Abstract
Three-amino-acid-loop-extension (TALE) homeodomain proteins including Meis and Pbx families are generally recognized for their roles in growth and differentiation during vertebrate embryogenesis and tumorigenesis. Whereas genetic studies indicate that Pbx1 regulates the development and function of insulin-producing pancreatic β-cells, the role of Meis family members in β-cells is still unknown. Here we show that Meis3 is abundantly expressed in pancreatic islets and β-cells and that it regulates β-cell survival. We further identify the 3-phosphoinositide–dependent protein kinase 1 (PDK1), a well-known kinase involved in the PI3K–Akt signaling pathway, as a direct Meis3 target, which mediates its role in β-cell survival. This regulatory module appears to function broadly as we also identify Meis3 regulation of cell survival and PDK1 expression in ovarian carcinoma cells, suggesting a unique function for Meis3 beyond the traditional roles for TALE homeodomain factors during embryogenesis.
Keywords: diabetes, transcription, pancreas, apoptosis
Three-amino-acid-loop-extension (TALE) homeodomain proteins are recognized as transcription factors responsible for regulating growth and differentiation during vertebrate embryogenesis. In general, TALE homeodomain factors act via the formation of heterodimeric and -trimeric DNA-binding complexes with one another and/or Hox proteins to activate specific gene expression (1–5). The TALE homeodomain superclass is composed of Pbx (Pbx1, Pbx2, Pbx3, and Pbx4) and Meis (Prep1, Prep2, Meis1, Meis2, and Meis3) family members in mammals (6). The founding members of both families were identified through their association with tumorigenesis. The mammalian Meis (myeloid ecotropic viral insertion site) genes were first identified in a murine leukemia model (7). Subsequent studies report that Meis proteins are extensively expressed in a large set of ovarian carcinomas and neuroblastoma (8, 9). Further, Meis1 and Meis2 are expressed in 10 different “cancer modules” within gene expression profiles of 22 diverse tumor types, raising the expectation that more data on Meis members in oncogenesis will emerge (10). However, apart from its role as a Hox cofactor, the function and underlying molecular mechanisms of Meis genes in cancer cells are still poorly understood.
Genetic studies indicate the role of TALE homeodomain protein Pbx1 in the development and function of pancreatic islets, which contain the insulin secreting β-cells required for maintaining normal glucose homeostasis. Pbx1−/− embryos exhibit pancreatic hypoplasia and marked defects in exocrine and endocrine cell differentiation before death at embryonic day (E) 15 or E16, whereas Pbx1+/− adults have pancreatic islet malformations, impaired glucose tolerance, and hypoinsulinemia. Analysis of transheterozygous Pbx1+/−Pdx1+/− mice revealed in vivo genetic interactions between Pbx1 and Pdx1 that are essential for postnatal pancreatic function. Consequently, these mice develop age-dependent overt diabetes mellitus (11). Similarly, a Pbx interaction-defective Pdx1 transgene is unable to rescue normal glucose homeostasis in Pdx1 null mice (12). Although Meis/Pbx/Pdx1 complexes regulate gene expression in cultured ductal and acinar cells (2, 4), no transcriptional targets of Pbx or Meis have been identified in the β-cell, and whether Meis family members are involved in regulating β-cell development and/or function is still unknown.
Here, we show that Meis3 is abundantly expressed in pancreatic islets and cultured β-cells and is required for β-cell survival. We identify the 3-phosphoinositide–dependent protein kinase 1 (PDK1), a well-known kinase involved in the PI3K–Akt signaling pathway, as a direct Meis3 target that mediates its role in β-cell survival. A similar regulatory module operates in ovarian carcinoma cells, despite low levels of Meis3 expression and markedly higher expression of other Meis family members, Meis1 and Meis2. These results define a unique regulatory role for Meis3 in cell survival.
Results
Meis3 Is the Predominant Meis Family Member Expressed in Pancreatic β-cells and Meis3 Deficiency Induces Cell Apoptosis in Min6 β-Cells.
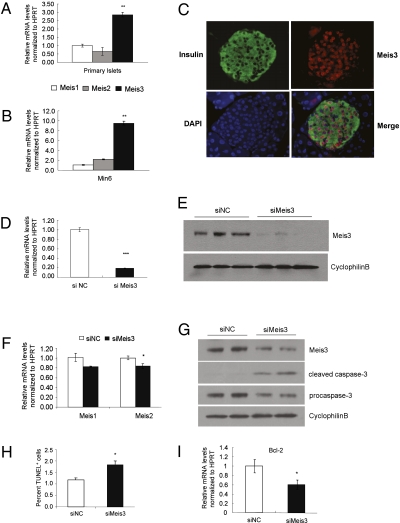
We initially evaluated the expression levels of mouse Meis family members in primary pancreatic islets. Expression of all three Meis genes was detectable, but the level of Meis3 expression was threefold higher than that of Meis1 or Meis2 (Fig. 1A). A similar profile of Meis3 enrichment was observed in cultured Min6 β-cells (Fig. 1B). Meis3 protein was easily detectable in the nuclei of adult pancreatic islets, in both insulin-expressing β-cells as well as non–β-cells (Fig. 1C). Meis1 protein was also nuclear in β- and non–β-cells of the islet, whereas Meis2 protein appeared to be expressed in the perinuclear region of β-cells only (Fig. S1).
Fig. 1.
Expression of Meis3 in mature pancreatic β-cells and its role in cell apoptosis in Min6 β-cells. Relative mRNA levels of Meis family members in wild-type primary islets isolated from adult C57/Bl6 mice (A) and cultured Min6 cells (B), as determined by quantitative RT-PCR. n = 3, **P < 0.01. (C) Representative image of adult mouse pancreatic islets following immunofluorescence for insulin (green), Meis3 (red), and DAPI (blue). D is a merged image of A, B, and C. (D) RT–QPCR analysis of relative Meis3 mRNA levels in siRNA-transfected Min6 cells. n = 3, ***P < 0.0001 compared with values of Meis3 mRNA levels in nontargeted control siRNA (siNC) transfected cells. (E) Representative Western blot analysis of Meis3 protein in transfected cells, performed in triplicate. (F) Relative Meis1 and Meis2 mRNA levels in siMeis3-transfected Min6 cells compared with control detected by RT-QPCR. n = 3. (G) Cleaved caspase-3 protein levels in siRNA-transfected cells assessed by Western blot analysis. Cyclophin B was detected as a loading control. (H) TUNEL staining assay of cell apoptosis after siRNA transfection. Percent adherent TUNEL-positive cells were quantified in 600 cell nuclei per group (n = 3, *P < 0.05 relative to siNC control). (I) RT-QPCR of relative Bcl2 mRNA levels in siRNA-transfected cells. n = 6, *P < 0.05.
To explore the functional role of Meis3 in pancreatic β-cells, siRNA-mediated silencing of Meis3 was performed in Min6 β-cells. Q-PCR analysis showed an 80% reduction of Meis3 transcript compared with nontargeting siRNA control (Fig. 1D) and a marked reduction in Meis3 protein was detected by Western blot analysis (Fig. 1E). In contrast, Meis1 and Meis2 transcripts were relatively unaffected (Fig. 1F), indicating the specificity of Meis3 silencing and the absence of compensatory up-regulation of other Meis family members. We observed increased numbers of floating cells in siMeis3-transfected groups, suggesting that Meis3 depletion results in cell death. Indeed, increased caspase-3 cleavage was observed in Meis3-deficient Min6 cells, accompanied by reduced procaspase-3 (Fig. 1G). Further, increased numbers of apoptotic cells were observed by TUNEL staining (Fig. 1H) and expression of the antiapoptotic gene Bcl-2 was reduced in Meis3-deficient cells (Fig. 1I). Thus, these findings indicate that Meis3 is abundantly expressed in mature pancreatic β-cells and plays a role in β-cell survival.
Meis3 Targets Pdpk1 and Regulates the PDK1/Akt Signaling Pathway in Pancreatic β-Cells.
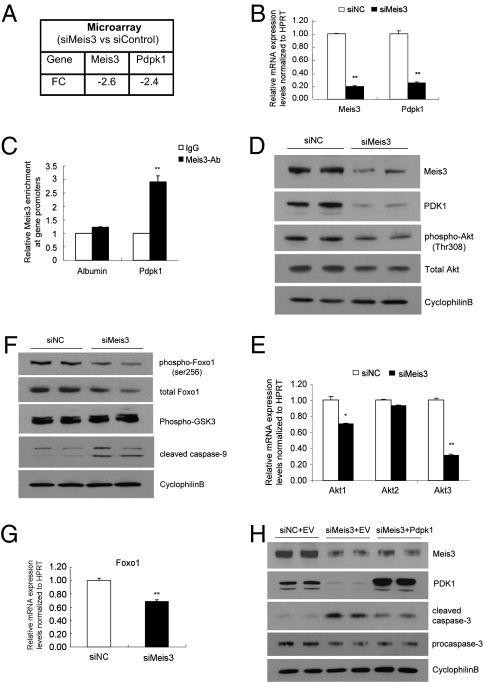
To identify genes regulated by Meis3, we performed Agilent cDNA microarray analysis comparing RNA samples isolated from Meis3-deficient or control Min6 cells. Of 21,609 genes detected, 143 genes were regulated by Meis3 with a fold change (FC) >2 and a 10% false discovery rate (FDR). Thirty-one targets were increased, whereas 112 genes were down-regulated in this profile. Considering the observed cell survival phenotype of Meis3 deficiency, we focused on 3-phosphoinositide–dependent protein kinase 1 (Pdpk1), which was down-regulated 2.4-fold in the setting of a 2.6-fold reduction in Meis3 expression (Fig. 2A). The reduction in Pdpk1 transcript observed by microarray was confirmed by Q-PCR analysis in which Pdpk1 mRNA levels were down-regulated by more than 70% when Meis3 was decreased by 80% (Fig. 2B). A similar regulation of Pdpk1 and caspase-3 cleavage was observed in another established mouse β-cell line, β-HC9 cells (Fig. S2). Other differentially regulated genes with potential roles in cell survival include Ndfip1, Bnip3, and Cyclin G1 (Ccng1). Q-PCR analysis confirmed Meis3 regulation of these genes (Fig. S3).
Fig. 2.
Meis3 directly targets Pdpk1 and regulates the PDK1/Akt signaling pathway in pancreatic β-cells. Down-regulation of Pdpk1 in Meis3-deficient Min6 cells observed by Agilent microarray analysis (A) and confirmed by quantitative RT-PCR (B) (n = 4; ***P < 0.001). (C) Meis3 directly binds to the Pdpk1 promoter as assessed by chromatin immunoprecipitation (ChIP) from Min6 cells. DNA enrichments are presented as fold change compared with control IgG binding. Three independent ChIP experiments were performed for both Meis3 antiserum and IgG, *P < 0.05. (D and F) Western blot analysis of siRNA-transfected Min6 cells with the indicated antisera. (E and G) QRT-PCR analysis of relative gene expression levels in siRNA-transfected Min6 cells, n = 3, **P < 0.01, *P < 0.05. (H) Overexpression of PDK1 partially rescues cell apoptosis in Meis3-deficient Min6 cells. siRNA-mediated Meis3 knockdown experiments performed in Min6 cells, with or without cotransfection of Flag-tagged Pdpk1 expression plasmid. Western blot analysis performed with the indicated antisera.
No direct Meis targets have been reported in pancreatic β-cells, although targets for Meis1 and Meis2 have been identified in acinar and ductal cells. We found a motif similar to the consensus binding site (tgacag) of the highly homologous Meis1 and Meis2 family members (13, 14) within the Pdpk1 promoter. To determine whether Meis3 directly bound the Pdpk1 promoter, chromatin immunoprecipitation (ChIP) assay was performed using our affinity-purified Meis3 antiserum. Q-PCR results revealed a three fold enrichment of Meis3 binding compared with IgG. In contrast, no significant Meis3 occupancy was found at the albumin promoter (Fig. 2C), indicating that Pdpk1 is a specific and direct target of Meis3. To further confirm the transcriptional regulation of Pdpk1 by Meis3, we performed ChIP assays to assess the effect of Meis3 deficiency on RNA polymerase II recruitment to the Pdpk1 proximal promoter. We observed an 11-fold enrichment of Pol-II occupancy at the Pdpk1 promoter in control siRNA-transfected Min6 cells, whereas recruitment was reduced by 52% in Meis3-deficient cells (Fig. S4), further demonstrating the role of Meis3 in the regulation of Pdpk1 transcription.
The Pdpk1 protein product, usually termed PDK1, was originally identified as a kinase critical for Akt/PKB activation in response to stimulation by insulin or other growth factors. Next, we evaluated the effect of Meis3 deficiency on Akt/PKB, which is phosphorylated by PDK1 at Thr308. Following the attenuation in Meis3 protein mediated by siRNA gene silencing in Min6 cells, we observed a dramatic reduction of PDK1 by Western blot, associated with inhibition of Akt/PKB Thr308 phosphorylation (Fig. 2D). Unexpectedly, we also observed a slight decrease in total Akt/PKB protein, which prompted us to examine Akt/PKB transcript levels including the three known Akt isoforms (Akt1/2/3 also called PKBα/β/δ). In Meis3-deficient Min6 cells, Akt1 and Akt3 mRNA levels were decreased by 30 and 68%, respectively, whereas no significant change was found in Akt2 transcript level (Fig. 2E). Thus, reduced Akt1 and Akt3 transcripts could be responsible for the decrease in total Akt protein levels.
The PI3K/PDK1/Akt pathway has multiple prosurvival effects on almost every aspect of apoptosis regulation, including mitochondrial and death receptor pathways, regulation of cell cycle, and regulation of transcriptional and protein translation events (15). We next evaluated the effect of the Meis3 on specific Akt/PKB downstream targets associated with cell survival. By Western blot analysis, both phospho- and total Foxo1 protein were found decreased moderately in parallel with a 32% reduction of Foxo1 mRNA levels in Meis3-deficient Min6 cells (Fig. 2 F and G). Although we did not examine caspase-9 phosphorylation directly, caspase-9 cleavage was visibly increased upon Meis3 depletion (Fig. 2F), consistent with the role of Akt-mediated phosphorylation of caspase-9 (16) and with the observed induction of caspase-3 cleavage (Fig. 1G). In contrast, the phosphorylation of GSKα/β was not affected (Fig. 2F). Taken together, these results suggest that modulation of the PDK1/Akt pathway by Meis3 could be associated with its effect on β-cell survival.
To further evaluate whether PDK1 mediates survival effects of Meis3 in β-cells, Pdpk1 expression plasmids were constructed and cotransfected into Min6 cells in parallel with Meis3-targeted siRNA. Compared with cells cotransfected with nontargeted siRNA control and empty expression vector, PDK1 protein levels were dramatically reduced in siMeis3 and empty vector cotransfected cells, whereas levels were significantly elevated by cotransfection of Pdpk1 plasmid with siMeis3 (Fig. 2H). Further, overexpressed PDK1 attenuated the activation of caspase-3 observed in Meis3-deficient but not control cells (Fig. 2H). To determine whether the partial reduction of caspase-3 activation was due to poor transfection efficiency of expression plasmids as compared with siRNA duplexes, we examined the expression and distribution of endogenous and overexpressed PDK1 in transfected cells by inmmunofluorescence staining. Expression of endogenous PDK1 was effectively inhibited by siMeis3, whereas a small proportion of cells restored PDK1 expression in the case of cotransfection with siMeis3 and Flag-tagged Pdpk1 plasmids (Fig. S5). The limited distribution of overexpressed PDK1 could be responsible for its partial reduction of caspase-3 activity (Fig. 2H). It is also possible that in addition to Pdpk1, other Meis3 transcriptional targets contribute to the survival phenotype. Taken together, our data suggest that the direct downstream target PDK1 could mediate the effect of Meis3 on β-cell survival.
Role of Meis3 in Cell Survival Is Conserved in Primary Islets.
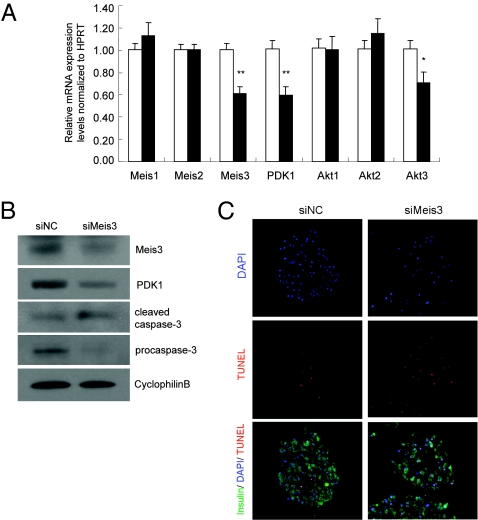
To determine whether the role of Meis3 to regulate Pdpk1 expression and cell survival extends to primary β-cells, we performed siRNA-mediated nucleofection of isolated mouse primary islets. Similar to the observation in Min6 cells, reduction in Meis3 transcripts was followed by a significant decrease in Pdpk1 transcript levels, but did not affect Meis1 or Meis2 gene expression in islets (Fig. 3A). These findings were paralleled by an increase in the abundance of the cleaved form of caspase-3 and a corresponding reduction in procaspase-3 (Fig. 3B). Further, an increase in TUNEL staining intensity in siMeis3-nucleofected primary mouse islets was observed (Fig. 3C and Fig. S6). The reduction in Akt3 transcript was also conserved in Meis3-deficient primary islets, although neither Akt1 nor Akt2 transcript was reduced (Fig. 3A). The significance of this regulatory pattern in β-cells remains unclear. Altogether, these data suggest that Meis3 regulates Pdpk1 expression and cell survival in primary pancreatic islets.
Fig. 3.
Meis3 regulates Pdpk1 gene expression and cell survival in mouse primary islets. Primary islets isolated from wild-type C57/Bl6 males were nucleofected with 1.0 nmol of siNC or siMeis3 and cultured for 4 d before harvest. (A) Quantitative RT-PCR analysis of gene expressions in siRNA-transfected primary islets. n = 5, **P < 0.01, *P < 0.05. (B) Western blot analysis of Meis3, PDK1, and cleaved caspase-3 protein in transfected primary islets. (C) TUNEL staining assay of cell apoptosis for islet sections following siRNA transfection and fixation.
Meis3 Regulates PDK1 and Cell Survival in Ovarian Carcinoma Cells.
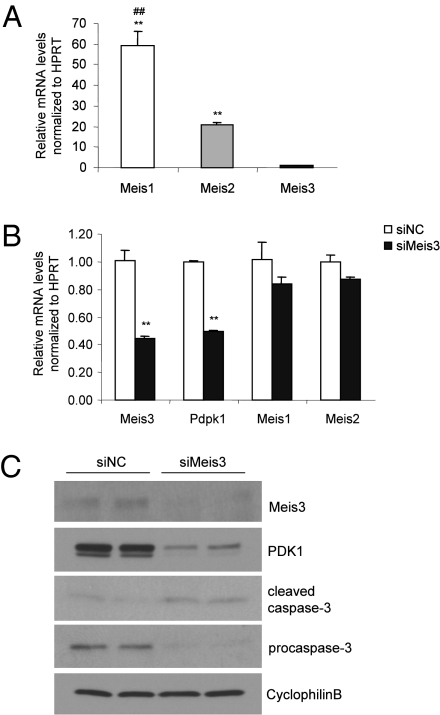
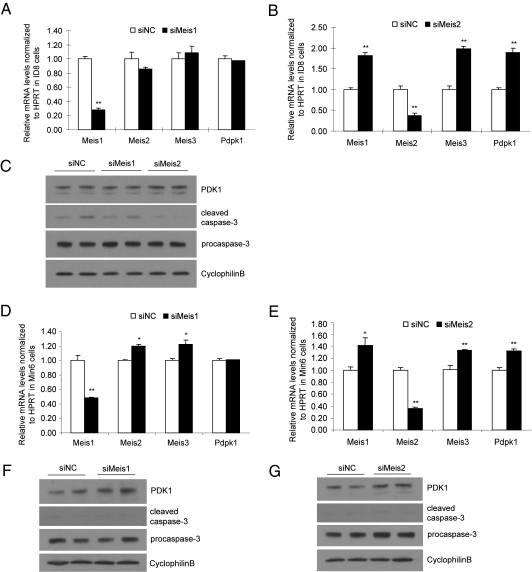
TALE homeodomain factors Meis1, Meis2, and Pbx are highly expressed in ovarian carcinomas. However, the molecular targets of these factors as well as of Meis3 in ovarian carcinogenesis are not known. The critical role of the PI3K–Akt signaling pathway in cell growth and its frequent disruption in human cancers (17) raises the interesting possibility that Meis3 regulation of PDK1 functions in tumor cell survival. We evaluated the expression levels of Meis family members in the mouse ovarian carcinoma ID8 cell. Whereas Meis3 expression was detectable, the levels of Meis1 and Meis2 expression were dramatically higher (Fig. 4A), consistent with previous reports that showed Meis1 is the highest expressed homeodomain factor in ovarian cancer cells compared with other cancer types (8). siRNA-mediated silencing of Meis3 was performed in the ID8 cells. Q-PCR analysis showed a 56% reduction of Meis3 transcript and a marked reduction of Meis3 protein compared with nontargeting siRNA control and no compensatory up-regulation of Meis1 or Meis2 (Fig. 4 B and C). Next we explored whether Meis3 depletion induced cell death in ovarian cancer cells. Indeed, increased caspase-3 cleavage and reduced procaspase-3 were observed in Meis3-deficient ID8 cells in parallel with significantly reduced expression of Meis3 target Pdpk1 at both the mRNA and protein levels (Fig. 4 B and C). These findings indicate that Meis3 is expressed in mouse ovarian carcinoma cells and plays a role in regulating Pdpk1 expression and cell survival.
Fig. 4.
The expression and functional role of Meis3 in ovarian carcinoma cells. (A) Relative mRNA levels of Meis family members in cultured ovarian carcinoma ID8 cells n = 3, **P < 0.001 relative to the Meis3 levels; ##P < 0.001 versus the Meis2 levels. (B) Quantitative RT-PCR analysis of gene expressions in siRNA-transfected ID8 cells. n = 3, **P < 0.01. (C) Western blot analysis of Meis3, PDK1, and cleaved caspase-3 protein in siRNA-transfected cells.
Meis1 or Meis2 Depletion Does Not Regulate Cell Survival.
Given that the binding site sequence of Meis3 on Pdpk1 is homologous to that identified for other Meis genes, Meis1 and Meis2, we hypothesized that Pdpk1 could be a common downstream target of all Meis family members that mediates their function in tumor cells. To our surprise, we observed a marked reduction of Pdpk1 and an increase in caspase-3 cleavage in Meis3-deficient murine ovarian carcinoma cells, despite its dramatically lower expression compared with Meis1 and Meis2 (Fig. 4A), suggesting that the high levels of Meis1 and Meis2 could not substitute for the function of Meis3 in tumor cells. Indeed, siRNA mediated silencing of Meis1 in ID8 cells did not affect Pdpk1 expression (Fig. 5 A and B). Accordingly, caspase-3 cleavage was also unaffected by Meis1 depletion (Fig. 5C). Silencing of Meis2 in ID8 cells was associated with a significant up-regulation of Meis1, Meis3, and Pdpk1 transcript and a subtle decrease in caspase-3 cleavage (Fig. 5 A–C). Thus, despite high levels of expression, Meis1 cannot substitute for Meis3 in the regulation of Pdpk1 and cell survival in ID8 cells. The role of Meis2 could not be definitively established due to compensation by Meis1 and Meis3 for Meis2 deficiency, although it is clear that Meis2 cannot uniquely regulate Pdpk1. A similar pattern of Meis and Pdpk1 gene expression and caspase-3 cleavage was observed upon silencing of Meis1 and Meis2 in Min6 cells (Fig. 5 D–G). In keeping with a unique role for Meis3 in cell survival and gene expression, the expression of Ndfip1, Bnip3, and Ccng1 was unaffected by Meis1 deficiency and up-regulated in concert with Meis3 transcripts in the setting of Meis2 deficiency in Min6 cells (Fig. S7).
Fig. 5.
Role of Meis1 and Meis2 in the regulation of Pdpk1 and cell survival. Relative mRNA levels of Meis1, -2, -3, and Pdpk1 after siRNA-mediated silencing of Meis1 (A and D) and Meis2 (B and E) in ID8 (A and B) and Min6 (D and E) cells. n = 3, *P < 0.05; **P < 0.01. Western blot analysis of PDK1, cleaved caspase-3, procaspase-3, and cyclophilinB after silencing of Meis1 or Meis2 in ID8 (C) and Min6 (F and G) cells.
Discussion
Pancreatic β-cell survival is crucial for the maintenance of β-cell mass and function, and impaired β-cell survival underlies the pathogenesis of both type 1 and type 2 diabetes. Here we unveil the role of Meis3 in regulating β-cell survival via its direct target Pdpk1. Notably, conditional deletion of Pdpk1 in β-cells demonstrated that PDK1 is important in maintenance of pancreatic β-cell survival, islet mass, and glucose homeostasis (18), whereas another study showed that PDK1 activity is also necessary to control proliferation, survival, and growth of developing pancreatic cells (19). These findings together with our present work suggest Meis3 may function in the maintenance of β-cell mass and glucose homeostasis.
Meis family members form heterodimeric and -trimeric complexes with Pbx and/or Pdx1 to regulate gene expression in acinar and ductal cells; however, the potential Meis binding site in the Pdpk1 promoter does not contain nearby consensus-binding sequences for Pbx or Pdx1. Further, Pdpk1 was not dysregulated in cDNA expression arrays examining Pdx1-deficient Min6 and primary islets cells (20). It will be interesting to determine whether Meis3 functions in concert with Pdx1 and/or Pbx family members or as an independent monomeric factor in regulating Pdpk1 gene expression. Notably, recent studies demonstrated that Meis3 and Pbx4 act in a complex to control specific gene expression in zebrafish (21, 22).
Evading apoptosis and self-sufficiency in growth signals have been recognized as important hallmarks of cancer (23). A number of reports reveal that Meis members are involved in oncogenesis (10, 24–26). Meis1 and Meis2 are highly expressed in ovarian cancer and neuroblastoma; however, the function and the molecular mechanism of Meis in cancer cells are still unclear. Our findings here suggest that Meis3 may uniquely regulate PDK1 expression and cell survival in tumor cells. In contrast to Meis3, deficiency of Meis1 or Meis2 did not result in a reduction in PDK1 expression or cell survival as measured by caspase-3 cleavage. Whether this reflects an intrinsic difference in binding specificity or unique interactions of Meis3 with other transcription factors and transcriptional coregulators merits further investigation.
In conclusion, the results presented here identify the role of Meis3, a TALE homeodomain family member, in regulating cell survival via its direct modulation of Pdpk1. This may broaden our understanding of the role of the TALE homeodomain family in mature tissues and cell types beyond their traditionally recognized roles during development. Further investigation of Meis functions and the underlying mechanisms will be relevant to both diabetes and cancer.
Materials and Methods
Antisera.
Meis3 (L-20) for immunostaining was obtained from Santa Cruz, whereas anti-Meis3 for Western blot analysis and chromatin immunoprecipitation was a rabbit polyclonal generated against synthetic peptide ([H]-CKSSAGEDEDLDLERRRN-[NH2], Covance). Anti-PDK1 was obtained from BD Biosciences; antisera directed against caspase-3, caspase-9, Thr308-phospho-Akt, total Akt, Ser256-phospho-Foxo1, total Foxo1, and Ser21/9-phospho-GSK3α/β were from Cell Signaling.
Islet Isolation.
Islet isolations from 10-wk-old wild-type C57/Bl6 males were performed using collagenase digestion similarly to previously described protocols (20). After anesthetizing each mouse, the pancreas was inflated with cold 1× Hank's buffer containing 0.2% collagenase (Crescent Chemical). Following collagenase digestion at 37 °C for 18 min, islets were purified through a Ficoll (GE Healthcare) gradient and then subjected to three rounds of handpicking under a light microscope.
RNA Isolation and Real-Time PCR.
Islet RNA was isolated using the RNeasy Mini kit (Qiagen) following the addition of TRIzol (Invitrogen) and then reverse transcribed with SuperScript II (Invitrogen) using oligo(dT) for priming. For cultured cell lines, cells were harvested and processed for RNA isolation using TRIzol followed by the precipitation with isopropyl alcohol, and the RNA pellets were dissolved in DEPC-treated water. Real-time PCR was performed in triplicate using a Bio-Rad iCycler. Primers sequence information is as follows:
Meis1-F: 5-CAGGAGACCCGACAATGAGT-3
R: 5-GCAGGAAAAGACCTGATTCG-3
Meis2-F: 5-GGGAGAAGTCTCCTTGGTTTG-3
R: 5-CATCAGTCTGCGCTCCAATA-3
Meis3-F: 5-CCACTACCCAGGCATTACAGA-3
R: 5-CCAGGGGCTCTAGGTACTGAA-3
Pdpk1-F: 5-CTCACAGAAGGGCCACATTT-3
R: 5-GCTTCTGGTCGGAGTTCTTG-3
Akt1-F: 5-GAAGCTGGAGAACCTCATGC-3
R: 5-CTTCATAGTGGCACCGTCCT-3
Akt2-F: 5-TTTGCACTCGAGAGATGTGG-3
R: 5-TTTGCACAAGCCAAAGTCAG-3
Akt3-F: 5-GAATCAAGCCATCAGGGAAA-3
R: 5-TGCATACATGTCCCCTCTCA-3
Foxo1-F: 5-GCTGGGTGTCAGGCTAAGAG-3
R: 5-TGGACTGCTCCTCAGTTCCT-3
HPRT-F: 5-GGCCAGACTTTGTTGGATTTG-3
R: 5-TGCGCTCATCTTAGGCTTTGT-3
Cell Culture and Transfection.
Min6 cells were maintained in DMEM containing 10% FBS and 25 mM glucose at 37 °C in 5% CO2. For gene silencing experiments, 1 nmol of Meis3-targeted siRNA duplexes (J-047533–11; Dharmacon), or nontargeting control siRNA duplexes (D-001210–03; Dharmacon) was introduced into ∼1 × 106 cells using nucleofection (Amaxa Biosystems). Cells were harvested at 48 h after nucleofection for RNA or at 72 h for total protein isolation. ID8 cells were cultured in DMEM containing 4% FBS and transfected with siRNA by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Meis1- and Meis2-targeted siRNA pools were also from Dharmacon (L-043400–01 and L-062671-01).
Microarray Analysis.
To identify Meis3 transcriptional targets, total RNA isolated from siNC or siMeis3-transfected Min6 cells (n = 4 independent transfections/group) was analyzed using an Agilent 2100 Bioanalyzer Lab-On-A-Chip Agilent 6000 Series II chip to determine the integrity of the samples. Approximately 400 ng of total RNA was amplified and labeled with Cy3 using the Low RNA Input Linear Amp Kit PLUS, One-Color (Agilent Technologies). After purification, 1.65 μg of cRNA was hybridized to the 44,000 array spotted with Agilent-designed 60-mer oligonucleotides, representing 21,609 known genes represented by 33,661 transcripts. Array analysis was performed using Genespring software (Agilent) and the data file contains all genes that were called differentially expressed by SAM analysis, with a FDR of 10% and a fold change >2. Data have been deposited to ArrayExpress (E-MTAB-273).
ChIP Assay.
The protocol was essential as described previously (20). Rabbit anti-Meis3 antiserum and normal rabbit serum (sc-2338, Santa Cruz Biotechnology) were used for the pull-downs. Data were analyzed quantitatively in duplicate by real-time PCR referenced to a dilution series of the relevant input to account for different efficiencies of primer sets. Forward and reverse primer sequences were as follows:
Pdpk1- F: 5-GAA CAG ACT GGC CTC CTA CTT G-3
R: 5-TGA CAA CTA AAG GAG GAT GTG G-3
Albumin-F: 5-TGGGAAAACTGGGAAAACCATC-3
R: 5-CACTCTCACACATACACTCCTGCTG-3
siRNA Delivery in Isolated Primary Islets.
Primary islets were isolated from wild-type C57/Bl6 males as above, and ∼120 size-matched islets were transfected with 1 nmol of siNC or siMeis3 using the Basic Nucleofector Kit for Primary Mammalian Epithelial Cells (VPI-1005; Lonza) and Program S-005. The transfected islets were cultured for 4 d and then harvested for RNA isolation or total protein extract.
Apoptosis Assay.
siRNA-transfected Min6 cells plated onto chamber slides were fixed by 4% paraformaldehyde (PFA) at 72 h after nucleofection and assessed by TUNEL assay using the Apoptag Peroxidase in Situ Apoptosis Detection Kit (Chemicon) according to manufacturer's instructions. Percent adherent TUNEL-positive cells were quantified in 600 cell nuclei per group. For the apoptosis assay in primary β-cells, siRNA-transfected mouse primary islets were fixed with 4% paraformaldehyde and then transferred into 2% agarose. Agarose plugs were embedded in paraffin for sectioning. Islet sections were costained with guinea pig anti-insulin (Linco) and TUNEL as described above. Images were captured with iVision software (BioVision Technologies) using constant exposure parameters for each fluorescence channel.
Statistical Analysis.
All data represent the mean ± SE. For all comparisons, including qPCR data, ChIP data, and β-cell apoptosis, we assessed statistical significance using a two-tailed Student's t test. Differences were considered significant if P < 0.05.
Supplementary Material
Acknowledgments
We thank David Groff for expert technical assistance and Gary Swain, Technical Director of the Morphology Core of the Center for Molecular Studies in Digestive and Liver Diseases [National Institutes of Health (NIH) P30DK50306], for guidance with immunofluorescence and confocal microscopy. This work was also supported by grants from the NIH (P01 DK049210 and R01 DK068157 to D.A.S.), and by the Functional Genomics Core of Pennsylvania Diabetes Endocrinology Research Center (P30DK19525).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Data have been deposited with ArrayExpress, accession no. E-MTAB-273.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007001107/-/DCSupplemental.
References
- 1.Jaw TJ, et al. Direct interaction of two homeoproteins, homothorax and extradenticle, is essential for EXD nuclear localization and function. Mech Dev. 2000;91:279–291. doi: 10.1016/s0925-4773(99)00316-0. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, MacDonald RJ, Swift GH. DNA binding and transcriptional activation by a PDX1.PBX1b.MEIS2b trimer and cooperation with a pancreas-specific basic helix-loop-helix complex. J Biol Chem. 2001;276:17985–17993. doi: 10.1074/jbc.M100678200. [DOI] [PubMed] [Google Scholar]
- 3.Berkes CA, et al. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 4.Deramaudt TB, et al. The PDX1 homeodomain transcription factor negatively regulates the pancreatic ductal cell-specific keratin 19 promoter. J Biol Chem. 2006;281:38385–38395. doi: 10.1074/jbc.M605891200. [DOI] [PubMed] [Google Scholar]
- 5.Sarno JL, Kliman HJ, Taylor HS. HOXA10, Pbx2, and Meis1 protein expression in the human endometrium: formation of multimeric complexes on HOXA10 target genes. J Clin Endocrinol Metab. 2005;90:522–528. doi: 10.1210/jc.2004-0817. [DOI] [PubMed] [Google Scholar]
- 6.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 7.Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crijns AP, et al. MEIS and PBX homeobox proteins in ovarian cancer. Eur J Cancer. 2007;43:2495–2505. doi: 10.1016/j.ejca.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 9.Jones TA, Flomen RH, Senger G, Nizetić D, Sheer D. The homeobox gene MEIS1 is amplified in IMR-32 and highly expressed in other neuroblastoma cell lines. Eur J Cancer. 2000;36:2368–2374. doi: 10.1016/s0959-8049(00)00332-4. [DOI] [PubMed] [Google Scholar]
- 10.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 11.Kim SK, et al. Pbx1 inactivation disrupts pancreas development and in Ipf1-deficient mice promotes diabetes mellitus. Nat Genet. 2002;30:430–435. doi: 10.1038/ng860. [DOI] [PubMed] [Google Scholar]
- 12.Dutta S, et al. PDX:PBX complexes are required for normal proliferation of pancreatic cells during development. Proc Natl Acad Sci USA. 2001;98:1065–1070. doi: 10.1073/pnas.031561298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang CP, et al. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, et al. Pax6 is regulated by Meis and Pbx homeoproteins during pancreatic development. Dev Biol. 2006;300:748–757. doi: 10.1016/j.ydbio.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 15.Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardone MH, et al. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 17.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: Its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto N, et al. Ablation of PDK1 in pancreatic beta cells induces diabetes as a result of loss of beta cell mass. Nat Genet. 2006;38:589–593. doi: 10.1038/ng1774. [DOI] [PubMed] [Google Scholar]
- 19.Westmoreland JJ, Wang Q, Bouzaffour M, Baker SJ, Sosa-Pineda B. Pdk1 activity controls proliferation, survival, and growth of developing pancreatic cells. Dev Biol. 2009;334:285–298. doi: 10.1016/j.ydbio.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachdeva MM, et al. Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proc Natl Acad Sci USA. 2009;106:19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.diIorio P, Alexa K, Choe S-K, Etheridge L, Sagerström CG. TALE-family homeodomain proteins regulate endodermal sonic hedgehog expression and pattern the anterior endoderm. Dev Biol. 2007;304:221–231. doi: 10.1016/j.ydbio.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choe S-K, Lu P, Nakamura M, Lee J, Sagerström CG. Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev Cell. 2009;17:561–567. doi: 10.1016/j.devcel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 24.Geerts D, Revet I, Jorritsma G, Schilderink N, Versteeg R. MEIS homeobox genes in neuroblastoma. Cancer Lett. 2005;228:43–50. doi: 10.1016/j.canlet.2005.01.047. [DOI] [PubMed] [Google Scholar]
- 25.Wong P, et al. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argiropoulos B, Yung E, Humphries RK. Unraveling the crucial roles of Meis1 in leukemogenesis and normal hematopoiesis. Genes Dev. 2007;21:2845–2849. doi: 10.1101/gad.1619407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.