Abstract
Mitochondria and peroxisomes execute some analogous, nonredundant functions including fatty acid oxidation and detoxification of reactive oxygen species, and, in response to select metabolic cues, undergo rapid remodeling and division. Although these organelles share some components of their division machinery, it is not known whether a common regulator coordinates their remodeling and biogenesis. Here we show that in response to thermogenic stimuli, peroxisomes in brown fat tissue (BAT) undergo selective remodeling and expand in number and demonstrate that ectopic expression of the transcriptional coactivator PGC-1α recapitulates these effects on the peroxisomal compartment, both in vitro and in vivo. Conversely, β-adrenergic stimulation of PGC-1α−/− cells results in blunted induction of peroxisomal gene expression. Surprisingly, PPARα was not required for the induction of critical biogenesis factors, suggesting that PGC-1α orchestrates peroxisomal remodeling through a PPARα-independent mechanism. Our data suggest that PGC-1α is critical to peroxisomal physiology, establishing a role for this factor as a fundamental orchestrator of cellular adaptation to energy demands.
Keywords: organelle biogenesis, adaptive thermogenesis, energy metabolism
Peroxisomes differ substantially from mitochondria both from a structural and evolutionary perspective, with the former derived from the endoplasmic reticulum and the latter originated through endosymbiosis (1). Yet, these two organelles exert converging functions on fatty acid degradation and reactive oxygen species (ROS) detoxification. Peroxisomal β-oxidation enzymes such as acyl-CoA oxidase 1 (ACOX1), enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase (PBFE), and 3-ketoacyl-CoA thiolase (PTHIO) reduce fatty acids with carbon chains longer than 22 (2), whereas mitochondrial β-oxidation enzymes only oxidize fatty acids with less than 22 carbon atoms. This substrate specificity implies a potential dependency of mitochondria on peroxisomes. The close interrelationship between these two organelles is further displayed in peroxisomal diseases such as Zellweger syndrome, characterized by absence of functional peroxisomes, or X-linked adrenoleukodystrophy, caused by a defect in the gene responsible for transport of very long fatty acids into peroxisomes. Affected tissues in these patients show profound abnormalities in both mitochondrial morphology and function (3).
In the past few years much effort has been spent in understanding the regulatory mechanisms governing mitochondrial function. PPARγ coactivator-1α (PGC-1α) has emerged as a critical regulator of energy homeostasis. In a variety of tissues, PGC-1α controls oxidative phosphorylation through expression of genes involved in the mitochondrial respiratory chain by cooperating with ERRα (4, 5) and NRF2 (6) and activates the β-oxidation gene expression program by synergizing with PPARα (7). Originally identified as a PPARγ-interacting molecule in brown fat tissue (BAT) (8), PGC-1α is induced in response to cold exposure and has been implicated in adaptive thermogenesis through its role in controlling mitochondrial remodeling and biogenesis (6, 9–11).
Increased energy demands, especially during periods of adaptive thermogenesis, require rapid and coordinated cellular responses to maximize efficient utilization of fatty acids as substrates. The similarities and interrelationship of mitochondria and peroxisomes with regard to energy utilization suggest the existence of a broader regulatory connection. We hypothesized that during periods of increased energy need, the transcriptional coactivator PGC-1α orchestrates not only mitochondrial remodeling but also peroxisomal specialization and biogenesis. Here we show that PGC-1α coordinates peroxisomal remodeling and abundance and that this function is via a PPARα-independent mechanism.
Results
Brown Fat Peroxisomal Compartment Undergoes Remodeling and Biogenesis During Terminal Differentiation and Thermogenic Responses.
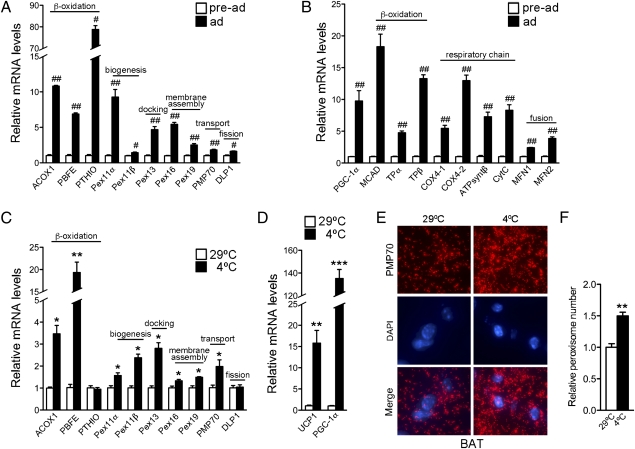
Differentiation of brown fat cells is accompanied by mitochondrial remodeling and biogenesis (11). The close functional interrelationship between mitochondria and peroxisomes suggested the possibility that peroxisomes may similarly undergo remodeling and/or biogenesis during brown fat differentiation. Fig. 1A shows that genes involved in peroxisomal β-oxidation and biogenesis were induced during adipogenesis. These changes were paralleled in mitochondria with the induction of genes involved in mitochondrial β-oxidation and oxidative phosphorylation (Fig. 1B).
Fig. 1.
Peroxisomes are induced in brown fat during differentiation and in cold exposure, along with mitochondria. mRNA expression of peroxisomal (A) and mitochondrial (B) genes in undifferentiated and differentiated brown adipocytes. All values were normalized using 36B4. (C and D) mRNA expression of peroxisomal genes in brown fat tissue of 7-wk-old C57BL/6J female mice maintained at 29 °C or after cold exposure at 4 °C for 7.5 h (n = 3). All values were normalized using 18S. (E) Immunohistochemistry of brown fat tissue obtained from 9-wk-old C57BL/6J female mice kept at 29 °C or cold exposed at 4 °C for 7.5 h, using antibodies against PMP70 and Alexa Fluor 568 goat anti-rabbit (red). The samples were counterstained with the nuclear dye DAPI (blue) (magnification 100×). (F) Quantification of peroxisomal abundance in brown fat tissue at 29 °C or after cold exposure (at 4 °C). Results are shown as relative peroxisomal number per cell ± SEM (n = 3). Statistical significance was determined by unpaired two-tailed Student's test. #P < 0.01, ##P < 0.001, *P < 0.05, **P < 0.01, ***P < 0.001.
BAT assists in adaptation to cold challenges through futile substrate-energy cycles. Morphological studies conducted in the early 1970s using electron microscopy on rodent brown fat showed that the abundance of peroxisomes in this tissue changes during cold exposure (12). To examine whether peroxisomal gene expression is regulated during cold adaptation, we determined whether the expression of peroxisomal enzymes, biogenesis, membrane assembly, and docking factors is affected by cold exposure. We found that peroxisomal genes are induced during thermogenic responses (Fig. 1C) in parallel to PGC-1a and UCP1 (Fig. 1D). To assess whether these alterations in peroxisomal gene expression are accompanied by organelle biogenesis, we quantified the number of peroxisomes within each cell by immunohistochemistry using antibodies to PMP70, a peroxisomal membrane protein that has been reliably used as a marker for peroxisomes. The number of peroxisomes in BAT of cold-exposed mice was significantly higher than that of mice kept at thermoneutrality (Fig. 1 E and F).
PGC-1α Promotes Peroxisomal Remodeling and Biogenesis.
The coordinated responses of peroxisomes and mitochondria to cellular differentiation and cold exposure suggest a common regulator. To assess whether PGC-1α could play such a role, we asked whether PGC-1α could regulate peroxisomal function and biogenesis. First, we overexpressed PGC-1α in several cell lines to measure its effects on the levels of functional and structural peroxisomal genes. Ectopic expression of PGC-1α in fully differentiated mouse brown fat cells, mesenchymal stem cells (10T1/2), and rat hepatoma (FAO) cells (Fig. 2 A–C) induced peroxisomal β-oxidation genes, including ACOX1 and PBFE, and critical peroxisomal biogenesis factors, such as peroxins Pex11α, -11β, -13, and -16 (13–16). As shown in Fig. 2D and Fig. S1, this transcriptional regulation was accompanied by increases in protein levels for many of these factors. Next, to test whether our observations were species specific, we ectopically expressed PGC-1α in human cells. The expression of genes involved in β-oxidation and peroxisomal assembly was induced by PGC-1α in the human U2OS cell line, whereas genes involved in other peroxisomal enzymatic functions, such as those catalyzed by the insulin-degrading enzyme (IDE) and xanthine dehydrogenase (XDH), involved in the degradation of oxidized proteins and in oxidative metabolism of purines, were not affected (Fig. 2E).
Fig. 2.
PGC-1α is sufficient to induce peroxisomal gene expression, protein, and number in vitro. mRNA expression of peroxisomal genes in brown fat (A), 10T1/2 (B), and FAO (C) cells infected with control or PGC-1α adenovirus for 72 h. Values were normalized using 36B4 (A) or 18S (B and C). Results are shown as the mean of three independent experiments ± SEM. (D) Western blot analysis of whole-cell lysates from 10T1/2 and FAO cells infected with control or PGC-1α adenovirus for 72 h. (E) mRNA expression of peroxisomal genes in U2OS cells infected with control or PGC-1α adenovirus for 48 h. All values were normalized using 36B4. Results are shown as the mean of three independent experiments ± SEM. (F) Differences in fluorescent peroxisomes in U2OS cells stably expressing RFP–SKL (RFP–SKL–U2OS) after 72-h infection with GFP or PGC-1α adenovirus, costained with DAPI (magnification 63×). (G) Peroxisome quantification in RFP–SKL–U2OS cells infected with GFP or PGC-1α adenovirus for 48 or 72 h. Results are shown as relative peroxisomal number per cell ± SEM. Statistical significance was determined by unpaired two-tailed Student's test. *P < 0.05, **P < 0.01, ***P < 0.001.
Our results suggest that PGC-1α promotes peroxisomal specialization through selective induction of genes. To determine whether this specialization was accompanied by an increase in peroxisomal number, we quantified the number of peroxisomes in cells that overexpressed PGC-1α. To examine organelle biogenesis, we generated a cell line that constitutively expresses a red fluorescent protein (RFP) fused to the peroxisomal import signal SKL (RFP–SKL) (17). These reporter cells (RFP–SKL–U2OS) were transduced either with adenoviral particles expressing an empty vector or PGC-1α. PGC-1α expression alone was sufficient to increase the number of peroxisomes per cell (Fig. 2 F and G).
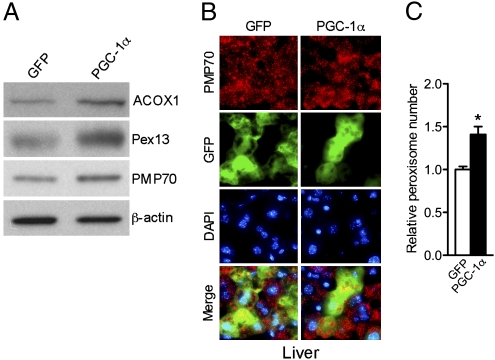
Given our observations showing that PGC-1α can induce peroxisomal remodeling and biogenesis in vitro, we wished to test the effects of PGC-1α expression in vivo. Liver represents a convenient target tissue system for transduction of exogenous genes. To benefit from this model, we used tail vein injections in mice to transduce their livers with adenoviral particles containing either PGC-1α or empty control vectors. Posttransduction, we monitored the levels of peroxisomal proteins and measured organelle biogenesis. Ectopic expression of PGC-1α in mouse livers increased the levels of ACOX1, Pex13, and PMP70 (Fig. 3A and Fig. S2). To evaluate organelle biogenesis, we used immunostaining on biopsy samples obtained from transduced livers. Fig. 3B shows immunohistochemical analysis of tissue samples obtained from mice transduced with PGC-1α adenovirus or control. Using antibodies to PMP70 we found increased numbers of peroxisomes in PGC-1α transduced livers. Fig. 3C shows quantification of organelle numbers using fluorescent microscopy, confirming that there were significantly more peroxisomes per cell in liver samples transduced with PGC-1α. These data support a role for PGC-1α in inducing peroxisomal biogenesis in vivo.
Fig. 3.
PGC-1α is able to induce the peroxisomal program in vivo. (A) Western blot analysis of whole-cell lysates of liver samples obtained from mice injected with GFP or PGC-1α adenovirus for 5 d. (B) Immunohistochemistry of liver cryosections of 8-wk-old male C57BL/6J mice injected with GFP or PGC-1α adenovirus for 5 d using the PMP70 and the Alexa Fluor 568 goat anti-rabbit antibodies (red) and counterstained with the nuclear dye DAPI (blue) (magnification 63×). (C) Quantification of peroxisome abundance in mouse livers injected with GFP and PGC-1α adenovirus for 5 d. Results are shown as relative number of peroxisomes per cell ± SEM. Statistical significance was determined by unpaired two-tailed Student's test. *P < 0.05.
Peroxin Expression Depends on PGC-1α.
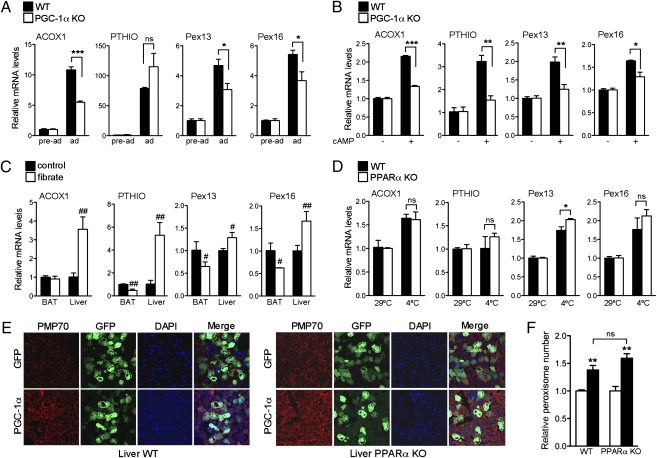
We have shown that PGC-1α is sufficient for peroxisomal biogenesis and remodeling. To assess whether these processes require PGC-1α, we examined peroxisomal function in PGC-1α knockout (KO) cells. As previously demonstrated (11), PGC-1α KO cells differentiate into brown fat without impediment, and, when properly stimulated, PGC-1α KO preadipocytes differentiate into phenotypically identifiable adipocytes, expressing markers of differentiation. Induction of brown fat differentiation in wild-type (WT) cells led to significant increase in ACOX1 and Pex13 and -16, whereas the induction of these genes was blunted in PGC-1α KO cells (Fig. 4A).
Fig. 4.
PGC-1α is required for the induction of peroxisomal genes. (A) mRNA expression of peroxisomal genes in preadipocytes or in fully differentiated WT and PGC-1α KO brown adipocytes. Results are presented as average of three independent experiments ± SEM. Values were normalized using 36B4. (B) mRNA expression of peroxisomal genes in differentiated WT and PGC-1α KO brown adipocytes treated with vehicle or with cAMP for 4 h. Data are shown as average of three independent experiments ± SEM. Values were normalized using 36B4. (C) mRNA expression of peroxisomal genes in brown fat and liver tissues from 10-wk-old male C57BL/6J mice fed with regular chow in the presence or absence of 0.2% fenofibrate for 2 wk (n = 3). Values were normalized using 36B4. (D) mRNA expression of peroxisomal genes in brown fat tissue obtained from 8-wk-old male C57BL/6J WT and PPARα KO mice kept at 29 °C or in the cold (4 °C) for 5 h (n = 3). Values were normalized using 36B4. Results are shown as mean ± SEM. (E) Confocal imaging of liver cryosections from 8-wk-old male C57BL/6J WT and PPARα KO mice injected with GFP or PGC-1α adenovirus for 5 d stained with PMP70 and the Alexa Fluor 594 goat anti-rabbit antibodies (red) and counterstained with DAPI (blue) (magnification 63×). (F) MetaMorph quantification of peroxisomes in mouse livers from WT and PPARα KO mice injected with GFP (open bar) and PGC-1α (filled bar) adenovirus for 5 d. Results are shown as relative number of peroxisomes per cell ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.05, ##P < 0.01. Statistical significance was determined by two-way ANOVA followed by Bonferroni posttest in A, B, D, and F and by unpaired two-tailed Student's test in C.
Although true thermogenic responses cannot be fully replicated in vitro, cAMP stimulation is often used to simulate the adrenergic activation of BAT during cold exposure. We compared the effects of cAMP treatment in PGC-1α KO cells to those obtained in WT cells. As shown in Fig. 4B, the induction of both Pex13 and -16 was significantly reduced in PGC-1α KO cells compared with WT, suggesting that PGC-1α is required for induction of peroxins in response to β-adrenergic stimuli. Interestingly, only a subset of peroxin genes appeared to be regulated by PGC-1α.
The nuclear receptor PPARα plays a pivotal role in regulating hepatic peroxisomal proliferation (18, 19), therefore we wished to examine whether PGC-1α influenced the peroxisomal compartment through this pathway. To determine whether PPARα played a significant role in the activation of peroxisomal gene expresssion in BAT, we assessed induction of β-oxidation and peroxisomal genes in brown fat tissue of mice treated with fibrates. As shown in Fig. 4C, known PPARα target genes such as ACOX1 and PTHIO were not induced in brown fat in response to fenofibrate treatment, suggesting that PPARα is not involved in the regulation of these genes in BAT. Conversely, fenofibrate significantly induced liver peroxisomal gene expression in these mice. To determine whether PGC-1α requires PPARα to regulate Pex13 and Pex16 during cold exposure, we assessed whether these genes can be induced in cold exposure in mice lacking PPARα. Interestingly, both peroxisomal genes (Fig. 4D) were induced at equal levels in BAT of WT and PPARα KO mice, suggesting that PGC-1α regulates biogenesis through transcriptional cooperation with other DNA binding factor(s). These data show that PPARα, required for the regulation of hepatic peroxisomal gene expression in response to fibrates, is dispensable for the induction of peroxisomal genes in BAT in response to cold. To determine whether other known PGC-1α interacting factors could be involved in PGC-1α–mediated peroxin gene induction, we assessed the mRNA levels of Pex13, Pex16, and Pex19 in 10T1/2 cells expressing PGC-1α when ERRα, NRF2, and Foxo1 were knocked down. Despite a significant reduction in the endogenous levels of these factors achieved by siRNA expression, no reduction in gene expression in any of the peroxins examined was observed (Fig. S3), suggesting that PGC-1α regulates these peroxins via a yet uncharacterized transcription factor.
To assess whether PGC-1α induction of peroxisomal biogenesis requires PPARα in the context of the liver, we ectopically expressed PGC-1α in livers of WT and PPARα KO mice and measured peroxisomal proliferation by confocal microscopy of PMP70 immunostained biopses. As shown in Fig. 4 E and F, peroxisomes increase in number in the livers of both WT and PPARα KO mice when PGC-1α is ectopically expressed, suggesting that PGC-1α is a critical regulator of peroxisomal biogenesis.
Discussion
In response to metabolic challenges, cells reorganize their organelles by changing their enzymatic composition, their membrane structure, and by altering the total number of select organelles (1). Through these modifications, cells cope with hormonal cues or nutrient challenges; for instance, in response to β-adrenergic stimulation, brown fat cells specifically increase the total numbers of mitochondria as well as their repertoire of respiratory enzymes, thereby increasing their capacity for thermogenesis (11).
In this study, we demonstrate a prominent role for PGC-1α in orchestrating peroxisomal remodeling and biogenesis, suggesting significantly more coordination in cellular responses to metabolic stimuli than previously appreciated. Gain-of-function experiments show PGC-1α sufficiency in inducing both peroxisomal remodeling and biogenesis. This link appears to be quite robust because our findings apply to both rodent and human cells, which have generally been considered to have divergent responses to fibrates. In support of our results in cell lines, we have also demonstrated similar changes in in vivo conditions.
Peroxisomes generated via overexpression of PGC-1α are specialized in a subset of functions related to β-oxidation, whereas expression of genes encoding for other peroxisomal enzymes is not affected. These changes in enzyme composition and in peroxisomal number induced by PGC-1α are reminiscent of the effects observed in yeast during the shift in their growth to oleic acid as a sole carbon source. Under those growth conditions, expression of genes encoding for enzymes involved in β-oxidation are accompanied by expansion of the peroxisomal compartment, producing a net increase in number of these specialized organelles per cell (20).
Our studies suggest that the peroxisomal program induced by PGC-1α is PPARα independent. Given the central role of fibrates and PPARα in regulating peroxisomal abundance, this result was a surprise. We further show that fibrates, which are highly effective in inducing peroxisomal gene activation in liver, do not regulate the expression of peroxisomal genes in BAT. We show here that PGC-1α induces a series of genes in BAT that have not been previously described as PPARα target genes and that lack PPRE sites in their promoters. Specifically, PGC-1α induces Pex11β, a gene known to be a potent peroxisomal biogenesis factor in the absence of fenofibrate (13). Others have demonstrated that the ectopic expression of Pex11β promotes peroxisome proliferation in cells that are peroxisome deficient due to absence of functional Pex5 (21). Given the relevance of Pex11β in peroxisomal proliferation, the induction of this specific peroxin by PGC-1α during cold exposure in BAT is significant. PGC-1α appears to be activating a PPARα-independent pathway of organelle biogenesis in response to very specific conditions of high fuel and energy demand. In addition to Pex11β, we found that the expression of Pex13 and -16, whose mutations lead to Zellweger syndrome (15, 16, 22) require PGC-1α. These genes are dramatically reduced in PGC-1α KO cells. No PPRE sites have been previously identified in the Pex13 and -16 promoters, suggesting that these proteins are regulated in a PPARα-independent way.
In conclusion, our data show a critical role for PGC-1α in the regulation of peroxisomal function and biogenesis. Furthermore, we demonstrate a unique transcriptional network governing peroxisome function that is independent of PPARα. It is of interest to examine whether PGC-1α effects on peroxisomes described in BAT and liver could extend to tissues such as the brain, where PGC-1α function has also been shown to be critical. Novel chemical regulators of PGC-1α that may lead to peroxisomal remodeling and biogenesis could be promising for the treatment of disorders such as Zellweger syndrome (23).
Materials and Methods
Constructs and Reagents.
Adenoviruses expressing GFP and PGC-1α were a kind gift of B. M. Spiegelman (Harvard Medical School, Boston, MA). The RFP–SKL plasmid was a generous gift of J. Lippincott-Schwartz (National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD). The primary antibodies against PMP70, ACOX1, and β-actin were obtained, respectively, from Invitrogen, Abgent, and Sigma. Anti–PGC-1α, -Pex13, and -DLP1 antibodies were purchased from Santa Cruz. The secondary antibodies ECL Rabbit IgG, HRP-linked, and ECL mouse IgG, HRP-linked were purchased from GE Healthcare. Alexa Fluor 568 and 594 goat anti-rabbit secondary antibodies were purchased from Invitrogen. Insulin, T3, IBMX, dexamethasone, indomethacin, dibutyryl cAMP, and fenofibrate were purchased from Sigma-Aldrich and geneticin from Invitrogen.
Cell Culture.
U2OS and 10T1/2 were purchased from ATCC and FAO cells were purchased from Sigma. Immortalized WT and PGC-1α null mouse brown preadipocytes were a generous gift of B. M. Spiegelman (Harvard Medical School, Boston, MA). U2OS, 10T1/2 cells and brown preadipocytes were maintained in DMEM (Cellgro) containing 10% FBS (HyClone) and 1% pen/strep (Cellgro). FAO cells were maintained in F12 medium with l-glutamine (Gibco) supplemented with 10% FBS and 1% pen/strep. Adipocyte differentiation of brown preadipocytes was performed as previously described (24).
To stimulate the thermogenic program, fully differentiated brown adipocytes were incubated with dibutyryl cAMP for 4 h. A clonal U2OS cell line stably expressing RFP–SKL was generated by transfecting the RFP–SKL plasmid with FuGene (Roche), followed by geneticin selection (500 μg/mL) and isolation of a single colony using a cloning cylinder. For adenoviral infections, RFP–SKL–U2OS cells were infected with 400 multiplicity of infection (MOI) of PGC-1α or GFP adenovirus in 0.5% FBS-containing medium. Cells were harvested for RNA analysis 48 h after infection or fixed for peroxisome staining 72 h after infection. 10T1/2 cells and FAO cells were plated at 70% confluency and infected with PGC-1α or control adenovirus in 0.5% FBS containing medium and harvested for mRNA or protein analysis 72 h postinfection. Fully differentiated brown adipocytes were infected with 4,000 MOI of PGC-1α or control adenovirus in 10% FBS medium after preincubation of the viral particles with lipofectamine (Invitrogen) to improve adenoviral infection efficiency. Cells were collected for RNA analysis 72 h later. For knockdown experiments, 10T1/2 cells (0.25 × 106) were transfected with 100 nM siRNA (siGENOME SMARTpool, Dharmacon) using the 96-well Amaxa Shuttle (Lonza). Forty-eight hours after transfection, cells were infected with 600 MOI of PGC-1α or GFP adenovirus in 0.5% FBS medium and collected for RNA analysis 48 h later.
Gene Expression Analysis.
Total RNA was extracted from cultured cells or tissues with TRIzol (Invitrogen) and treated with DNase (Ambion). mRNA was reverse transcribed using the High Capacity cDNA Archive kit (Applied Biosystems). Real-time PCR was performed with the ABI PRISM 7900HT sequence detection system (Applied Biosystems) using SYBR green (Roche). Gene expression levels were determined using the delta Ct method, after normalization to 18S or 36B4 expression. Sequences of primers used for real-time PCR are listed in Table S1.
Western Blot Analysis.
Whole-cell extracts were prepared using lysis buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1mM EDTA, 1% Triton X-100, 1% sodium pyrophosphate and protease inhibitor mixture (Roche). Tissue whole-cell extracts were prepared by grinding samples in lysis buffer. Western blots were performed using polyacrylamide 4–12% Bis-Tris gradient gels (Invitrogen) and PVDF membranes (Pall). Detection and autoradiography were performed using ECL (Pierce) and B-Plus films (Ewen-Parker X-Ray). Blots were quantified using Adobe Photoshop CS2 9.0 software.
Immunofluorescence.
RFP–SKL–U2OS cells were plated at 50% confluency on Lab-Tek slides (Nalge Nunc International) and fixed, after infections, in 4% formaldehyde (Invitrogen) for 15 min at 37 °C. Tissues were dissected and fixed in 4% paraformaldehyde (Electron Microscopy Sciences) overnight at 4 °C, rinsed with PBS, cryopreserved in 30% sucrose, and embedded in OCT (Electron Microscopy Sciences). Cryosections of liver (6 μm) and brown fat (8 μm) were air dried for 45 min, stained for PMP70 with the peroxisome labeling kit (Invitrogen), according to manufacturer protocol, mounted with DAPI-containing solution (Invitrogen), and analyzed with an Axioplan 2 imaging microscope (Carl Zeiss). Images were captured using a CoolSNAP HQ camera (Photometrics) and processed with IP Lab 3.9 software (Scanalytics). The number of peroxisomes per cell was quantified by counting the number of PMP70 positive organelles per cell in at least 20 randomly selected cells for each condition, using Adobe Photoshop CS2 9.0 software.
Confocal images were acquired using a Zeiss LSM 510 META microscope with a 63× NA = 1.4 oil immersion objective (Carl Zeiss MicroImaging). Peroxisome quantification of confocal images was performed using the Granularity Application Module of the MetaMorph software (64-bit, version 7.6.5.0, Molecular Devices).
Animal Experiments.
All animal experiments were performed according to guidelines of the National Institute of Diabetes and Digestive and Kidney Diseases’ Animal Care and Use Committee. C57BL/6J mice were purchased from The Jackson Laboratory and Taconic and PPARα null mice were obtained from Taconic. Adenoviruses expressing GFP and PGC-1α (0.5–2 × 109 PFU) were delivered by tail-vein injection. Livers were collected 5 d after injection: a portion was fixed in 4% paraformaldehyde for immunofluorescence, and the remaining tissue was snap frozen and stored at −80 °C. For cold-exposure experiments, mice individually caged and kept overnight at 29 °C were exposed to cold temperatures (4 °C) with free access to water for 5 or 7.5 h. Interscapular brown fat was divided into two parts, one of which was used for immunofluorescence and the other for RNA processing. For fibrate treatment, mice were fed powdered chow with or without fenofibrate (at 0.2% wt/wt) for 2 wk.
Statistical Analysis.
Data were compared by t test or ANOVA analysis followed by Bonferroni posttest, as indicated, using Prism 5 software (GraphPad Software). Differences were considered significant with P < 0.05. Results are shown as means ± SEM, with n as the number of independent experiments.
Supplementary Material
Acknowledgments
We are grateful to Richard Proia and to members of the laboratory for reading the manuscript and for their helpful suggestions and to Pasha Sarraf for critical discussions and comments. We thank Bruce Spiegelman (Harvard Medical School, Boston, MA) for the kind gift of PGC-1α and GFP adenoviruses and for the WT and PGC-1α KO brown preadipocyte cell lines. We are indebted to Jennifer Lippincott-Schwartz (National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD) for providing us with the RFP–SKL plasmid. We also thank Rick Dreyfuss (NIH Medical Arts, Bethesda, MD) for microscopy pictures, Morgan Gallazzini for confocal imaging, and Deborah Simon and Huiyan Lu for performing tail vein injections.This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009176107/-/DCSupplemental.
References
- 1.Schrader M, Yoon Y. Mitochondria and peroxisomes: Are the ‘big brother’ and the ‘little sister’ closer than assumed? Bioessays. 2007;29:1105–1114. doi: 10.1002/bies.20659. [DOI] [PubMed] [Google Scholar]
- 2.Reddy JK, Mannaerts GP. Peroxisomal lipid metabolism. Annu Rev Nutr. 1994;14:343–370. doi: 10.1146/annurev.nu.14.070194.002015. [DOI] [PubMed] [Google Scholar]
- 3.Goldfischer S, et al. Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science. 1973;182:62–64. doi: 10.1126/science.182.4107.62. [DOI] [PubMed] [Google Scholar]
- 4.Mootha VK, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreiber SN, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci USA. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 7.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 9.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uldry M, et al. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Cannon B, Alexson S, Nedergaard J. Peroxisomal beta-oxidation in brown fat. Ann N Y Acad Sci. 1982;386:40–58. doi: 10.1111/j.1749-6632.1982.tb21406.x. [DOI] [PubMed] [Google Scholar]
- 13.Schrader M, et al. Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J Biol Chem. 1998;273:29607–29614. doi: 10.1074/jbc.273.45.29607. [DOI] [PubMed] [Google Scholar]
- 14.Maxwell M, et al. Pex13 inactivation in the mouse disrupts peroxisome biogenesis and leads to a Zellweger syndrome phenotype. Mol Cell Biol. 2003;23:5947–5957. doi: 10.1128/MCB.23.16.5947-5957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honsho M, et al. Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am J Hum Genet. 1998;63:1622–1630. doi: 10.1086/302161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 19.Dreyer C, et al. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68:879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 20.Rottensteiner H, et al. Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J. 1996;15:2924–2934. [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Gould SJ. PEX11 promotes peroxisome division independently of peroxisome metabolism. J Cell Biol. 2002;156:643–651. doi: 10.1083/jcb.200112028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Dirbashi OY, et al. Zellweger syndrome caused by PEX13 deficiency: Report of two novel mutations. Am J Med Genet A. 2009;149A:1219–1223. doi: 10.1002/ajmg.a.32874. [DOI] [PubMed] [Google Scholar]
- 23.Santos MJ, et al. Peroxisomal proliferation protects from beta-amyloid neurodegeneration. J Biol Chem. 2005;280:41057–41068. doi: 10.1074/jbc.M505160200. [DOI] [PubMed] [Google Scholar]
- 24.Tseng YH, Kriauciunas KM, Kokkotou E, Kahn CR. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol Cell Biol. 2004;24:1918–1929. doi: 10.1128/MCB.24.5.1918-1929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.