Abstract
Phosphatidylinositol 3,4,5-triphosphate (PIP3) plays a key role in neuronal polarization and axon formation. PIP3-containing vesicles are transported to axon tips by the kinesin KIF13B via an adaptor protein, centaurin α1 (CENTA1). KIF13B interacts with CENTA1 through its forkhead-associated (FHA) domain. We solved the crystal structures of CENTA1 in ligand-free, KIF13B-FHA domain-bound, and PIP3 head group (IP4)-bound conformations, and the CENTA1/KIF13B-FHA/IP4 ternary complex. The first pleckstrin homology (PH) domain of CENTA1 specifically binds to PIP3, while the second binds to both PIP3 and phosphatidylinositol 3,4-biphosphate (PI(3,4)P2). The FHA domain of KIF13B interacts with the PH1 domain of one CENTA1 molecule and the ArfGAP domain of a second CENTA1 molecule in a threonine phosphorylation-independent fashion. We propose that full-length KIF13B and CENTA1 form heterotetramers that can bind four phosphoinositide molecules in the vesicle and transport it along the microtubule.
Keywords: kinesin, forkhead-associated domain, vesicle transport, phosphatidylinositol triphosphate, neuronal development
Neurons are highly polarized cells that typically feature one long axon and several shorter dendrites. Newborn neurons initially develop immature neurites that undergo constant, random growth and retraction regulated by positive and negative feedback signals (1), and become polarized when this balance is shifted so that one neurite becomes an axon and the others become dendrites. Accumulation of phosphatidylinositol (3,4,5)-trisphosphate (PIP3), which is synthesized by phosphoinositide 3-kinase (PI3K) and catabolized by the phosphatase PTEN (2) at the distal end of one neurite, induces axonal development (2, 3). However, an alternative pathway for PIP3 accumulation is the transport of PIP3-containing vesicles by KIF13B and the PIP3-binding protein centaurin α1 (CENTA1) (4).
KIF13B is a kinesin-3 subfamily member containing an N-terminal motor domain with ATPase activity that drives movement along the microtubule and a forkhead-associated (FHA) domain, the only known phosphothreonine (pThr)-specific recognition domain (5–8). KIF13B also contains a long coiled-coil domain and a C-terminal glycine-rich microtubule-binding domain (9), both of which function in dimer formation (10). KIF13B does not interact with PIP3-containing vesicles directly; rather, it binds via its FHA domain to CENTA1 (4), which contains an ArfGAP domain that inactivates Arf6 (11) and two pleckstrin homology (PH) domains that interact with PIP3 (12–15). CENTA1 is associated with presynaptic vesicle structures (16), and its expression is elevated in the neurons of Alzheimer’s patients (17).
To understand how CENTA1 and PIP lipids interact, we solved the structures of lipid-free and lipid-bound forms of CENTA1 and confirmed binding specificity using a PIP array assay. We found that the first PH domain of CENTA1 binds PIP3, while the second binds phosphatidylinositol (3,4)-bisphophate (PI(3,4)P2) and PIP3. We also solved the structure of the KIF13B-FHA domain bound to full-length CENTA1, which interacts with KIF13B-FHA mainly through its first PH domain in a phosphorylation-independent manner. Our result provides structural evidence that FHA can function as a non-pThr-binding module. The structural findings suggest that KIF13B-mediated cargo transport happens by two KIF13B and two CENTA1 molecules forming a heterotetramer that binds up to four PIP molecules in each vesicle.
Results
Crystal Structure of Free and IP4-Bound Centaurin α1.
We solved the crystal structures of CENTA1 in lipid-free and PIP3 head group (IP4)-bound conformations (Fig. 1A and Table S1). CENTA1 is an L-shaped protein with three domains linked by two short helices: an N-terminal ArfGAP domain and PH1 and PH2 domains. The lipid-binding pockets of the two PH domains are on the same, nearly flat surface (Fig. 1 A and B). The ArfGAP domain contains a C4-type zinc finger, and has a contact interface of 517 Å2 with the PH1 domain, but is far from the PH2 domain. The PH1 and PH2 domains both adopt a typical PH fold (18) consisting of a β-barrel of orthogonal three- and four-strand β-sheets, with one end blocked by a C-terminal α-helix and the other end open for binding phosphoinositides. The PH1 and PH2 domains have an average contact area of 523 Å2, with multiple hydrophobic and polar interactions. Residues 360–370 are invisible in the lipid-free structure, but appear as a three-turn α-helix in the IP4-bound structure (Fig. 1A), the C terminus of which points to the IP4-binding surface. The Cα atoms of the ArfGAP domain have an rmsd difference of 0.19 Å between the lipid-free and the IP4-bound structures, while the rmsds of PH1 and PH2 are 0.38 Å and 0.53 Å, respectively. The flexibility of the PH2 domain is consistent with its high B-factor (Fig. S1).
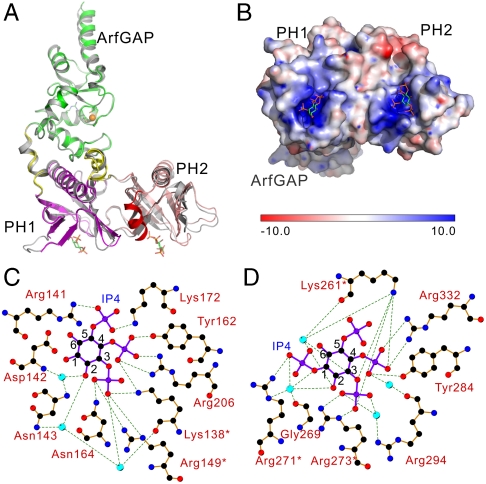
Fig. 1.
Structures of CENTA1 in apo- and IP4-bound forms. (A) Cartoon diagram of CENTA1 in apo-form (gray) aligned with IP4-bound form: ArfGAP (1–119, green), PH1 (131–238, magenta), PH2 (253–359, salmon red), interdomain linkers (120–130, 239–252, yellow), C-terminal helix (360–370, red). IP4 molecules and CENTA1 zinc-finger cysteines are shown in sticks. Zinc is shown as an orange sphere. (B) Bottom-up view of the ± 10.0 kT/e electrostatic potential surface of CENTA1 in the IP4-bound structure. (C) and (D) Ligplot representations of the interaction between IP4 and PH1, PH2 domains respectively. Residues from the KXn(K/R)XR motif are marked with stars.
In the CENTA1-PIP3 structure, the IP4 moiety of the dioctanoyl-PIP3 occupies the phosphoinositide-binding pocket of both PH domains, but the diacylglycerol group is not visible in the electron density map. We hereinafter refer to IP4 as the ligand bound to CENTA1. The phosphate groups of IP4 make polar contacts with similar sets of residues around the binding pockets of both PH domains (Fig. 1 C–D). However, while PH2 contains the canonical KXn(K/R)XR motif in the β1-β2 loop important for high-affinity 3-phosphoinositide-binding (18–20), PH1 has a leucine (Leu147) in place of the (K/R) residue, which is not seen in any other known phosphoinositide-binding PH domains. The invariant lysine residue in all canonical polyphosphoinositide-binding PH domains, Lys138 in PH1 and Lys261 in PH2, interacts with both the 3- and 4- phosphates of IP4. The invariant arginine residue, Arg149 in PH1 and Arg273 in PH2, interacts with the 3-phosphate of IP4. In both cases, the axial 2-hydroxyl group points to the β1-β2 loop and forms a water bridge with residues in the sequence motif, thus fixing the inositol ring in the β-barrel cleft.
Variability among other residues results in differential phosphoinositide-binding specificity of the PH domains. The guanidinium group of Arg271 in PH2 interacts directly with the 1-phosphate, and indirectly through a water bridge (Fig. 1D). This water molecule also forms a strong hydrogen bond with the main chain carboxyl group of Gly269 and with the 2-hydroxyl group of the inositol ring. In contrast, Leu147 of PH1 (the position equivalent to Arg271) does not contact IP4, and the 1-phosphate of IP4 is not visible. Tyr162 and Arg206 in PH1 and their equivalent residues, Tyr284 and Arg332, in PH2 interact directly with a 4-phosphate group. Strikingly, the 5-phosphate of PH2-bound IP4 does not directly contact PH2, while that of PH1-bound IP4 forms two strong hydrogen bonds with the Arg141 and Lys172 side chains of PH1. The equivalent residues in PH2 are Pro264 and Arg294. While the side and main chains of Arg294 interact with the 3- and 4-phosphates of IP4 bridged by two water molecules, Pro264 is oriented such that the side chain of Lys265 is far from the IP4 5-phosphate, preventing 5-phosphate interaction with the PH2 domain. These structural data (Fig. 1 C–D) suggest that the two PH domains have different phosphoinositide-binding specificity, with PH1 binding PIP3 and PH2 interacting with both PIP3 and PI(3,4)P2.
PIP Array Assay and Thermostability.
To test the binding of CENTA1 to all eight naturally occurring phosphoinositides, we performed a membrane-based phosphoinositide array assay. Consistent with the structure-based prediction, CENTA1 interacts with both PIP3 and PI(3,4)P2, though more weakly with the latter (Fig. 2A), and does not bind to other phosphoinositides.
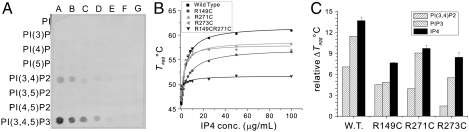
Fig. 2.
Biochemical and biophysical characterization of phosphoinositides-CENTA1 interaction. (A) PIP array assay. Lanes A–G, each spot contains 100, 50, 25, 12.5, 6.25, 3.13, and 1.56 pmol of corresponding phosphoinositides on the membrane. (B) Tagg titration curve of wild-type and mutant CENTA1 at different IP4 concentrations. (C) Relative aggregation temperature of wild-type and mutant CENTA1 to that of R149CR271C double mutant. Values for IP4 are fitted ΔTmax results from titration. Values for PIP3 and PI(3,4)P2 are ΔTagg in the presence of 100 μg/mL phosphoinositides (109 and 123 μM, respectively).
To test the effect of phosphoinositides on CENTA1 stability and of key protein residues on lipid-binding affinity, we used differential static light scattering (21, 22) to measure the thermal aggregation of wild-type and mutant CENTA1 in the presence of phosphoinositides (Fig. 2 B–C). Phosphoinositide-interacting residues were mutated: Arg149 in PH1, and Arg271, Arg273 in PH2. Three single-site (R149C, R271C, and R273C) and one double-site (R149CR271C) mutants were generated, and the thermal aggregation temperatures (Tagg) of the proteins were measured.
Not surprisingly, Tagg decreases rapidly with increasing salt concentration (Fig. S2A). We measured the Tagg of wild-type and mutant CENTA1 with varying IP4 concentrations in low salt buffer [20 mM Tris, pH 8.0, 50 mM sodium chloride, 1 mM tri(2-carboxyethyl)phosphine], then fitted the IP4 titration data with a pseudo one-site binding model to extrapolate maximal aggregation temperature increase (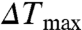 ) (Fig. 2B). Although both PH1 and PH2 interact with three phosphate groups of the phosphoinositides (Fig. 1 C–D), PH1 has a slightly higher affinity for IP4 than does PH2 (
) (Fig. 2B). Although both PH1 and PH2 interact with three phosphate groups of the phosphoinositides (Fig. 1 C–D), PH1 has a slightly higher affinity for IP4 than does PH2 (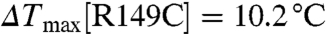 ,
, 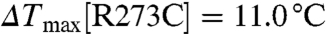 ), consistent with the observation that IP4 appears to be bound to PH1 but not PH2 in the CENTA1/KIF13B-FHA/IP4 complex (see below). In the PH2 domain, Arg273 contributes more than Arg271 to protein stabilization upon IP4 binding, because the
), consistent with the observation that IP4 appears to be bound to PH1 but not PH2 in the CENTA1/KIF13B-FHA/IP4 complex (see below). In the PH2 domain, Arg273 contributes more than Arg271 to protein stabilization upon IP4 binding, because the 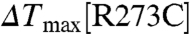 (11.0 °C) is less than
(11.0 °C) is less than 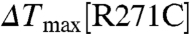 (12.3 °C). Double-point mutations in both PH1 and PH2 (R149CR271C) greatly reduced IP4 binding (
(12.3 °C). Double-point mutations in both PH1 and PH2 (R149CR271C) greatly reduced IP4 binding (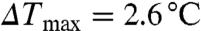 ).
).
Unexpectedly, we observed a decrease in the Tagg of CENTA1 mutants with phosphoinositides bearing long aliphatic side chains, such as dioctanyl-PIP3 and PI(3,4)P2, so simple models do not fit the data (Fig. S2B). To determine the effect of phosphoinositides on the thermal stability of PH domains, we calculated the relative aggregation temperature (Tr,agg) of wild-type and mutant CENTA1, to the double-mutant R149CR271C, which shows near-complete loss of inositol phosphate binding. Tr,agg offsets the effect of phosphoinositide aliphatic chain binding to thermally denatured protein, and is thus a measure of the binding affinity of the head groups to PH domains. When Arg149 is mutated (R149C), phosphoinositide binding occurs mainly in the PH2 domain. The Tr,agg of R149C with PIP3 and PI(3,4)P2 is similar, indicating that PH2 binds equally well to both. In contrast, proteins with mutation in PH2 (R271C and R273C) show higher Tr,agg in the presence of PIP3 than of PI(3,4)P2, indicating that PH1 binds preferentially to PIP3. Thus, the PIP array assay and thermostability measurements confirmed our structure-based prediction that PH1 is PIP3-specific, while PH2 binds to both PIP3 and PI(3,4)P2.
Crystal Structures of CENTA1/FHA and CENTA1/KIF13B/IP4 Complexes.
KIF13B has previously been shown by yeast two-hybrid and GST-pull-down assays to interact directly through its FHA domain with CENTA1 (4, 23). We verified the direct interaction of the KIF13B-FHA domain (aa 440–545) with CENTA1 by gel filtration (Fig. S3A). A longer KIF13B construct containing both the motor and FHA domains (aa 1–550) also interacts directly with CENTA1 by GST-pull-down and gel filtration (Fig. S3B), consistent with an earlier observation that the KIF13B motor domain does not interfere with KIF13B-FHA interactions with CENTA1 (4).
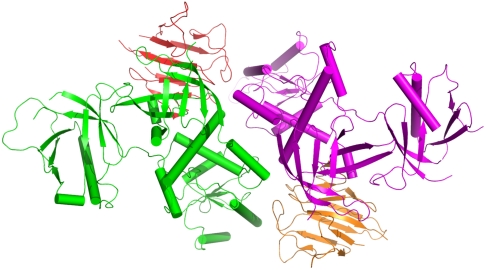
We solved the crystal structure of the CENTA1/KIF13B-FHA complex at 2.3 Å resolution (Table S1). The asymmetric unit contains two CENTA1 and two KIF13B-FHA molecules (Fig. 3), and the CENTA1 in the complex has an overall Cα rmsd of 0.74 Å compared to the lipid-free CENTA1 structure, mainly due to interdomain movement of the PH2 domain upon FHA binding (Table S2). Other significant conformational changes in the complex include disordering of the PH1 β4-β5 and PH2 β5-β6 loops and the appearance of the C-terminal α-helix (aa 360–370). The FHA domain of KIF13B is a typical 11-stranded β-fold common in other FHA structures (5, 24, 25). In the asymmetric unit, FHA residues located mainly on strands β7 and β10 and in the β1-β2 loop interact with a surface of PH1 orthogonal to the phosphoinositide-binding pocket, which consists of residues at the N terminus of β2, the β5-β6 loop, and the β7−α-helix loop. The FHA domain and PH1 interface has an average buried surface area of 663 Å2 and contains multiple polar and hydrophobic interactions (Fig. S4A). In addition, the interface contains three bridging water molecules. The KIF13B FHA domain also interacts with the ArfGAP domain of the second CENTA1 molecule in the asymmetric unit through its Asn536 and Met486 side chains (Fig. S4A). Thus, KIF13B-FHA interacts mainly with the PH1 domain of one CENTA1 molecule, and less extensively with the ArfGAP domain of the second CENTA1 molecule. The CENTA1-CENTA1 interface has a buried surface of 667 Å2 and contains multiple polar and hydrophobic interactions, and three bridging water molecules (Fig. S4B). The interactions mainly happen between the first interdomain linker of one CENTA1 molecule with the first interdomain linker and PH1 domain of the second CENTA1 molecule.
Fig. 3.
Structure of CENTA1 bound to KIF13B-FHA domain in the asymmetric unit.
The structure of the CENTA1/KIF13B-FHA/IP4 ternary complex was also solved at 2.95 Å resolution (Table S1). The overall structure is almost the same as that of the binary complex, with an rmsd of 0.21 Å for all the Cα atoms of both KIF13B and CENTA1. In contrast to the CENTA1/IP4 binary complex where both the PH1 and PH2 domains were occupied by IP4 ligands, however, a IP4 ligand occupies only one of the two PH1 domains of the heterotetramer in the ternary complex, but not the other PH1 domain and the PH2 domains. Compared to the structure of CENTA1/IP4, the most obvious conformational change is the visibility of the 1-phosphate of the IP4 bound to PH1 in the ternary complex. Thus, no major conformational change is seen when IP4 is bound to the CENTA1/KIF13B-FHA complex.
CENTA1/KIF13B Interaction Specificity.
To confirm that the interface between KIF13B-FHA and CENTA1 observed in the crystal structure is biologically relevant, we introduced single-site mutations in both CENTA1 and KIF13B-FHA and measured the thermodynamic parameters using isothermal titration calorimetry (ITC). Wild-type proteins interact at a Kd of 0.8 μM, while single conservative mutations result in enthalpy-entropy compensation (Table S3 and Fig. S5 A and B). For example, the Y211F CENTA1 mutation eliminates interactions between the Tyr211 hydroxyl group and the Asn454 and Asn537 side chains of KIF13B-FHA, leading to a ΔH increase of 3.4 kcal/mol due to loss of the polar interactions, and an entropy increase (-TΔS) of 2.98 kcal/mol. The net free energy change is only 0.4 kcal/mol. Mutation of Tyr211 to glycine or ariginine caused CENTA1 to precipitate at the temperature used for ITC measurement. The interactions of CENTA1 Y211G, and Y211R mutants with KIF13B-FHA were thus tested using size exclusion chromatography, and their complex formation was not detectable (Fig. S5C).
To test the binding specificity between CENTA1 and the FHA domains of different kinesin-3 family members, we carried out GST-pull-down experiments. CENTA1 interacts with a GST-tagged KIF13B motor+FHA construct and a GST-tagged KIF13A motor+FHA construct, but not with the GST control (Fig. S6). Furthermore, the FHA domains of KIF1B, KIF1C, KIF14, and KIF16B do not interact with GST-CENTA1, while KIF13B-FHA does (Table S4). Thus, CENTA1 interacts with KIF13A and 13B, but not with other kinesin-3 family members.
Discussion
CENTA1 Binds to both PIP3 and PI(3,4)P2.
Our data indicate that the PH1 domain of CENTA1 preferentially binds PIP3, while the PH2 domain binds both PIP3 and PI(3,4)P2. Although the 3- and 4- but not the 5-phosphate groups are recognized by the PH2 domain of lipid-bound CENTA1, the binding affinity of PH2 to PIP3 is comparable to that of PH1, due to coordination of the 1-phosphate by the Arg271 side chain and by solvent-mediated interactions with Gly269.
PI(3,4)P2 and PIP3 are two major PI3K kinase products that regulate intracellular signaling (26, 27). While PIP3 is mainly in the plasma membrane, PI(3,4)P2 is found in the plasma membrane, endoplasmic reticulum, and multivesicular endosomes (28, 29). In developing neurons, the balance of PIP3 generation and dephosphorylation into PI(4,5)P2 leads to constant outgrowth and collapse of immature neurites (1). The role of PI(3,4)P2 is not clear, but it is a direct, positive regulator of Akt/PKB (30, 31) and is insensitive to PTEN inactivation (32, 33). Our data suggest that the CENTA1 PH2 domain may mediate transport of PI(3,4)P2-containing vesicles to the distal end of developing neurons. Thus, delivery of PI(3,4)P2 to neurite tips may provide an alternative signaling pathway to tip the balance of PIP3-mediated feedback loops and regulate neurite outgrowth.
Conformational Change of CENTA1 upon Ligand Binding.
The binding pockets of both PH domains are located on the same surface of CENTA1, allowing them to bind the same vesicle. Upon PIP3 or FHA binding, previously invisible residues C-terminal to the PH2 domain (aa 360–370) of CENTA1 appeared as a short α-helix. The C terminus of the helix points to the lipid-binding surface of the molecule, bringing the last four residues of the molecule (KHKP) close to the vesicle. The basic lysine residues can then bind nonspecifically to the head group of the phosphoinositide-containing vesicle (34), further enhancing the affinity of CENTA1 for the vesicle.
The most significant conformational change of CENTA1 upon PIP3 binding occurs in the PH2 domain. The rmsd of Cα atoms in PH2 with and without lipid-binding is larger than that of the rmsd of the three domains combined (Table S2). However, in the CENTA1/KIF13B-FHA and CENTA1/KIF13B-FHA/IP4 complexes, the rmsds of the Cα atoms in the individual CENTA1 domains are all much smaller than the overall rmsd of the three domains combined when compared to the apo-form of CENTA1. Upon binding of the FHA domain, PH1 shows the largest conformational change among the three domains in the binary complex. In the ternary complex, even though only one IP4 is bound to PH1, PH2 undergoes additional conformation changes (Table S2), suggesting that not only phosphoinositide binding causes local conformational changes in PH domains, but that KIF13B-FHA binding to PH1 of CENTA1 causes PH2 to adjust its orientation, possibly promoting assembly of the PIP3-vesicle/CENTA1/KIF13B transport machinery.
CENTA1 and KIF13-FHA Interaction Is Specific and Phospho-Threonine-Independent.
The discovery that KIF13B interacts with CENTA1 via its FHA domain and transports PIP3 vesicles suggested a cargo-binding role for the FHA domain in kinesins (4). Phosphorylation-independent binding of CENTA1 to KIF13B-FHA was first proposed based on GST-pull-down experiments (4). We have also individually mutated each of the serines, threonines, and tyrosines in the ArfGAP domain of CENTA1 to alanine, tested the binding of the mutants to KIF13B-FHA by GST-pull-down, and found no effect. The structure of KIF13B-FHA-bound CENTA1 and our ITC measurements of CENTA1 and KIF13B-FHA mutant interactions confirmed that KIF13B-FHA binds to both the PH1 and ArfGAP domains of CENTA1 via a unique interface in a phosphorylation-independent manner. Indeed, KIF13B-FHA lacks the conserved phosphate-interacting residues (Fig. S7) and the positively charged surface (Fig. S8) of pThr-binding FHA domains.
KIF13A is a close homolog of KIF13B and differs in the CENTA1-interacting regions by only two residues. Phe514 and Met486 of KIF13B, which interact hydrophobically with Pro192, and the ArfGAP domain of CENTA1 respectively, are a cysteine and a hydrophobic isoleucine in KIF13A (Fig. S4 and Fig. S7). Our GST-pull-down data suggest that CENTA1 binds to KIF13A-FHA and KIF13B-FHA with similar affinity, but does not bind the FHA domain of other kinesin-3 family members. As coimmunoprecipitation data from COS-7 cells (4) showed no interaction between KIF13A and CENTA1, alternative phosphorylation or KIF13A regulation mechanisms may exist in intact cells. KIF13A is known to transport the mannose-6-phosphate receptor via AP1/β1-adaptin complex through its C-terminal tail domain (35) and is expressed in the central nervous system during early mouse development (36). The binding of the FHA domain of KIF13A to CENTA1 suggests that KIF13A may also play a role in PIP3 vesicle transport and neuronal development.
Based on the structure of FHA bound to a pThr-containing peptide (24) and evolutionary trace analysis (37), pThr-binding-independent functions of FHA were proposed. Phosphorylation-independent interactions of FHA with c-Myc have also been reported for SNIP1, a candidate transcriptional regulator (38). The interface residues in our CENTA1/KIF13B-FHA structure lie on strands β7 and β10 of KIF13B-FHA, consistent with the evolutionary trace analysis (37). Structure-based sequence alignment (Fig. S7) suggests that a glycine (Gly474 in KIF13B) following strand β3 is the only invariant residue in FHA domains. All known pThr-binding FHA domains have an arginine after this glycine, while the KIF13B-FHA has a serine. Other proteins, such as MLLT4, PHLDB1, and RADIL have an FHA domain without residues necessary for phosphate group binding. Our CENTA1/KIF13B-FHA structure provides atomic details of how the FHA domain interacts with another protein in a pThr-independent manner. Our results indicate that the FHA domain could function through mechanisms other than phosphothreonine recognition.
PIP3 Vesicle Transport and Cargo Loading.
KIF13B was proposed to function as a dimer based on chemical cross-linking and fragment pull-down data (10). KIF13B dimerizes through its C-terminal domain, unlike conventional kinesins, which dimerize through the coiled-coil domain following the motor domain. In the asymmetric unit of the CENTA1/KIF13B-FHA crystal structure, two CENTA1 molecules and two FHA domains formed a heterotetramer. The two FHA domains do not interact with each other. Instead, each FHA domain interacts with both the PH1 domain of one CENTA1 molecule and the ArfGAP domain of the second CENTA1 molecule, as fewer interactions were seen with the ArfGAP domain (Fig. S4A). In addition, the two CENTA1 molecules form a homodimer with an interface containing multiple hydrogen bonds and salt bridges (Fig. S4B), although CENTA1 homodimer is not detectable in solution (Fig. S3). Together, we predict that each of the two FHA domains of full-length KIF13B dimer may interact with two CENTA1, promoting CENTA1 homodimerization and thereby forming the heterotetramer.
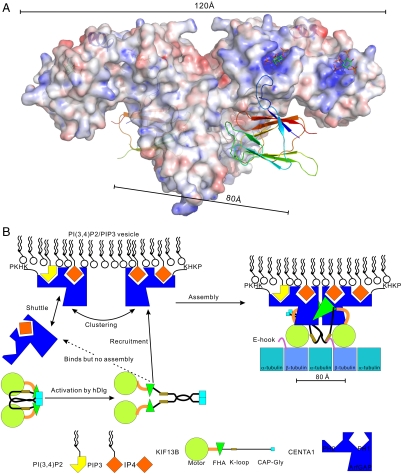
The crystallographic heterotetramer is likely biologically relevant for the following reasons: (i) interaction between the ArfGAP domain of CENTA1 with the N terminus of the FHA-containing KIF13B stalk domain has been verified both by pull-down and in vivo colocalization data (23); (ii) gel filtration of KIF13B-motor-FHA with CENTA1 (Fig. S3B) shows that the complex size approaches heterotetramer size with increased salt concentration, suggesting the asymmetric unit of CENTA1/KIF13B-FHA is primed for heterotetramerization in the context of full-length KIF13B; (iii) the distance between the N termini of the two FHA domains in the asymmetric unit is 80 Å, similar to the distance between adjacent β-tubulin subunits in microtubules, suggesting that the configuration of the crystallographic heterotetramer is compatible with a model of two motor heads bound to two neighboring β-tubulins.
Heterooligomerization of KIF13B with CENTA1 doubles the number of PH domains exposed to the PIP3 vesicle, thus increasing the affinity between the cargo and KIF13B. However, PIP3-containing lipid vesicles may also help to cluster the CENTA1/KIF13B complex into a heterotetramer (Fig. 4A). Dimerization of another kinesin-3 family member, UNC-104, with the aid of PI(4,5)P2-containing liposomes renders the kinesin highly processive and triggers membrane transport (39, 40). Endogenous KIF13B exists in an autoinhibited state until activated by membrane-associated guanylate kinase family member hDlg (10). CENTA1 can shuttle easily between the cytosol and PIP-containing lipid-membrane (41). We propose that CENTA1 is recruited to the membrane, independent of KIF13B activation. Once KIF13B is activated by membrane-bound hDlg, it is recruited to the CENTA1-bound PIP-vesicle through interaction of the FHA domain of KIF13B and the PH1 domain of CENTA1. Clustering of membrane-bound CENTA1 and additional conformational changes of CENTA1 upon KIF13B binding complete the assembly of the vesicle transport machinery (Fig. 4B).
Fig. 4.
Model of the KIF13B-CENTA1 transport machinery. (A) Structural model of KIF13B/CENTA1/IP4 complex. Electrostatic potential surface of centaurin molecules was calculated from IP4-bound CENTA1 coordinate. KIF13B-FHA domains are shown as ribbons. IP4s bound to the PH domains were modeled from the CENTA1/IP4 complex structure. The C-terminal helix of CENTA1 that appeared upon FHA or IP4 binding is shown as a blue ribbon. (B) Assembly of KIF13B/CENTA1 transport machinery.
How nonconventional kinesins achieve processivity has been a matter of debate (42–44). A model based on the crystal structures of the KIF1A motor domain in multiple intermediate states during ATP hydrolysis (44, 45) suggests that the neck linker is the key regulatory element of kinesin motility, and that a stretch of positively charged residues (K-loop) in the KIF1A motor domain interacts with the negatively charged E-hook at the C terminus of tubulins, tethering the kinesin molecule to the microtubule and preventing its premature dissociation from the microtubule (46). A stretch of positively charged residues has also been found in the kinesin CENP-E and is important for its processivity (47). Interestingly, KIF13 kinesins also contain a stretch of positively charged residues not within the motor domain, but after the FHA domain (residues KKKKK, 546-550 for KIF13B, residues KRKRR, 545-549 for KIF13A). A KIF13B construct containing these residues (1–557) can transport PIP3 vesicles processively in the presence of CENTA1 (4). In our model, heterotetramerization of CENTA1/KIF13B does not necessarily regulate processivity; rather, it may provide a mechanism for promoting cargo assembly and coordinating motor head movement.
In summary, our results show the first molecular details of the interaction between a kinesin cargo-binding domain and an adaptor protein. The unusual modular structure of KIF13 subfamily members adds a new twist to the processive transport mechanism of cargo by kinesin motor proteins. The confirmation of phosphorylation-independent binding of the FHA domain to CENTA1 reveals another potential function of what has long been considered a pThr recognition structural module. Finally, the unexpected finding that CENTA1 can recognize PI(3,4)P2 raises questions about the role of this previously largely ignored phosphoinositide in signaling and axon development.
Materials and Methods
We made DNA constructs of CENTA1 and KIF13B with an N-terminal His6 tag into pET28-MHL and purified the expressed proteins by affinity and size exclusion chromatography. Diffraction datasets of the crystals of CENTA1 in various conformational states were collected at the Advanced Photon Source and their structures were determined by using the crystallography programs. Assays of PIP array (Echelon Biosciences) and differential static light scattering using StarGazer (Harbinger Biotechnology and Engineering) were performed by following manufacture’s instructions. ITC measurement was performed by using a VP-ITC MicroCalorimeter by following manufacture’s instructions. We used glutathione Sepharose beads (Novagen) for GST-pull-down assays. The detailed methods are described in the online SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Haizhong Zhu for initial work on KIF13B-FHA domain, and Dr. Toshihiko Hanada for the first proposal and demonstration of phosphorylation-independent binding of KIF13B FHA domain with CENTA1. This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant 371633-09 (to H.-W.P.) and an NIH Grant CA094414 (to A.H.C.). The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck and Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.A.A. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009008107/-/DCSupplemental.
References
- 1.Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- 2.Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- 3.Menager C, Arimura N, Fukata Y, Kaibuchi K. PIP3 is involved in neuronal polarization and axon formation. J Neurochem. 2004;89:109–118. doi: 10.1046/j.1471-4159.2004.02302.x. [DOI] [PubMed] [Google Scholar]
- 4.Horiguchi K, Hanada T, Fukui Y, Chishti AH. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J Cell Biol. 2006;174:425–436. doi: 10.1083/jcb.200604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahajan A, et al. Structure and function of the phosphothreonine-specific FHA domain. Science Signaling. 2008;1:re12. doi: 10.1126/scisignal.151re12. http://www.ncbi.nlm.nih.gov/pubmed/19109241. [DOI] [PubMed] [Google Scholar]
- 6.Liang X, Van D., Sr. Mechanistic insights into phosphoprotein-binding FHA domains. Acc Chem Res. 2008;41:991–999. doi: 10.1021/ar700148u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammet A, et al. FHA domains as phospho-threonine binding modules in cell signaling. IUBMB Life. 2003;55:23–27. doi: 10.1080/1521654031000070636. [DOI] [PubMed] [Google Scholar]
- 8.Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- 9.Steinmetz MO, Akhmanova A. Capturing protein tails by CAP-Gly domains. Trends Biochem Sci. 2008;33:535–545. doi: 10.1016/j.tibs.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Yamada KH, Hanada T, Chishti AH. The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry. 2007;46:10039–10045. doi: 10.1021/bi701169w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkateswarlu K, Brandom KG, Lawrence JL. Centaurin-α1 is an in vivo phosphatidylinositol 3,4,5-trisphosphate-dependent GTPase-activating protein for ARF6 that is involved in actin cytoskeleton organization. J Biol Chem. 2004;279:6205–6208. doi: 10.1074/jbc.C300482200. [DOI] [PubMed] [Google Scholar]
- 12.Hammonds-Odie LP, et al. Identification and cloning of centaurin-α. A novel phosphatidylinositol 3,4,5-trisphosphate-binding protein from rat brain. J Biol Chem. 1996;271:18859–18868. doi: 10.1074/jbc.271.31.18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka K, et al. A target of phosphatidylinositol 3,4,5-trisphosphate with a zinc finger motif similar to that of the ADP-ribosylation-factor GTPase-activating protein and two pleckstrin homology domains. Eur J Biochem. 1997;245:512–519. doi: 10.1111/j.1432-1033.1997.00512.x. [DOI] [PubMed] [Google Scholar]
- 14.Shirai T, et al. Specific detection of phosphatidylinositol 3,4,5-trisphosphate binding proteins by the PIP3 analogue beads: an application for rapid purification of the PIP3 binding proteins. Biochim Biophys Acta. 1998;1402:292–302. doi: 10.1016/s0167-4889(98)00014-7. [DOI] [PubMed] [Google Scholar]
- 15.Venkateswarlu K, Oatey PB, Tavare JM, Jackson TR, Cullen PJ. Identification of centaurin-α1 as a potential in vivo phosphatidylinositol 3,4,5-trisphosphate-binding protein that is functionally homologous to the yeast ADP-ribosylation factor (ARF) GTPase-activating protein, Gcs1. Biochem J. 1999;340:359–363. [PMC free article] [PubMed] [Google Scholar]
- 16.Kreutz MR, et al. Expression and subcellular localization of p42IP4/centaurin-α, a brain-specific, high-affinity receptor for inositol 1,3,4,5-tetrakisphosphate and phosphatidylinositol 3,4,5-trisphosphate in rat brain. Eur J Neurosci. 1997;9:2110–2124. doi: 10.1111/j.1460-9568.1997.tb01378.x. [DOI] [PubMed] [Google Scholar]
- 17.Reiser G, Bernstein HG. Altered expression of protein p42IP4/centaurin-α1 in Alzheimer's disease brains and possible interaction of p42IP4 with nucleolin. Neuroreport. 2004;15:147–148. doi: 10.1097/00001756-200401190-00028. [DOI] [PubMed] [Google Scholar]
- 18.DiNitto JP, Lambright DG. Membrane and juxtamembrane targeting by PH and PTB domains. Biochim Biophys Acta. 2006;1761:850–867. doi: 10.1016/j.bbalip.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Isakoff SJ, et al. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 1998;17:5374–5387. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 21.Vedadi M, et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci USA. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senisterra GA, et al. Screening for ligands using a generic and high-throughput light-scattering-based assay. J Biomol Screen. 2006;11:940–948. doi: 10.1177/1087057106294699. [DOI] [PubMed] [Google Scholar]
- 23.Venkateswarlu K, Hanada T, Chishti AH. Centaurin-α1 interacts directly with kinesin motor protein KIF13B. J Cell Sci. 2005;118:2471–2484. doi: 10.1242/jcs.02369. [DOI] [PubMed] [Google Scholar]
- 24.Durocher D, et al. The molecular basis of FHA domain: phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang P, et al. II. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides. J Mol Biol. 2000;302:927–940. doi: 10.1006/jmbi.2000.4095. [DOI] [PubMed] [Google Scholar]
- 26.Leevers SJ, Vanhaesebroeck B, Waterfield MD. Signalling through phosphoinositide 3-kinases: the lipids take center stage. Curr Opin Cell Biol. 1999;11:219–225. doi: 10.1016/s0955-0674(99)80029-5. [DOI] [PubMed] [Google Scholar]
- 27.Toker A. Phosphoinositides and signal transduction. Cell Mol Life Sci. 2002;59:761–779. doi: 10.1007/s00018-002-8465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watt SA, et al. Detection of novel intracellular agonist responsive pools of phosphatidylinositol 3,4-bisphosphate using the TAPP1 pleckstrin homology domain in immunoelectron microscopy. Biochem J. 2004;377:653–663. doi: 10.1042/BJ20031397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivetac I, et al. The type Iα inositol polyphosphate 4-phosphatase generates and terminates phosphoinositide 3-kinase signals on endosomes and the plasma membrane. Mol Biol Cell. 2005;16:2218–2233. doi: 10.1091/mbc.E04-09-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheid MP, et al. Phosphatidylinositol (3,4,5)P3 is essential but not sufficient for protein kinase B (PKB) activation; phosphatidylinositol (3,4)P2 is required for PKB phosphorylation at Ser-473: studies using cells from SH2-containing inositol-5-phosphatase knockout mice. J Biol Chem. 2002;277:9027–9035. doi: 10.1074/jbc.M106755200. [DOI] [PubMed] [Google Scholar]
- 31.Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 32.Leslie NR, Batty IH, Maccario H, Davidson L, Downes CP. Understanding PTEN regulation: PIP2, polarity and protein stability. Oncogene. 2008;27:5464–5476. doi: 10.1038/onc.2008.243. [DOI] [PubMed] [Google Scholar]
- 33.Downes CP, Perera N, Ross S, Leslie NR. Substrate specificity and acute regulation of the tumor suppressor phosphatase, PTEN. Biochem Soc Symp. 2007;74:69–80. doi: 10.1042/BSS0740069. [DOI] [PubMed] [Google Scholar]
- 34.Roth MG. Phosphoinositides in constitutive membrane traffic. Physiol Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa T, et al. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell. 2000;103:569–581. doi: 10.1016/s0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 36.Jamain S, Quach H, Fellous M, Bourgeron T. Identification of the human KIF13A gene homologous to Drosophila kinesin-73 and candidate for schizophrenia. Genomics. 2001;74:36–44. doi: 10.1006/geno.2001.6535. [DOI] [PubMed] [Google Scholar]
- 37.Lee GI, Ding Z, Walker JC, Van D., Sr. NMR structure of the forkhead-associated domain from the Arabidopsis receptor kinase-associated protein phosphatase. Proc Natl Acad Sci USA. 2003;100:11261–11266. doi: 10.1073/pnas.2031918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujii M, et al. SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol Cell. 2006;24:771–783. doi: 10.1016/j.molcel.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 40.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stricker R, et al. Oligomerization controls in tissue-specific manner ligand binding of native, affinity-purified p42IP4/centaurin α1 and cytohesins-proteins with high affinity for the messengers D-inositol 1,3,4,5-tetrakisphosphate/phosphatidylinositol 3,4,5-trisphosphate. Biochim Biophys Acta. 2003;1651:102–115. doi: 10.1016/s1570-9639(03)00241-3. [DOI] [PubMed] [Google Scholar]
- 42.Valentine MT, Gilbert SP. To step or not to step? How biochemistry and mechanics influence processivity in Kinesin and Eg5. Curr Opin Cell Biol. 2007;19:75–81. doi: 10.1016/j.ceb.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirokawa N, Nitta R, Okada Y. The mechanisms of kinesin motor motility: lessons from the monomeric motor KIF1A. Nat Rev Mol Cell Biol. 2009;10:877–884. doi: 10.1038/nrm2807. [DOI] [PubMed] [Google Scholar]
- 45.Nitta R, Okada Y, Hirokawa N. Structural model for strain-dependent microtubule activation of Mg-ADP release from kinesin. Nat Struct Mol Biol. 2008;15:1067–1075. doi: 10.1038/nsmb.1487. [DOI] [PubMed] [Google Scholar]
- 46.Kikkawa M, Okada Y, Hirokawa N. Resolution model of the monomeric kinesin motor, KIF1A. Cell. 2000;100:241–252. doi: 10.1016/s0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- 47.Rosenfeld SS, et al. The ATPase cycle of the mitotic motor CENP-E. J Biol Chem. 2009;284:32858–32868. doi: 10.1074/jbc.M109.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.