Abstract
Several proteins of the mitochondrial intermembrane space are targeted by internal targeting signals. A class of such proteins with α-helical hairpin structure bridged by two intramolecular disulfides is trapped by a Mia40-dependent oxidative process. Here, we describe the oxidative folding mechanism underpinning this process by an exhaustive structural characterization of the protein in all stages and as a complex with Mia40. Two consecutive induced folding steps are at the basis of the protein-trapping process. In the first one, Mia40 functions as a molecular chaperone assisting α-helical folding of the internal targeting signal of the substrate. Subsequently, in a Mia40-independent manner, folding of the second substrate helix is induced by the folded targeting signal functioning as a folding scaffold. The Mia40-induced folding pathway provides a proof of principle for the general concept that internal targeting signals may operate as a folding nucleus upon compartment-specific activation.
Keywords: NMR, oxidative protein folding, protein folding intermediates
Folding processes have been extensively characterized in vitro, mostly starting from artificially unfolded proteins. A number of studies have shown that some unfolded polypeptides require interactions with specific chaperones to facilitate the folding process, by increasing the efficiency of the overall process—i.e., by reducing the probability of competing nonphysiological conformational pathways or by accelerating steps in the folding process that can otherwise be extremely slow (1). The elementary steps of a folding process, starting from the newly synthesized polypeptide and following its various states to the final one, on the contrary, have never been characterized at the atomic level. Here we describe such a process as a function of compartment-specific protein components that facilitate this process. The oxidative protein folding in the intermembrane space (IMS) of mitochondria represents one of such processes.
Mitochondrial proteins sharing a coiled coil-helix-coiled coil-helix (CHCH) domain, like Cox17 and the small Tims, are reduced in the cytoplasm where they are released once synthesized, then they move to the IMS through the translocase of the outer membrane protein channel (2, 3), and here they are trapped through a compartment-dependent oxidation of four cysteines that bridge the CHCH domain (4, 5). Formation of disulfide bonds in vivo does not occur spontaneously but requires dedicated protein catalysts, which usually are part of an oxidative machinery able to introduce disulfide bonds in the final protein target (6). Specifically, the oxidoreductase Mia40 is responsible for disulfide bond formation in CHCH proteins after they are targeted to the IMS (7). Mia40 has an active CPC redox site as shown by mutagenesis experiments in an in vitro reconstituted system (8). The CPC motif participates in the formation of a mixed disulfide bonded intermediate (8–10) that is crucial for the oxidation of the reduced substrates. The solution structure of human Mia40 (9) and the crystal structure of yeast Mia40 (10) revealed the presence of a shallow hydrophobic cleft adjacent to the CPC motif. This shallow cleft was proposed to bind the substrates on the basis of extensive mutagenesis of the hydrophobic residues and a combination of substrate binding studies and phenotypic analysis in vivo as reported by two independent studies (9, 10). Furthermore, in vitro and in vivo data suggested that Mia40 recognizes the substrates depending on the presence of an internal targeting signal [named ITS by Sideris et al. (11) or MISS by Milenkovic et al. (12)], located in different parts of the polypeptide depending on the substrate, either near the N terminus for the small Tims (11, 12) or near the C end for Cox17 (11). In all cases it binds to the cleft of Mia40 in such a way as to orient one specific cysteine of the substrate (the docking cysteine) for disulfide pairing with the active site cysteine of Mia40 (11). Studies from several groups have reported that Mia40 is sufficient for the complete oxidation of substrates in vitro. In particular, Banci et al. showed that complete oxidation of human Cox17 can occur in vitro by incubation with oxidized human Mia40 alone (9), whereas more recently Bien et al. (13) showed that this is also the case for yeast Cox19 (a homologue of Cox17). The possible participation of Erv1 in the full oxidation reaction for the substrate in vivo has been proposed by Stojanovski et al. (14) on the basis of the observation of a ternary complex containing Mia40, the substrate, and Erv1 (a FAD-linked oxidase shown to recycle Mia40 to its oxidized state).
The mechanistic details of the Mia40-dependent pathway are not yet understood, in particular because of the complete absence of the structures at atomic resolution of both substrate states and Mia40–substrate complexes occurring during this process. Similar studies on other protein translocation systems have been hampered by the difficulty to isolate the relevant transient complexes between the translocase and the imported protein. In the few cases where this was achieved (15, 16), only a complex of the synthetic peptide with the translocase was isolated, not allowing one to draw any conclusions on the overall folding process and the factors affecting it.
In this study we used the Mia40–substrate interaction as an example of a compartment-specific folding process. The Mia40 substrates here used are the CHCH proteins Cox17 and small Tims (i.e., Tim10 and Tim9) that have unrelated functions and differ in the spacing between the conserved cysteines forming the two disulfide bonds catalyzed by Mia40. Cox17 contains a twin CX9C motif and facilitates mitochondrial copper incorporation in the CuA site of cytochrome c oxidase (17). Small Tims contain a twin CX3C motif and act as a chaperone for mitochondrial membrane proteins (18). We analyzed in vitro these proteins and their crucial folding intermediates taking into account their cellular localization and also directly in living cells. In particular, an artificially arrested Mia40–substrate intermediate was achieved by using cysteine-mutated forms of the substrate and Mia40 previously shown in vivo and in vitro to result in the formation of a stable Mia40–substrate (covalently bonded) complex (8, 9, 19, 20) and thus structurally characterized. The data illustrate the actual role of Mia40 as a chaperone hub that folds specifically the targeting signal, which then operates as a nucleus for subsequent cooperative folding steps resulting in the native conformation for the substrate. The Mia40-dependent example studied here provides a proof of principle for the concept that some internal targeting signals operate as a folding nucleus upon compartment-specific activation.
Results
In Vitro and In-Cell NMR Characterization of the Folding State of Reduced ApoCox17.
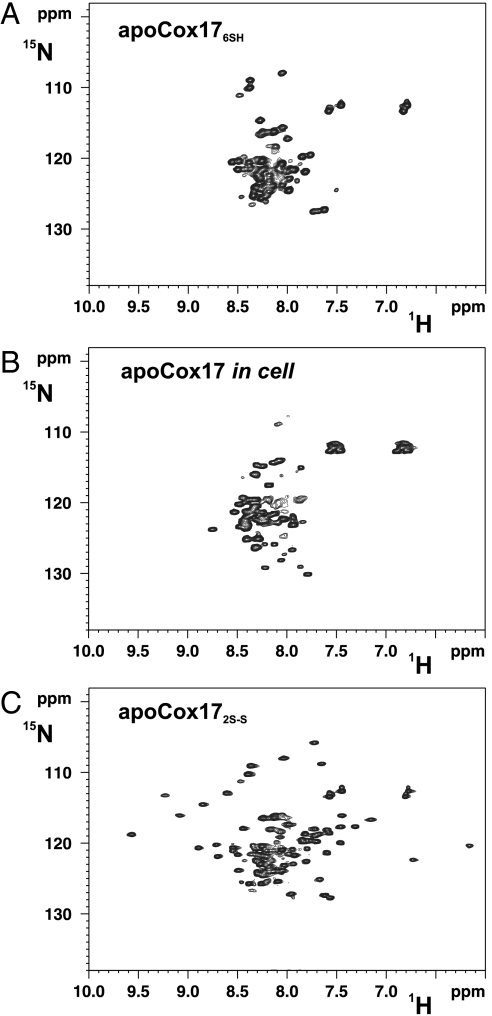
In-cell 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectrum (Fig. 1) recorded on Escherichia coli cells overexpressing cytoplasmically Cox17 (see SI Text for details) indicate that the protein is in an unstructured state with minimal 1H chemical shift dispersion, characteristic of proteins with no spatially defined conformation. The spectra are essentially identical (i.e., with minimal dispersion for 1H chemical shifts) to those acquired on the completely reduced, unfolded protein in vitro and different with respect to that of oxidized, folded Cox17 with two disulfide bonds (i.e., high dispersion for 1H chemical shifts) (Fig. 1). In conclusion, Cox17 is fully reduced and unfolded in the cytoplasm. Once these CHCH proteins are imported in the IMS, they interact with Mia40, which catalyzes the formation of the disulfide bonds in the twin CX9C or CX3C motifs (9). The latter process is fast, with the complex between the two proteins formed only transiently, which therefore does not accumulate in solution to an extent to be characterized (9, 21, 22). This transient complex, which was detected in vivo (7), involves the formation of an intermolecular disulfide bond between Cys55 of Mia40 and the third cysteine (Cys45) of the twin CX9C motif in Cox17 or the first cysteine of the twin CX3C motif in Tims (9, 11, 19, 20). The folding events that lead to the adoption of the final folded structure of the substrate with two disulfides and the role of Mia40 in these folding events remain completely unknown.
Fig. 1.
1H-15N HSQC spectra of apoCox17. (A) Fully reduced (Cox176SH); (B) in-cell Cox17; (C) oxidized with two disulfide bonds (Cox172S─S).
NMR Analysis of the Covalent Cox17–Mia40 Intermediate Complex Reveals Induced Folding of One Helix Through a Chaperone Function of Mia40.
To understand the possible functional role of Mia40 in the substrate folding process, it was critical to isolate the covalent complex that has remained elusive because of its transient nature. On the basis of recent work from us (11, 19) and other groups (20), we selected cysteine mutants that allowed us to stably trap the Cox17–Mia40 covalent complex. In particular, the cysteines engaged in the intermolecular disulfide bond between Cox17 and Mia40 were maintained (i.e., Cys55, which is the second Cys of the CPC motif of Mia40, and Cys45, which is the third Cys of the twin CX9C motif of Cox17), whereas the others involved in the acquisition of the CHCH fold of Cox17 and Cys53 of Mia40 were mutated to serine, thus blocking the progression of the redox reaction.
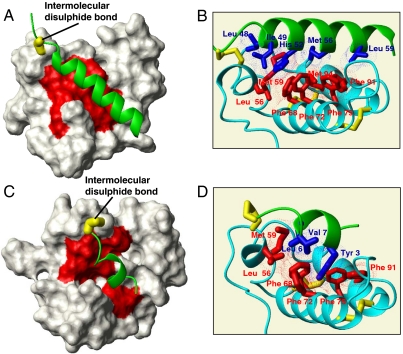
Backbone NMR chemical shift variations of Mia40 in this complex showed that only the residues spatially surrounding the catalytic CPC site of Mia40 and those forming the adjacent hydrophobic cleft are largely affected by the interaction with Cox17 upon covalent complex formation (Fig. 2A). This finding provides a structural basis to the previously reported biochemical studies (9, 23). More significantly, the NMR structural analysis shows that, although the secondary structure in Mia40 is not affected upon Cox17 binding, that of Cox17 is drastically perturbed downstream of the docking Cys45 (Fig. S1). Residues 48–62 form indeed an α-helix that is tightly packed against the hydrophobic cleft of Mia40 (Fig. 2B). This stretch therefore selectively undergoes folding from an essentially disordered state when it is isolated in solution (or in the cytosol), to an α-helix upon disulfide bond formation and docking with Mia40. In striking contrast, all other residues of Cox17 remain unfolded, highlighting a specific and very localized structural organization induced by Mia40 on the substrate (Fig. S1). From the structure of the complex, it appears that the interactions underpinning the folding mechanism involve the hydrophobic cleft of Mia40 (9, 10) and Leu48, Ile49, His52, Met56, and Leu59 of Cox17, all clustered on the same side of the “Mia40-induced” amphipathic helix (Fig. 2B). All these residues are crucial for Cox17 binding to Mia40 in organello as well (9, 10). In particular, we showed previously that Leu48, Ile49, and His52 are part of the ITS that guides Cox17 to Mia40, because point mutations in these residues affected binding to Mia40 (11). Additionally, residues Met56 and Leu59 of Cox17, which in the Cox17–Mia40 complex interact with the hydrophobic cleft of Mia40, are also important in organello because deletion of these residues abolishes the interaction with Mia40. In particular, Leu59 establishes hydrophobic interactions with Phe91 and Val88 of Mia40 and with Ile63 of Cox17 to determine one turn α-helical extension with respect to the mature Cox17 form. From these data, therefore, it emerges that the length of the hydrophobic ITS segment can be extended up to about 10 residues from the intermolecular disulfide. We conclude that the Cox17 fragment recognized by Mia40 folds upon covalent binding with Mia40, thus illustrating a coupled folding and binding event. The fact that the involved interactions are mainly hydrophobic in nature is reminiscent of the type of interactions that underpin the recognition by chaperones of unfolded client proteins (24).
Fig. 2.
Structural characterization of substrate-Mia40 covalent adducts. (A) The solution structure of the Cox17–Mia40 complex: The Mia40-induced α-helix in Cox17 is in green, the NHs chemical shift variations of Mia40 residues upon complex formation are mapped in red, and the intermolecular disulfide bond is in yellow. (B) Hydrophobic residues involved in the protein–protein recognition between Cox17 (in green) and Mia40 (in cyano) are shown in blue and red, respectively. Inter- and intramolecular disulfide bonds are in yellow; van der Waals contacts are shown in blue and red dots. (C) Experimental data-driven docking model of Tim9 peptide–Mia40 complex: The Mia40-induced α-helix in Tim9 peptide is in green, the NHs chemical shift variations of Mia40 residues upon complex formation are mapped in red, and the intermolecular disulfide bond is in yellow. (D) Hydrophobic residues involved in the protein–protein recognition between Tim9 peptide (in green) and Mia40 (in cyano) are shown in blue and red, respectively. Inter- and intramolecular disulfide bonds are also shown in yellow; van der Waals contacts are shown in blue and red dots.
The above data therefore allowed us to discover the following key points of the role of Mia40: (i) It induces folding of the substrate selectively on one of its helices (the ITS-containing segment), and (ii) it shares a common folding mechanism with molecular chaperones based on hydrophobic interactions.
Covalent Binding to Mia40 Is Essential for the Folding of the First Helix of the Substrate.
When Cox17 was mixed with Mia40 in a reducing environment (10 mM DTT), which prevents the formation of the intermolecular disulfide bond, no spreading of amide protons (NHs) chemical shift was observed (Fig. S2), in contrast to what was found in an oxidative environment (5 mM ferricyanide; Fig. S2), indicating that no observable folding occurs upon noncovalent protein–protein interaction. The chemical shift perturbation analysis shows that only the NHs of the residues in the second CX9C motif are slightly affected by Mia40 addition (backbone-weighted average chemical shift difference less than 0.06 ppm) or broadened beyond detection, indicating a molecular recognition in the ITS region of the substrate but with a low affinity (Fig. S2). Covalent binding is therefore thermodynamically essential to induce the α-helical folding of Mia40 substrates and stabilize the protein–protein interactions. The formation of an α-helix is by itself thermodynamically unfavored in agreement with the lack of an α-helical conformation in the free state (Fig. S1), but once the intermolecular S─S bond is formed, entropic contributions do not prevent anymore the folding and do not counterbalance the enthalpy of interaction.
The Second Helix Folding Is Induced Using the First Helix as a Scaffold Independently of Mia40.
After the formation of the first helix in the substrate upon Mia40 binding, the next step of the oxidative folding process consists of the formation of the first, inner intramolecular disulfide bond within Cox17 (connecting the inner cysteines) with its subsequent release from Mia40. This intermediate was structurally characterized by mutating the cysteine residues responsible for the formation of the second disulfide bond in Cox17. Previous reports indicated that the mutated substrates of Mia40 with only one disulfide could still be oxidized by Mia40 in organelle, in the case of Cox17 (9), Cox19 (13), or Tim10 (19), but no information on the folding of these intermediates is available. We found that the formation of the first intramolecular disulfide bond induces the formation of the second helix (Fig. S3) with a mechanism similar to that observed for the formation of the first helix. Indeed, once the first disulfide bond is formed in Cox17, thus releasing Cox17 from Mia40, hydrophobic interactions between the hydrophobic residues of the first formed helix (the same ITS segment responsible for the recognition with Mia40) and the hydrophobic residues of the second CX9C stretch are established, thus inducing α-helix formation of the latter stretch. Also in this step, the α-helix formation is coupled to the formation of a disulfide bond—i.e., the intramolecular one. The two α-helices of the Cox17 folding intermediate acquire the same length as found for the wild-type mature Cox17. Folding of the substrate therefore occurs via two consecutive induced folding events, each one coupled to one disulfide bond formation: (a) In the first one, the first ITS helix folding is induced by the substrate cleft of Mia40, coupled to the formation of the intermolecular disulfide; (b) the second helix is induced by the now-folded ITS first helix, coupled to the intramolecular disulfide formation between the inner cysteines of Cox17. Whereas the folding/chaperone action of Mia40 in the first step is absolutely vital, the second folding step occurs independently of Mia40. Under aerobic in vitro conditions, oxygen can rapidly form the second disulfide bond in an already-folded protein (9); without kinetic folding barriers, or, under anaerobic in vitro conditions in the presence of an excess of Mia40, another molecule of Mia40 is also able to rapidly oxidize the second disulfide bond (13). These possibilities or others for the completion of the reaction in vivo could apply.
Induced Substrate Folding by Mia40 Is a General Mechanism.
This surprisingly localized induction of folding of the substrate by Mia40 is quite general because it occurs also for other CHCH proteins that contain a twin CX3C motif. NMR spectra analysis showed that 10 residues upstream of the first docking cysteine of Tim10 are affected by the complex formation with Mia40, in agreement with previous import assays that identified this region as the ITS of this class of Mia40 substrates (11). Because degradation of the C- and N-terminal segments of Tim10 covalently bound to Mia40 [also observed for Tim10 overexpressed alone in bacteria (25)] did not allow a detailed structural characterization of the complex, a 10-residue peptide corresponding to the Tim9-ITS stretch including the docking Cys35 (RLYSNLVERC, Cys10 in the peptide numbering, and Cys35 in the Tim9 sequence) was synthesized and its covalent complex with Mia40 was isolated and investigated. Similarly to the Cox17–Mia40 complex, the NHs chemical shifts of the residues constituting the hydrophobic cleft of Mia40 are affected, indicating that the peptide is interacting with the same substrate binding region of Mia40 (Fig. 2C). The peptide, similarly to Tim10 and Cox17 proteins, is unfolded in the free state as shown by NMR and CD analysis (Fig. S4 and SI Text for details); however, upon covalent interaction with Mia40, through an intermolecular disulfide bond between Cys35 of the Tim9 peptide and Cys55 of Mia40, it adopts an α-helical conformation (Fig. S5). Therefore, the peptide shows a conformational transition from an unstructured state to a folded α-helical state upon Mia40 oxidative binding. The hydrophobic residues of the Tim9-ITS motif face the same side of the α-helix and pack into the hydrophobic cleft of Mia40, as it occurs in the Cox17–Mia40 complex (Fig. 2D). The coupled binding-folding mechanism is therefore a common mechanism for these CHCH substrates. Experiments in organello support that the folding coupled to binding of the ITS segments holds true also in vivo, because positioning the crucial cysteine of the substrate either upstream or downstream of its wild-type position, thus spanning a full turn of a helix, abolished dramatically the capacity for the substrate to interact in vivo with Mia40 (11).
Discussion
Disulfide bond formation in mitochondria has been studied intensely in recent years and is intimately linked to Mia40-dependent oxidative trapping of the substrates in the IMS. Recent efforts by several groups have elucidated important aspects mainly of the mechanism of electron transfer or the initial recognition of the substrates; however, the actual protein folding events in this process and the role of Mia40 on these have remained elusive.
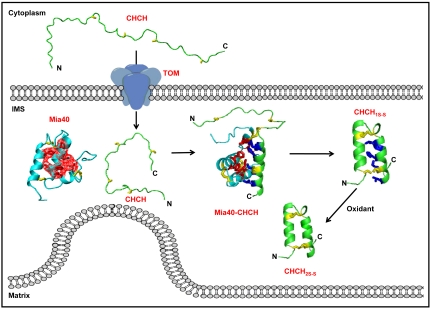
By dissecting and monitoring the elementary steps of the folding process we were able to unravel in a very comprehensive way the Mia40-dependent oxidative folding mechanism (Fig. 3). The starting point was the production of the substrate in the E. coli cytosol and the in-cell NMR analysis that showed the substrate is completely unfolded. This condition represents well the unfolded state of the protein in the eukaryotic cytosol, which is also a reducing environment.
Fig. 3.
Compartment-dependent folding mechanism of CHCH proteins. Unfolded CHCH proteins enter mitochondria via a translocase of the outer membrane (TOM) channel. To do so, they have conformational plasticity in order both to sneak in and to optimize the interactions with TOM. Once in the IMS, they must be trapped there by acquiring the mature, folded form. The following steps along the compartment-dependent folding process have been structurally characterized: the cytoplasmatic form of CHCH, CHCH–Mia40 covalent intermediate, the quasi-mature form CHCH1S─S, and the mature CHCH2S─S form (5). Side chains of the hydrophobic residues of Mia40 cleft, of the hydrophobic ITS residues of CHCH, and of cysteine residues are in red, blue, and yellow sticks, respectively; van der Waals contacts of the side chains constituting the hydrophobic cleft of Mia40 are shown as red dots surface.
The isolation of the complex between Cox17 and Mia40 allowed us to obtain structural information on the interaction between a translocase component and a full-length substrate. Key to this effort was the use of previously described cysteine mutants of the substrate and Mia40 (9, 19, 20) that allow their interaction but block further progression of the redox reaction, hence trapping the complex in a stable state. Previous reports on the structures between a synthetic mitochondrial presequence peptide and the cytosolic domain of the outer membrane receptor Tom20 (15) and between a synthetic bacterial signal peptide and SecA (16) indicated in both cases a bimodal type of interaction involving both hydrophobic and ionic interactions. Mia40 interacts with its substrate in a different manner: Exclusively hydrophobic interactions coupled with the formation of disulfide pairing induce the folding of the substrate. Mia40 therefore functions as a chaperone hub with a foldase action on the targeting signal of the substrate, determining induced folding of only one helix of the substrate. The disulfide bond formation is essential to fold the ITS, whereas Mia40 itself does not undergo any large structural changes upon binding of the substrate. The fact that the same type of induced folding is observed on both the ITS of Tim9 (which, in the full-length protein, is located near the N terminus) and the ITS segment of Cox17 (which is located at the C terminus of the protein) argue for a clear and surprising localized effect on the folding of the ITS by Mia40, irrespective of its exact location within the polypeptide chain. In support of this notion, in vivo data showing interaction with Mia40 upon import of a deleted version of Tim10 containing only the ITS helix but not the second helix have been reported (11). We have also showed that Mia40 weakly interacts with the ITS region before the formation of the intermolecular disulfide but without significant α-helical folding induction. This first step in the Mia40-substrate recognition process is essential to select and to correctly position the substrate cysteine forming the intermolecular disulfide. The so-driven disulfide pairing then determines ITS α-helical formation thanks to the Mia40–substrate hydrophobic interactions. In support to this mechanism, we previously showed that removal of hydrophobic residues of Mia40 cleft or substrate-ITS prevents the formation of the covalent intermediate (9–11).
After the helical folding of the ITS segment kept covalently attached to Mia40 via the intermolecular disulfide bond, nucleofilic attack of the second inner cysteine of the substrate to this bond releases the substrate that now undergoes folding of its second helix through interactions with the same hydrophobic residues that were engaged in the Mia40 substrate binding cleft. In essence, the now released and folded ITS functions as a scaffold for the folding of the second helix. The entire reaction can therefore be described as an α-helical folding chain reaction—i.e., in step 1 the first ITS helix in Mia40 substrates is formed coupled to the covalent interaction with Mia40, and in step 2 the ITS helix of the substrate released from Mia40 with one formed disulfide bond induces the α-helical folding of the other CX9C segment not interacting with Mia40 (Fig. S3). Our results highlight that only the first folding step is Mia40-dependent, whereas the second one is dependent on the sequence of the substrate. Both events, however, employ a common mechanism: Helix formation is induced by the creation of a hydrophobic folding nucleus coupled to the formation of a disulfide bond. The difference is that the formation of the first helix requires the intermolecular disulfide bond with the catalyst Mia40, whereas that of the second helix requires the intramolecular disulfide bond (Fig. 3). Also the hydrophobic side of the substrate ITS has a pivotal role first to make the contact with the Mia40 cleft and then to interact with the hydrophobic side chains of the second helix, thus inducing its folding.
The generation of the second disulfide is still open and could follow different routes depending on the substrate and on different cellular conditions (for example, anaerobic or hypoxic growth). Indeed, the fact that foldase activity of Mia40 for the substrate is no longer needed at this final stage could indicate that the second disulfide bond on the substrate may be introduced by different small oxidants (oxygen and metals). On the other hand, Bien et al. (13) have recently presented 4-acetamido-4'-maleimidylstilbene-2,2'-disulfonic acid-thiol trapping assays as proof for the capacity of Mia40 to completely oxidize the substrate under anaerobic conditions. The latter finding is in complete agreement with our previous suggestion that Mia40 is the only protein necessary and sufficient for the complete oxidation of the substrate (9). In this report, we had also shown that in the case of Cox17, it is the inner disulfide that is first recognized by Mia40 by using a mutant lacking either the inner or the outer disulfide of this substrate (9). Bien et al. (13) extended this finding by using the Cox19 protein that is homologous to Cox17. Bien et al. have also suggested a “proofreading” role for glutathione in the full oxidation reaction for the substrate, but this proposal was based solely on in vitro data.
The previously unknown function of Mia40 reported here is to induce the folding specifically of the ITS (in an event coupled to the intermolecular disulfide pairing). This folded segment represents the key initial folding unit that triggers complete folding of the rest of the substrate, which is usually a highly cooperative process (26). The second folding step of a Mia40–substrate complex may differ for different substrates (for example, involving different kinds of residues able to make hydrophobic and H bonds and/or salt bridges interactions), all of them being Mia40-independent and not necessarily requiring the formation of a second disulfide bond. The formation of the first Mia40-induced helix can therefore differently influence the second folding steps of the rest of the protein depending on the substrate topology. This mechanism allows us therefore to rationalize the function of Mia40 even for substrates that contain only one disulfide bond. We propose that the essential function of Mia40 is that of a chaperone foldase that assists specifically the folding of the targeting signal (through an assisted hydrophobic collapse). The now-folded targeting signal can operate as the crucial folding nucleus in subsequent cooperative steps that complete folding of the substrate to its native state.
Is there a relevance of these data for other (mitochondria- and Mia40-independent) protein targeting and folding events in eukaryotic cells? There are many proteins that are targeted to intracellular compartments by internal targeting signals that are still poorly defined and whose function is not understood. The case of Mia40-dependent folding represents an example that provides a proof of principle that internal targeting signals may function as the key initiating folding units for a protein after they get “activated” as folding units in the right compartment. Activation of the internal signal in each case should be compartment-specific (in terms of the environment and components present in the target compartment). Such a role of the internal targeting signals would be a fundamental underlying property and provide an efficient way for the cell to ensure correct protein folding in various intracellular compartments.
Materials and Methods
Protein Production.
Cysteine to serine mutations of two or three of the four cysteines involved in the formation of the two structural disulfide bonds of human Cox17 (hCox17) or of yeast Tim10 (yTim10) as well as the cysteine to serine mutation of one of the two cysteines of the CPC motif of human Mia40 (hMia40) were generated by PCR-based site-directed mutagenesis from available pETG-30A, pDEST-MBP, and pGEX plasmids (5, 9, 27) containing, respectively, hMia40, hCox17, and yTim10 genes. hCox17- and hMia40-mutated proteins were expressed in E. coli BL21(DE3) gold cells following the same protocol reported for the wild-type proteins (5, 9). hCox17- and hMia40-mutated proteins were purified as previously described for the wild-type proteins (5, 9). Wild-type and mutated yTim10 were similarly obtained (see details in SI Text). All expressed proteins contain four or two additional amino acids (GSFT or GS), corresponding to Tobacco Etch Virus or thrombin protease recognition sites, respectively, at the N terminus. The numbering of all constructs, however, follows the corresponding human or yeast sequences as deposited at the National Center for Biotechnology Information RefSeq sequence database.
Oxidative Coupling Reaction.
The hCox17–hMia40, yTim10–hMia40, and yTim9 peptide–hMia40 complexes were obtained and stabilized through the production of specific cysteine mutants, according to the in vivo identification of the cysteines responsible of the formation of the intermolecular disulfide bond between the two partners (11, 19). In the case of hCox17 and yTim10, three Cys of the twin CX9C or CX3C motif, respectively, were mutated while maintaining the cysteine that forms the intermolecular disulfide bond (Cys45 for hCox17 and Cys40 for yTim10) (11, 19). Similarly, Cys53 of the active CPC motif of hMia40 was mutated, still maintaining Cys55, which is essential in vivo and crucial for the formation of the mixed disulfide bond with the substrate (9). Consistently, the complexes are covalently formed through a disulfide bond between Cys55 of C53S hMia40 and Cys45 of C26/36/55S hCox17 or of Cys40 of C44/61/65S yTim10, as monitored from the 13C chemical shift values of the Cβ of cysteines (28). The yTim9 peptide contains only the cysteine residue responsible for the mixed disulfide bond formation with hMia40 (11, 20).
Purified C26/36/55S hCox17 or C44/61/65S yTim10 mutants were first fully reduced by 100 mM DTT overnight at room temperature under anaerobic conditions and then exchanged into degassed phosphate buffer (KPi 50 mM, pH 7.0, EDTA 0.5 mM) by using a PD-10 desalting column (Amersham Biosciences). The oxidative coupling reaction between C26/36/55S hCox17 and C53S hMia40 or between C44/61/65S yTim10 and C53S hMia40 to obtain C26/36/55S hCox17-C53S hMia40 or C44/61/65S yTim10-C53S hMia40 complexes, respectively, was then performed in the presence of 5 mM ferricyanide [Fe(CN)6]3-, with the proteins ratio of 2∶1 kept for 2 h at 4 °C (Fig. S6). The unreacted proteins were removed from the mixtures by loading the sample, after its concentration, in a 16/60 Superdex 75 chromatographic column (Amersham Biosciences), previously equilibrated in phosphate buffer. The fractions, showing a single component at the molecular weight of the covalent complex in the SDS-PAGE performed in nonreducing conditions, were collected and concentrated by ultrafiltration for NMR analysis (Fig. S6). The same experimental conditions were used for the oxidative reaction between yTim9 peptide and C53S hMia40, with the exception that a 3∶1 peptide∶protein ratio was used. The unreacted peptide was removed by a PD-10-desalting column.
NMR Spectroscopy and Structure Calculations.
NMR experiments and backbone, side-chain assignments were performed following standard procedures that are reported in SI Text. The solution structure of the C26/36/55S hCox17-C53S hMia40 complex was solved by using distance constraints derived from intra- and intermolecular (Fig. S7) NOE cross-peaks, the presence of a disulfide bond formed between Cys55 of C53S hMia40 and Cys45 of C26/36/55S hCox17, and ϕ and ψ dihedral angle constraints (Table S1) derived from the chemical shift index (29) and PECAN programs (30). The program CYANA (31) was used for structure calculations. The best 30 conformers were then energy minimized in explicit water by using the AMBER10 program (32). The conformational, energetic analysis and the stereochemical quality of the final family of 30 structures is reported in Table S1.
The solution structure of yTim9 peptide in the complex with Mia40 was solved through the CYANA program by using distance constraints derived from intramolecular NOE cross-peaks, integrated in the 2D NOESY map with 1H-15N filtering in the two dimensions.
The docking model of the C53S hMia40-yTim9 peptide adduct was calculated through the HADDOCK 2.0 program (33) by using as ambiguous interacting restraints the residues of both C53S hMia40 and yTim9 peptide showing a significant chemical shift perturbation upon complex formation complemented by specific site-directed mutagenesis data (9, 11). Moreover, the intermolecular disulfide bond between the two molecules was included by adding a distance restraint. The structures of C53S hMia402S─S (obtained from homology modeling on wild-type protein, Protein Data Bank ID code 2k3j) and yTim9 peptide (obtained from the above CYANA calculations) were used as a starting point. Structural statistics of the C53S hMia40-yTim9 peptide docking model are reported in Table S2.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by SPINE-II-COMPLEXES Contract LSHG-CT-2006-031220, EU-NMR Contract 026145 “European Network of Research Infrastructures for Providing Access and Technological Advancements in Bio-NMR,” the Italian MIUR-FIRB PROTEOMICA-RBRN07BMCT, the IMBB-FORTH, the University of Crete, and the European Social Fund and National Resources.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structural restraints for the human Cox17–Mia40 complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2L0Y). Resonance assignments are also available at the BioMagResBank www.bmrb.wisc.edu (accession no. 17067).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010095107/-/DCSupplemental.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Lu H, et al. Functional TIM10 chaperone assembly is redox-regulated in vivo. J Biol Chem. 2004;279:18952–18958. doi: 10.1074/jbc.M313045200. [DOI] [PubMed] [Google Scholar]
- 3.Curran SP, Leuenberger D, Oppliger W, Koehler CM. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 2002;21:942–953. doi: 10.1093/emboj/21.5.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webb CT, et al. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed alpha-propeller. Mol Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Banci L, et al. A structural-dynamical characterization of human Cox17. J Biol Chem. 2008;283:7912–7920. doi: 10.1074/jbc.M708016200. [DOI] [PubMed] [Google Scholar]
- 6.Riemer J, Bulleid N, Herrmann JM. Disulfide formation in the ER and mitochondria: Two solutions to a common process. Science. 2009;324:1284–1287. doi: 10.1126/science.1170653. [DOI] [PubMed] [Google Scholar]
- 7.Mesecke N, et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Grumbt B, et al. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J Biol Chem. 2007;282:37461–37470. doi: 10.1074/jbc.M707439200. [DOI] [PubMed] [Google Scholar]
- 9.Banci L, et al. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Mol Biol. 2009;16:198–206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- 10.Kawano S, et al. Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc Natl Acad Sci USA. 2009;106:14403–14407. doi: 10.1073/pnas.0901793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sideris DP, et al. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J Cell Biol. 2009;187:1007–1022. doi: 10.1083/jcb.200905134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milenkovic D, et al. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol Biol Cell. 2009;20:2530–2539. doi: 10.1091/mbc.E08-11-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bien M, et al. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol Cell. 2010;37:516–528. doi: 10.1016/j.molcel.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Stojanovski D, et al. Mitochondrial protein import: Precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J Cell Biol. 2008;183:195–202. doi: 10.1083/jcb.200804095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe Y, et al. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- 16.Gelis I, et al. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobine PA, Pierrel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim Biophys Acta. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Koehler CM, Merchant S, Schatz G. How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem Sci. 1999;24:428–432. doi: 10.1016/s0968-0004(99)01462-0. [DOI] [PubMed] [Google Scholar]
- 19.Sideris DP, Tokatlidis K. Oxidative folding of small Tims is mediated by site-specific docking onto Mia40 in the mitochondrial intermembrane space. Mol Microbiol. 2007;65:1360–1373. doi: 10.1111/j.1365-2958.2007.05880.x. [DOI] [PubMed] [Google Scholar]
- 20.Milenkovic D, et al. Biogenesis of the essential Tim9-Tim10 chaperone complex of mitochondria: Site-specific recognition of cysteine residues by the intermembrane space receptor Mia40. J Biol Chem. 2007;282:22472–22480. doi: 10.1074/jbc.M703294200. [DOI] [PubMed] [Google Scholar]
- 21.Chacinska A, et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naoe M, et al. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J Biol Chem. 2004;279:47815–47821. doi: 10.1074/jbc.M410272200. [DOI] [PubMed] [Google Scholar]
- 23.Terziyska N, Grumbt B, Kozany C, Hell K. Structural and functional roles of the conserved cysteine residues of the redox-regulated import receptor Mia40 in the intermembrane space of mitochondria. J Biol Chem. 2009;284:1353–1363. doi: 10.1074/jbc.M805035200. [DOI] [PubMed] [Google Scholar]
- 24.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 25.Vial S, et al. Assembly of Tim9 and Tim10 into a functional chaperone. J Biol Chem. 2002;277:36100–36108. doi: 10.1074/jbc.M202310200. [DOI] [PubMed] [Google Scholar]
- 26.Portman JJ. Cooperativity and protein folding rates. Curr Opin Struct Biol. 2010;20:11–15. doi: 10.1016/j.sbi.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Vergnolle MAS, et al. Distinct domains of small Tims involved in subunit interaction and substrate recognition. J Mol Biol. 2005;351:839–849. doi: 10.1016/j.jmb.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Sharma D, Rajarathnam K. 13C NMR chemical shifts can predict disulfide bond formation. J Biomol NMR. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- 29.Wishart DS, Sykes BD, Richards FM. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 30.Eghbalnia HR, et al. Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J Biomol NMR. 2005;32:71–81. doi: 10.1007/s10858-005-5705-1. [DOI] [PubMed] [Google Scholar]
- 31.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 32.Case DA, et al. San Francisco: University of California; 2008. AMBER 10 (8.0) [Google Scholar]
- 33.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.