-signal proteins could be of medical relevance (pathogenicity, implications in Chagas disease clinical diversity, drug and vaccine resistance, etc.). Thus, our data might provide significant information for identifying drug or vaccine targets specific for given T. cruzi genotypes.
Among the DTU-specific spots identified, six of nine correspond to structural proteins for the microtubules α- and β-tubulin and elongation factor (Table 1). Microtubules are important components of the cytoskeleton and are involved in essential processes such as cellular division, mobility, and intracellular transport (35). The basic synthesis of microtubules is based on heterodimer proteins such as the α- and β-tubulins. Furthermore, these proteins are known to undergo posttranscriptional modification. Their various isotypes are related to biological functions involved in the structure and function of microtubules (35). The two other identified proteins correspond to heat-shock protein-like, which are stress proteins (Table 1). There is only one metabolic protein, namely, superoxide dismutase. These observations would suggest that the above-cited proteins might be significant DTU-specific taxonomic biomarkers.
When host–parasite interactions are considered, a main assumption is that, over evolutionary timescales, virulence is genetically settled at the onset of the interaction (36, 37). Environmental factors are traditionally viewed as not having a relevant role in the crosstalk. However, it is common to find in populations of a parasite species substantial variation in virulence, even when parasites are collected in the same environment and at the same time. When a character such as virulence is variable for both genetic and environmental reasons, two individuals may differ because they differ in genotype, because they have undergone different environmental experiences, or both. The extent to which different individual parasites and parasite ecotypes display different virulence abilities is poorly documented or ascertained. Our survey suggests differential proteomic expression among T. cruzi DTUs. These differences open up the way to study the variation of virulence within and between DTUs linked or not to environmental conditions (20). Although the basic blueprint of life is encoded in DNA, the execution of the genetic plan is carried out by the activities of proteins. The fabric of biological diversity is therefore protein-based, and natural selection acts at the protein and phenotypic level. Population proteomics could be a promising avenue to settle relevant issues specific to the metapopulation biology and adaptive processes of T. cruzi DTUs.
Conclusion
Our study establishes a definite link between T. cruzi phylogenetic diversity and quantified proteomic expression. Accordingly, we are proposing proteomics as an informative biomarker because it could reveal processes involved in different biological pathways of the parasite. Our results also suggest that some proteins, or their differential levels of expression, could be specifically associated with different DTUs. In particular, this could be the case for structural proteins, such as α- and β-tubulins, as well as for some metabolic proteins, such as superoxide dismutase. Broadening the current focus of research so as to include other proteins that might be differently expressed in different DTUs, might conceivably lead to new specific therapies associated with T. cruzi phylogenetic diversity.
Materials and Methods
Parasite Stocks Under Study.
Table S1 gives the origin of the 28 stocks surveyed in the present study. All stocks were laboratory-cloned with verification of cloning efficiency under the microscope. They have been previously characterized by MLEE (22 loci; ref. 8) and most of them by random-primer DNA as well (9, 28, 29). All of the stocks were grown and sampled according to protocols previously described (8), adjusted to strictly synchronize the cultures.
Epimastigote forms were maintained at 27 °C in liver-infusion tryptose medium supplemented with 10% heat-inactivated FCS. Stocks were all collected at the end of the exponential growth phase (determined by the growth curve) so that they were exactly at the same physiological state. They were harvested by centrifugation (1,000 × g, 10 min, 4 °C). Pellets were resuspended in a minimal volume of buffer S (7 M urea, 2 M thiourea, 1.2% CHAPS, 0.5% Triton X-100, and 16 mM DTT) plus the protease inhibitor mixture. This suspension was lightly sonicated in an ice bath to disrupt membranes and centrifuged at 2,500 × g for 10 min. The protein concentration of each sample was determined by the Bradford method (Bio-Rad). In each of the two proteomic experiments (described below), all stocks were cultured twice.
Proteomic 2D Electrophoresis.
Two sets of experiments were performed. The first one concerned 9 stocks. The second one concerned 26 stocks, including the 9 stocks of the first experiment, and 2 stocks of the sister taxon T. cruzi marinkellei. The fact that the same 9 stocks were considered in the two experiments made it possible to ascertain the reproducibility of our protocols: the results were virtually identical between the two experiments. This level of reproducibility after two experiments is the one that is routinely used at the proteomic technological platform at the Institut de Recherche pour le Développement center of Montpellier.
In the first experiment, 261 spots were taken into account. In the second experiment, we aimed at eliminating more spots that could be artifactual by visual examination (because they appeared weak) and kept only 172 spots. However, we made the same statistical analyses with all 261 spots in the second experiment and reached identical results.
Computing Phylogenetic Clustering from Multilocus Enzyme Electrophoresis Data.
To estimate the genetic divergence among the stocks, Jaccard's genetic dissimilarity index (38) was used. It measures the proportion of band mismatches between a pair of stocks according to the formula D = 1 − (a/(a + b+ c)), where a is the number of bands that are common to the two compared genotypes, b is the number of bands present in the first genotype and absent in the second, and c is the number of bands absent in the first genotype and present in the second. The neighbor-joining method (39) was used to cluster the stocks from the Jaccard's distance matrix. The package PHYLIP (40) was used to design the neighbor-joining trees.
2D-DIGE, Image Scanning, and Statistical Analysis.
The proteins were labeled according to the Ettan DIGE minimal labeling protocol (GE Healthcare). For each stock, 50 μg of protein were labeled with 150 pmol of CyDye. Two biological replicates were made for each stock. To determine and exclude nonspecific labeling between stocks in both biological replicates, a “forward” and a “reverse” labeling was done. An internal standard consisting of aliquots of all stocks was labeled with Cy2. The Cy2 internal standard was used to normalize protein abundances across gels and to control for gel-to-gel variation (41).
For isoelectric focusing, internal standard labeled with Cy2, one sample labeled with Cy3, and another sample labeled with Cy5 were loaded into 24-cm Immobiline Drystrip (GE Healthcare) and were rehydrated in passive conditions at 20 °C in a solubilization solution (7 M urea, 2 M thiourea, 4% CHAPS, 0.5% Triton X-100, 40 mM Tris, 12 mg/mL DeStreak Reagent, and 0.75% IPG buffer). Isoelectric focusing was performed with an IPGphor apparatus (GE Healthcare). The run conditions were as follows: 60 V for 3 h, 1,000 V (gradient) for 5 h, 8,000 V (gradient) for 4 h, and 8,000 V constant for 5 h.
Equilibrated IPG strips (SDS, 0.2%) were placed on top of a SDS polyacrylamide gel to initiate the 2D electrophoresis and run at 15 mA per gel for 12 h until the bromophenol blue front reached the end of the gel.
2D-DIGE gels were scanned with a Typhoon 9400 imager set according to the manufacturer's instructions (GE Healthcare). Images were preprocessed with ImageQuant software (GE Healthcare). All 2D-DIGE images were analyzed by using Progenesis SameSpots 3.1 software (Nonlinear Dynamics). This software allows background subtraction, spot detection, quantification, and spot matching across different gels. Three quantitative measurements are available for each spot: “Od,” the highest calibrated pixel intensity in the protein spot; “Area,” the protein spot's area in mm2; and “Volume,” the integration of Od over the spot's area. Intragel spot detection and intergel matching were performed by using the differential in-gel analysis mode and biological variation analysis mode of Progenesis SameSpots 3.1 software, respectively. Protein spot volumes were normalized to the internal standard as described above. One-way ANOVA statistical tests were used to reveal statistically significant protein expression differences between the samples. Significantly underexpressed or overexpressed proteins were identified by multiple comparisons using Student's t test.
For the two separate experiments, spots of interest, with significant altered expression of their normalized volume (P < 0.05), were used to create input-matrix data to perform a hierarchical cluster analysis (amalgamation rule: Ward's method; distance measure: Euclidean distance), and a “French PCA” in Statistica version 7 (http://www.statsoft.com) was run to determine the similarities between the different stocks studied.
The hierarchical cluster analysis uses the dissimilarities or distances between parasite stocks when forming the clusters. These distances can be based on multiple dimensions (i.e., each protein spot is a dimension). Because it is difficult to homologize loci among populations and/or species by using classical 2D electrophoresis or 2D-DIGE, we used the normalized volume of the protein spots of interest and the most straightforward way of computing distances between parasite stocks in a multidimensional space: the computing of the Euclidean distances. This is probably the most commonly chosen type of distance for the tree cluster analysis. It simply is the geometric distance in the multidimensional space. This method has certain advantages (e.g., the distance between any two parasite stocks is not affected by the addition of new parasite stocks to the analysis, which may be outliers). The PCA is an unsupervised technique (i.e., it does not use any knowledge of the groupings of the data) that provides a simplified graphical representation of the multidimensional data. It is useful for determining whether samples have the expected groupings or whether there are any outliers in the data. Finally, we used a Mantel test (32) to assess the correlation between genetic distances obtained from MLEE data (8) and proteomic Euclidian distances.
Identification of DTU-Specific Proteins by Mass Spectrometry.
Spots of interest were identified (Table 1) and excised. Protein spots were in-gel digested with Trypsin Gold (300 ng, Promega) as previously described (42). Peptide extract was dehydrated (by vacuum centrifuge), desalted using Zip Tips C18 (Millipore), and eluted in 10 μL of 0.1% trifluoroacetic acid in 50% acetonitrile solution. Then, 0.5 μL of sample peptide was mixed with 0.5 μL of α-cyano-4-hydroxy-transcinnamic acid and deposited on a 384-well MALDI anchorship target by using the dry-droplet procedure (43). Analyses were performed with an UltraFlex I MALDI TOF-TOF mass spectrometer (Bruker Daltonics) and calibrated internally with the autoproteolysis peptides of trypsin (m/z 842·51; 1,045·56; 2,211·10). Peptides were selected in the mass range of 900–3,000 Da. Peptide mass fingerprint identification of proteins was performed by searching against the Trypanosoma entries of either the SwissProt or TrEMBL databases (http://www.expasy.ch) and by using the Mascot v 2.2 algorithm (http://www.matrixscience.com) with trypsin enzyme specificity and one trypsin missed cleavage. Carbamidomethyl was set as fixed cysteine modification; oxidation was set as variable methionine modification for searches. A mass tolerance of 50 ppm was allowed for identification. Matching peptides with one missed cleavage were considered as pertinent when there were two consecutive basic residues or when arginine and lysine residues were in an acidic context. Mascot scores higher than 51 were considered significant (P < 0.05) for SwissProt and TrEMBL database interrogation.
Supplementary Material
Acknowledgments
We thank Dr. Stéphane Dussert (Institut de Recherche pour le Développement, Montpellier, France) for providing us with the software Statistica version 7.1.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015496107/-/DCSupplemental.
References
- 1.Barnabé C, Yaeger R, Pung O, Tibayrenc M. Trypanosoma cruzi: A considerable phylogenetic divergence indicates that the agent of Chagas disease is indigenous to the native fauna of the United States. Exp Parasitol. 2001;99:73–79. doi: 10.1006/expr.2001.4651. [DOI] [PubMed] [Google Scholar]
- 2.Herwaldt BL, et al. Use of polymerase chain reaction to diagnose the fifth reported US case of autochthonous transmission of Trypanosoma cruzi, in Tennessee, 1998. J Infect Dis. 2000;181:395–399. doi: 10.1086/315212. [DOI] [PubMed] [Google Scholar]
- 3.Moncayo A. Chagas disease: Current epidemiological trends after the interruption of vectorial and transfusional transmission in the Southern Cone countries. Mem Inst Oswaldo Cruz. 2003;98:577–591. doi: 10.1590/s0074-02762003000500001. [DOI] [PubMed] [Google Scholar]
- 4.Miles MA, et al. Do radically dissimilar Trypanosoma cruzi strains (zymodemes) cause Venezuelan and Brazilian forms of Chagas’ disease? Lancet. 1981;1:1338–1340. doi: 10.1016/s0140-6736(81)92518-6. [DOI] [PubMed] [Google Scholar]
- 5.Revollo S, et al. Trypanosoma cruzi: Impact of clonal evolution of the parasite on its biological and medical properties. Exp Parasitol. 1998;89:30–39. doi: 10.1006/expr.1998.4216. [DOI] [PubMed] [Google Scholar]
- 6.de Lana M, et al. Trypanosoma cruzi: Infectivity of clonal genotype infections in acute and chronic phases in mice. Exp Parasitol. 2000;96:61–66. doi: 10.1006/expr.2000.4552. [DOI] [PubMed] [Google Scholar]
- 7.da Silveira Pinto A, de Lana M, Britto C, Bastrenta B, Tibayrenc M. Experimental Trypanosoma cruzi biclonal infection in Triatoma infestans: Detection of distinct clonal genotypes using kinetoplast DNA probes. Int J Parasitol. 2000;30:843–848. doi: 10.1016/s0020-7519(00)00058-8. [DOI] [PubMed] [Google Scholar]
- 8.Barnabé C, Brisse S, Tibayrenc M. Population structure and genetic typing of Trypanosoma cruzi, the agent of Chagas disease: a multilocus enzyme electrophoresis approach. Parasitology. 2000;120:513–526. doi: 10.1017/s0031182099005661. [DOI] [PubMed] [Google Scholar]
- 9.Tibayrenc M, et al. Genetic characterization of six parasitic protozoa: Parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira RP, et al. Probing the genetic population structure of Trypanosoma cruzi with polymorphic microsatellites. Proc Natl Acad Sci USA. 1998;95:3776–3780. doi: 10.1073/pnas.95.7.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol Biochem Parasitol. 1996;83:141–152. doi: 10.1016/s0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- 12.Subileau M, Barnabé C, Douzery EJ, Diosque P, Tibayrenc M. Trypanosoma cruzi: New insights on ecophylogeny and hybridization by multigene sequencing of three nuclear and one maxicircle genes. Exp Parasitol. 2009;122:328–337. doi: 10.1016/j.exppara.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Tibayrenc M. In: Modelling Parasite Transmission and Control. Michael E, Spear RC, editors. Austin, TX: Landes Bioscience; 2009. pp. 200–211. [Google Scholar]
- 14.Tibayrenc M, Ward P, Moya A, Ayala FJ. Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proc Natl Acad Sci USA. 1986;83:115–119. doi: 10.1073/pnas.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaunt MW, et al. Mechanism of genetic exchange in American trypanosomes. Nature. 2003;421:936–939. doi: 10.1038/nature01438. [DOI] [PubMed] [Google Scholar]
- 16.Tibayrenc M, Kjellberg F, Ayala FJ. A clonal theory of parasitic protozoa: The population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tibayrenc M. Genetic epidemiology of parasitic protozoa and other infectious agents: the need for an integrated approach. Int J Parasitol. 1998;28:85–104. doi: 10.1016/s0020-7519(97)00180-x. [DOI] [PubMed] [Google Scholar]
- 18.Zingales B, et al. Second Satellite Meeting. A new consensus for Trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 19.El-Sayed NM, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 20.Biron DG, et al. Population proteomics: An emerging discipline to study metapopulation ecology. Proteomics. 2006;6:1712–1715. doi: 10.1002/pmic.200500423. [DOI] [PubMed] [Google Scholar]
- 21.Karr TL. Fruit flies and the sperm proteome. Hum Mol Genet. 2007;16 Spec No. 2:R124–R133. doi: 10.1093/hmg/ddm252. [DOI] [PubMed] [Google Scholar]
- 22.Cieslak A, Riberia I. Aplicaciones de proteómica en ecología y evolución. Ecosistemas. 2009;18:34–43. [Google Scholar]
- 23.Biron DG, Moura H, Marché L, Hughes AL, Thomas F. Towards a new conceptual approach to “parasitoproteomics”. Trends Parasitol. 2005;21:162–168. doi: 10.1016/j.pt.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Biron DG, et al. The pitfalls of proteomics experiments without the correct use of bioinformatics tools. Proteomics. 2006;6:5577–5596. doi: 10.1002/pmic.200600223. [DOI] [PubMed] [Google Scholar]
- 25.Pando Robles V, Ferreira Batista C. Proteómica: Hacia el entendimiento del lenguaje de las proteínas en. In: Lopez-Munguia A, editor. A Una Ventana al Quehacer Científico. Mexico: Instituto de Biotecnología; 2008. pp. 97–108. [Google Scholar]
- 26.Avise JC. Molecular Markers, Natural History and Evolution. 2nd Ed. New York, London: Chapman & Hall; 2004. [Google Scholar]
- 27.Parodi-Talice A, et al. Proteome analysis of the causative agent of Chagas disease: Trypanosoma cruzi. Int J Parasitol. 2004;34:881–886. doi: 10.1016/j.ijpara.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Brisse S, Barnabé C, Tibayrenc M. Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int J Parasitol. 2000;30:35–44. doi: 10.1016/s0020-7519(99)00168-x. [DOI] [PubMed] [Google Scholar]
- 29.Brisse S, Dujardin JC, Tibayrenc M. Identification of six Trypanosoma cruzi lineages by sequence-characterised amplified region markers. Mol Biochem Parasitol. 2000;111:95–105. doi: 10.1016/s0166-6851(00)00302-9. [DOI] [PubMed] [Google Scholar]
- 30.Telleria J, Barnabé C, Hide M, Bañuls AL, Tibayrenc M. Predominant clonal evolution leads to a close parity between gene expression profiles and subspecific phylogeny in Trypanosoma cruzi. Mol Biochem Parasitol. 2004;137:133–141. doi: 10.1016/j.molbiopara.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Barnabé C, Brisse S, Tibayrenc M. Phylogenetic diversity of bat trypanosomes of subgenus Schizotrypanum based on multilocus enzyme electrophoresis, random amplified polymorphic DNA, and cytochrome b nucleotide sequence analyses. Infect Genet Evol. 2003;2:201–208. doi: 10.1016/s1567-1348(02)00130-2. [DOI] [PubMed] [Google Scholar]
- 32.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 33.Ayala FJ, Escalante AA. The evolution of human populations: A molecular perspective. Mol Phylogenet Evol. 1996;5:188–201. doi: 10.1006/mpev.1996.0013. [DOI] [PubMed] [Google Scholar]
- 34.Baker JR, Miles MA, Godfrey DG, Barrett TV. Biochemical characterization of some species of Trypansoma (Schizotrypanum) from bats (Microchiroptera) Am J Trop Med Hyg. 1978;27:483–491. doi: 10.4269/ajtmh.1978.27.483. [DOI] [PubMed] [Google Scholar]
- 35.McKean PG, Vaughan S, Gull K. The extended tubulin superfamily. J Cell Sci. 2001;114:2723–2733. doi: 10.1242/jcs.114.15.2723. [DOI] [PubMed] [Google Scholar]
- 36.Bull JJ. Perspective: Virulence. Evolution. 1994;48:1423–1437. doi: 10.1111/j.1558-5646.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 37.Dieckmann U, Metz JAJ, Sabelis MW, Sigmund K, editors. The Adaptive Dynamics of Infectious Diseases: In Pursuit of Virulence Management. Cambridge, UK: Cambridge University Press; 2002. [Google Scholar]
- 38.Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat. 1908;44:223–270. [Google Scholar]
- 39.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Felsenstein J. PHYLIP, Phylogeny Inference Package. Seattle, WA: Department of Genetics, University of Washington; 1993. Version 3.5c. [Google Scholar]
- 41.Alban A, et al. A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 42.Wilm M, et al. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 43.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.