Abstract
Myc activation has been implicated in the pathogenesis of hepatoblastoma (HB), a rare embryonal neoplasm derived from liver progenitor cells. Here, microRNA (miR) expression profiling of 65 HBs evidenced differential patterns related to developmental stage and Myc activity. Undifferentiated aggressive HBs overexpressed the miR-371–3 cluster with concomitant down-regulation of the miR-100/let-7a-2/miR-125b-1 cluster, evoking an ES cell expression profile. ChIP and Myc inhibition assays in hepatoma cells demonstrated that both miR clusters are regulated by Myc in an opposite manner. We show that the two miR clusters exert antagonistic effects on cell proliferation and tumorigenicity. Moreover, their combined deregulation cooperated in modulating the hepatic tumor phenotype, implicating stem cell-like regulation of Myc-dependent miRs in poorly differentiated HBs. Importantly, a four-miR signature representative of these clusters efficiently stratified HB patients, and when applied to 241 hepatocellular carcinomas (HCCs), it identified invasive tumors with a poor prognosis. Our data argue that Myc-driven reprogramming of miR expression patterns contributes to the aggressive phenotype of liver tumors originating from hepatic progenitor cells.
Keywords: hepatoblastoma, hepatocellular carcinoma, Myc, signature, stemness
Micro-RNAs (miRs) are small noncoding RNAs that intervene in virtually all cellular processes and are considered to be epigenetic regulators (1). In addition to their well-characterized ability to bind target messenger RNAs and impede protein translation, miRs can regulate gene expression by modulating promoter activity through direct binding or DNA methylation (2–4). Deregulation of miR expression contributes to cancer development, because specific miRs can either promote or block tumorigenesis (5, 6), and altered miR expression has been correlated with clinical behavior, underlining the therapeutical potential of miRs in cancer (7).
Hepatoblastoma (HB) is the most frequent pediatric liver cancer, generally occurring before 3 y of age. HB presents heterogeneous epithelial histotypes evoking different steps of intrauterine liver development (8). Major differences in etiology and morphological patterns distinguish HB from hepatocellular carcinoma (HCC), the predominant form of adult liver cancer. Because HB develops in the absence of liver disease or viral infection, this tumor might have a genetic or epigenetic origin. Implication of the Wnt/β-catenin pathway was demonstrated by the high rate (50–90%) of mutations in CTNNB1 (9) and by the increased risk associated with familial adenomatous polyposis (10). We recently showed that interplay of Wnt/β-catenin and Myc signaling plays a critical role in poorly differentiated aggressive HBs and identified a 16-gene signature with strong prognostic significance (11). Myc oncoproteins are crucial players in stem cell biology and tumorigenesis (12), and aberrant activation of Myc induces prooncogenic changes in miR expression (13).
Here, we profiled miR expression in HB and investigated the impact of altered miR expression on liver tumor biology. Our studies underscore the value of miR expression changes for stratification of patients with liver tumors, including HB and HCC.
Results
miR Expression Profiling and HB Classification.
We conducted a miR array analysis of 49 HB tumor samples (Table S1), seven nontumor liver specimens, and two fetal livers. Supervised analysis comparing tumors and nontumor livers revealed significant deregulation of 18 miRs (fold change >1.5; P < 0.005) (Table S2). Strongly up-regulated miRs are implicated in proliferation and cell cycle progression, such as miR-221 and miR-222, which target the CDKN1B/p27 inhibitor (14), and miR-181b, which is overexpressed in undifferentiated HCCs (15). Conversely, miRs harboring tumor suppressor properties are strongly down-regulated, such as the liver differentiation marker miR-122, and miR-29b that targets DNMT3 methyltransferases (4, 16). No significant change was found in tumors from patients who received preoperative chemotherapy compared with untreated patients.
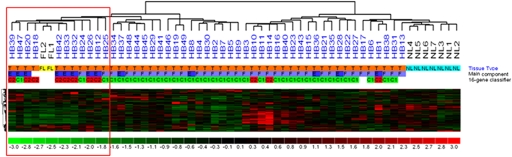
We reported previously that HBs can be classified by a 16-gene classifier into mild (C1) and aggressive (C2) subtypes, which differ in differentiation, proliferation, tumor stage, and outcome (11). Tumor samples were assigned to C1 and C2 subtypes by quantitative PCR (qPCR) using the 16-gene classifier (Table S3). We then investigated HB sample distribution by unsupervised hierarchical clustering using multiple miR lists based on coefficient of variation analysis and identified a subgroup that comprised almost exclusively C2 tumors and coclustered with fetal livers (Fig. 1).
Fig. 1.
miR expression in HB. Unsupervised clustering of HB tumor (T), fetal liver (FL), and normal liver (NL) samples according to their miR profile using 150 probe sets with the highest coefficient of variation. Tumor annotations include the main epithelial component (F, fetal; E, embryonal, crowded fetal and/or macrotrabecular) and molecular classification (C1 or C2) based on the 16-gene signature. A cluster comprising fetal livers and C2-type HBs is ringed by a red square.
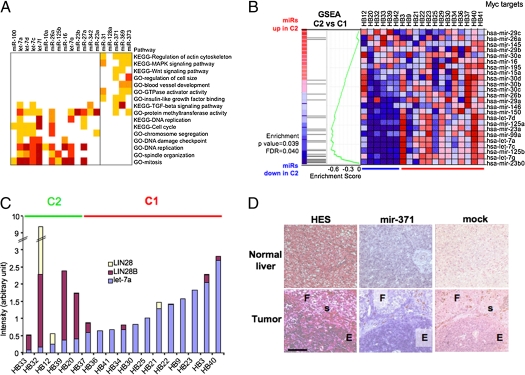
Supervised analysis of miRs differentially expressed between C1 and C2 subtypes was carried out in 19 HBs profiled by gene expression arrays. Distinct miR profiles were evidenced, with 20 miRs showing significant differential expression (fold change >1.5; P < 0.0005) (Table 1). To analyze miR/mRNA regulatory networks in C1 and C2 HB subtypes, we crossed mRNA and miR expression profiles and used seed sequence homology to identify candidate miR target genes. For each miR, negatively correlated genes were filtered by a list of predicted targets and relevant biological functions were analyzed (Fig. 2A). Inversely correlated target genes of miRs down-regulated in C2 HBs are involved in regulating cell cycle and checkpoints that ensure DNA integrity. Most putative target genes of miRs overexpressed in C2 HBs are involved in the regulation of cell growth, invasiveness, and survival. In addition, a search for enrichment of transcription factor-binding regions in miR target gene promoters revealed significant differences. For miRs overexpressed in C2, target gene promoters contained binding sites for the homeodomain, high-mobility group and forkhead families that play crucial roles in liver development. For miRs underexpressed in C2, target gene promoters possessed binding motifs for helix-loop-helix domain factors that regulate cell metabolism, proliferation, survival, and transformation (Fig. S1A and Table S4). Thus, altered miR expression may influence HB phenotypes through multiple mechanisms.
Table 1.
miRs differentially expressed between HB subtypes
| P value | FDR | C2/C1 ratio | Mature miR | Function |
| 2.0e-05 | 6.6e-04 | 2.62 | miR-373 | Tumor invasion, growth under hypoxia |
| 4.0e-04 | 0.0059 | 2.57 | miR-369 | |
| 3.3e-05 | 8.5e-04 | 2.29 | miR-371–5p | |
| 2.9e-04 | 0.0049 | 1.57 | miR-128a | |
| 4.3e-04 | 0.0062 | 1.52 | miR-31 | Antimetastatic |
| 8.9e-05 | 0.0018 | 0.55 | miR-23a | Differentiation, glutamine metabolism |
| 4.3e-05 | 0.0010 | 0.55 | miR-342–3p | Down-regulated in colon cancer |
| 7.2e-05 | 0.0016 | 0.54 | miR-27a | Fat metabolism regulation |
| 2.2e-04 | 0.0038 | 0.51 | miR-30e | Sumoylation |
| 1.2e-05 | 4.9e-04 | 0.50 | miR-23b | Glutamine metabolism |
| 3.0e-05 | 8.5e-04 | 0.46 | let-7g | Differentiation, tumor suppressor |
| 1.7e-06 | 8.1e-05 | 0.36 | miR-16 | Proapoptotic |
| 1.9e-05 | 6.6e-04 | 0.35 | miR-125b | Tumor suppressor, tissue differentiation |
| <1e-07 | <1e-07 | 0.33 | miR-26a | Cell cycle inhibitor, TGF-β and Akt pathways |
| 3.2e-04 | 0.0049 | 0.32 | miR-10a | Ribosomal protein translation |
| 1e-06 | 5.3e-05 | 0.32 | let-7f | |
| 6e-07 | 3.7e-05 | 0.28 | let-7c | Differentiation, tumor suppressor |
| 2e-07 | 2.1e-05 | 0.27 | let-7d | |
| 1e-07 | 1.4e-05 | 0.25 | let-7a | Differentiation, tumor suppressor |
| <1e-07 | <1e-07 | 0.20 | miR-100 | Mitotic check point, mTOR signaling |
miRs repressed by Myc are shown in bold. FDR, false discovery rate. Fold change cutoff = 1.5; P value cutoff <5.0e-04.
Fig. 2.
Specific miR profiles in two HB subtypes. (A) Comparative analysis of gene and miR expression in HB molecular subclasses. For each miR differentially expressed between C1 and C2 HBs, genes with negatively correlated expression profile (r < −0.45) were filtered with candidate miR targets defined by four predictive algorithms. Functional modules or pathways significantly associated with miR-related gene lists are shown as a heat map. Dark yellow, P < 0.05; orange, P < 0.01; light red, P < 0.005; dark red, P < 0.001. (B) GSEA analysis of HB miR data with a list of miRs repressed by Myc, visualized as a heat map in which the red to blue color code indicates high to low miR expression. FDR, false discovery rate. (C) Analysis of let-7, LIN28, and LIN28B expression in HB samples by qPCR. Each bar represents juxtaposed normalized data of gene and miR expression. (D) ISH assay on paraffin-embedded HB and liver samples. F, fetal; E, embryonal; s, siderophage foci. (Scale bar, 100 μm.)
Impact of Myc on miR Expression Profiles in HB.
Because most miRs down-regulated in C2 HBs are known Myc targets (Table 1), we investigated the impact of Myc activation on miR expression in C2 HB. Gene set enrichment analysis (GSEA) using a list of miRs known to be repressed by Myc (13) evidenced a significant association of the down-regulation of these miRs with C2 tumors (Fig. 2B).
The Myc target genes LIN28 and LIN28B contribute to regulate miR expression, notably by inhibiting biogenesis of let-7 family miRs (17). qPCR analysis revealed strong and often mutually exclusive overexpression of LIN28 and LIN28B in C2 HBs and a negative correlation with expression of several miRs of the let-7 family, including let-7a (Fig. 2C).
Overexpression of the miR-371–3 Cluster in Immature HBs.
Up-regulation of miR-371 and miR-373 in undifferentiated HBs was confirmed by Taqman miR qPCR assays (Table S5A). This analysis showed similar expression profiles of miR-372, indicating that all miRs of the miR-371–3 cluster were coregulated. Next, miR-371 expression was investigated by in situ hybridization (ISH) on fixed paraffin-embedded HB slices. Digoxygenin-labeled locked nucleic acid (LNA) probes for miR-371 strongly stained the poorly differentiated embryonal compartment, whereas the differentiated fetal pattern was weakly labeled and no signal was observed in normal liver (Fig. 2D). As a control, miR-122 expression showed consistent expression in normal liver and progressive fainting in fetal and embryonal HB compartments (Fig. S1B).
miR-371–3 Cluster Is an Myc Target.
Overexpression of the miR-371–3 cluster in aggressive HBs showing active Myc signaling prompted us to evaluate whether these miRs are regulated by Myc. By crossing data from miR arrays and Affymetrix gene expression arrays, miR-373 and miR-371 expression was found to be positively correlated with MYC expression in 19 HBs (Table S5B). Using siRNA-mediated inhibition of Myc, we found that miR-371 expression was strongly impaired in small interfering Myc (siMyc)-treated HepG2 cells compared with scrambled control (Fig. S2A). Conversely, the Myc target miR-26a was significantly up-regulated, as were miR-100, let-7a, and miR-125b, which belong to the same cluster and are strongly down-regulated in C2 HBs, confirming that this miR cluster is negatively regulated by Myc in hepatoma cells.
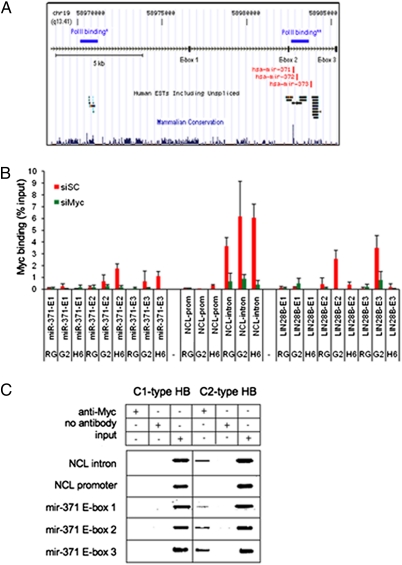
Direct control of miR-371–3 expression by Myc was investigated by quantitative ChIP in Huh6, HepG2, and HepaRG cells, which express miR-371–3 at different levels (Fig. S2B). Two putative transcriptional start sites for this cluster have been mapped about 17 and 0.5 kb upstream of miR-371 (18, 19). We scanned the genomic region and identified three canonical E-boxes (Fig. 3A). Cells were transfected with either Myc-specific siRNA or control scrambled siRNA. Myc binding to the miR-371–3 genomic region in Huh6 cells was enriched at E-box 2, located 200 bp upstream of miR-371, and, to a lesser extent, at E-box 3, located 3.2 kb downstream of miR-373. Myc occupancy correlated with expression of the miR cluster, with lower levels in HepG2 cells and no binding in HepaRG cells, which do not express the miR cluster (Fig. 3B). Moreover, binding was severely reduced in siMyc-treated cells. Occupancy of NUCL intron 1 and promoter was used as a positive control and a negative control, respectively (Fig. 3B and Fig. S2C). To evaluate ChIP assay sensitivity, we assessed Myc binding to LIN28B. We found Myc binding at LIN28B E-boxes 2 and 3 in HepG2 cells but only weak intensity at E-box 2 in Huh6 and HepaRG cells, correlating with relative LIN28B expression levels (Fig. S2D).
Fig. 3.
Expression of the miR-371–3 cluster is regulated by Myc. (A) Schematic representation of the chromosome 19q13 locus spanning the miR-371–3 cluster (adapted from the Genome Browser Web page). *PolII binding according to Ozsolak et al. (18); **PolII binding according to Crosby et al. (19). (B) Quantitative ChIP analysis of Myc binding to different genomic regions in HepaRG (RG), HepG2 (G2), and Huh6 (H6) cell lines treated with siMyc or control siRNA (siSC). The background threshold shown in gray was set as the highest signal in mock samples. Results are presented as the mean ± SD of at least three independent experiments. (C) Comparative tissue ChIP analysis of C1- and C2-type HB.
In vivo confirmation of Myc occupancy of miR-371–3 promoter was obtained by tissue ChIP on tumor samples representative of C1 and C2 subtypes. Myc binding to E-boxes 2 and 3 was detected only in C2-type HB, which strongly expresses the miR cluster, suggesting that Myc recruitment is associated with miR-371–3 expression in vivo (Fig. 3C).
Cooperation of miR-371–3 and miR-100/let-7a-2/miR-125b-1 Clusters in Huh6-Transformed Phenotype.
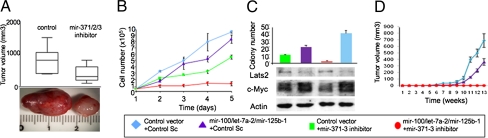
To investigate the role of miR-371–3 in liver carcinogenesis, we inhibited its expression in Huh6 cells using microRNA Mimics and Hairpin Inhibitors (miRIDIAN, Dharmacon) technology. Efficient decrease of mature miR production was verified by qPCR (Fig. S3A). Treatment of Huh6 or HepG2 cells with miR-371–3 inhibitors strongly compromised colony formation in soft agar (Fig. S3B), with a drastic effect on the part of the miR-371 inhibitor (Fig. S3C). The s.c. injection of cells transfected with miR-371–3 inhibitors into immunodeficient mice resulted into tumors considerably smaller than those from control cells (Fig. 4A).
Fig. 4.
Altered expression of miR-371–3 and miR-100/let-7a-2/miR-125b-1 clusters influences tumor cell behavior in vitro and in vivo. Huh6 cells were used in all experiments. (A) Box plot (10–90%) representing tumor sizes in mice injected with cells treated with miR-371–3 inhibitor or scrambled control (11 mice for each condition). Mean values are shown by internal horizontal bars, and SD by vertical bars. (B) Proliferation assay of cells treated as indicated. (C) Soft agar assay and Western blot analysis of cells treated as indicated. (D) Growth curve of tumors in nude mice injected with cells treated as indicated, using 5 mice for each condition. Bars indicate SD.
To evaluate the effects of the miR-100/let-7a-2/miR-125b-1 cluster in tumorigenesis, Huh6 cells were infected with a retroviral vector encoding the three miRs and efficient miR expression was verified by qPCR (Fig. S3D). Huh6 cells overexpressing the miR cluster showed slightly reduced proliferation compared with control cells (Fig. 4B). Next, we investigated the effects of simultaneous misregulation of miR-371–3 and miR-100/let-7a-2/miR-125b-1 clusters. Although miR-371–3 inhibition significantly decreased cell proliferation, the lowest proliferative rate was obtained by simultaneous inhibition of miR-371–3 and overexpression of miR-100/let-7a-2/miR-125b-1, suggesting functional cooperation of these clusters (Fig. 4B). Accordingly, combined deregulation of the two miR clusters affected colony formation in soft agar more dramatically than single cluster deregulation (Fig. 4C). In Western blot analysis, Huh6 cells treated with miR-371–3 inhibitors showed increased levels of Lats2, a direct target of miR-373 (Fig. 4C). Protein levels of Myc, a let-7a target, were reduced by overexpression of miR-100/let-7a-2/miR-125b-1. Strikingly, Myc protein was also down-regulated by inhibition of the miR-371–3 cluster, and an enhanced effect resulted from opposite regulation of the two miR clusters.
Functional cooperation was evaluated in vivo by injecting treated cells into immunodeficient mice. Overexpression of the miR-100/let-7a-2/miR-125b-1 cluster in Huh6 cells delayed tumor occurrence, and simultaneous inhibition of miR-371–3 completely blocked tumor formation, suggesting that antagonistic interplay of the two miR clusters decides cancer cell fate (Fig. 4D).
Myc-Dependent miR Signature Discriminates Aggressive HBs and HCCs.
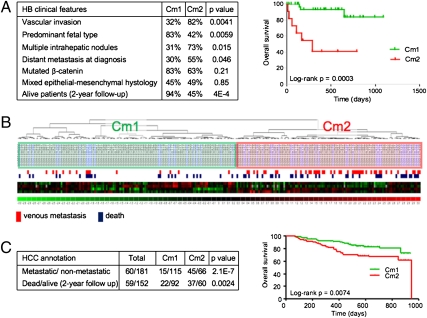
We evaluated the ability of the miR-371–3 and miR-100/let-7a-2/miR-125b-1 clusters to define clinically significant tumor subgroups. All miRs were individually analyzed by qPCR in 19 HBs previously used for supervised analysis, and we selected four miRs with highest correlation between microarray and qPCR expression, namely, miR-100, let-7a, miR-371, and miR-373 (Table S5A). The four-miR signature was used to classify HBs, employing microarray or qPCR data on the 19 HBs as a training set to classify a test set of 46 HBs, including 30 samples analyzed by miR microarray and 16 samples analyzed by qPCR (Table S3). Two subgroups, Cm1 and Cm2, were identified, and clinical correlations were evaluated in the test set. The HB Cm2 subgroup was associated with poorly differentiated, invasive, and metastatic tumors and shorter overall patient survival compared with the Cm1 subgroup (Fig. 5A).
Fig. 5.
Classification of HB and HCC with a four-miR signature. (A) Cm1 and Cm2 HB subclasses defined by differential expression of four miRs are significantly associated with clinical parameters. Kaplan–Meier analysis shows significant differences in overall survival probability between patients carrying Cm1- and Cm2-type HBs. (B) Unsupervised hierarchical clustering of 241 HCC samples by using the HB four-miR signature identifies two main tumor classes. (C) Cm1 and Cm2 HCC subclasses present significant differences in clinical annotations and overall patient survival.
Because previous studies indicate that aggressive HCCs harbor liver precursor/stem cell features (20, 21), we explored the performance of the four-miR signature in adult liver cancer using a dataset of 241 HCCs profiled on the same miR array platform (22) (Table S6). Unsupervised analysis promptly separated HCCs into two main subclasses with significantly different prometastatic phenotypes (Fig. 5B). Only 11% of Cm1 tumors displayed an invasive phenotype, whereas the frequency in Cm2 tumors was over 40%. Accordingly, patients with Cm2 tumors had lower survival probability in Kaplan–Meier plots, and this difference was highly significant (Fig. 5C).
Discussion
High-grade undifferentiated tumors are thought to be enriched in cancer stem/initiating cells that share with normal stem cells common self-renewal characteristics (23, 24). miRs have been integrated recently into regulatory networks that control stem cell identity as well as tumor pathogenesis. Here, we profiled miR expression in 65 HBs and identified a miR fingerprint of poorly differentiated tumors, including massive down-regulation of Myc target miRs, such as members of the let-7 family and the miR-100/let-7a-2/miR-125b-1 cluster and concomitant overexpression of the miR-371–3 cluster. We show that these two miR clusters exert opposite functions on liver cell phenotype, evoking self-reinforcing loops seen in ES cells (25).
miR-371–3 Cluster, a Stemness Marker and a Potential Oncogene Regulated by Myc.
The miR 371–3 cluster and its murine ortholog, the ES cell-specific cell cycle-regulating miR-290–5 cluster, are overexpressed in ES cells and down-regulated during differentiation (26, 27). These miRs play a crucial role in the maintenance of ES cell renewal (28). They have oncogenic potential in primary cells by preventing Ras-induced senescence and suppressing the CDK inhibitor Lats2, and they promote tumor invasion and metastasis in response to hypoxia (29, 30). We provide evidence for direct regulation of the miR-371–3 cluster by Myc. Moreover, as recently found for the murine ortholog miR-294 in ES cells (25), our data suggest that Myc is up-regulated by the miR-371–3 cluster. This might reflect either inhibition of an intermediate repressor or direct activation through binding of the miRs to the MYC genomic region, similar to the activation of CDH1 by miR-373 in prostate cancer (2). Such a positive regulatory loop might unveil a previously undescribed oncogenic mechanism in undifferentiated tumors.
Opposite Regulation of Two Antagonistic miR Clusters in Aggressive HB.
The most strongly down-regulated miRs in C2 HBs were miR-100, let-7a, and the Caenorhabditis elegans lin-4 ortholog miR-125b, which belong to the same cluster. These miRs are developmentally regulated, with increased levels in differentiated cells (27). Let-7 miRs have tumor suppressive activity by targeting KRAS, MYC, and the chromatin-remodeling factor HMGA2. Strikingly, cooperation between ectopic expression of the miR-100/let-7a-2/miR-125b-1 cluster and silencing of the miR-371–3 cluster in HB cells resulted in total shutdown of cell proliferation, anchorage-independent growth, and in vivo tumorigenicity. Thus, interplay of the two miR clusters strongly affects oncogenic processes, underscoring remarkable analogies between the regulation of cancer stem/progenitors and the ES cell switch.
Clinical Relevance of miR Expression Profiles in Childhood and Adult Liver Tumors.
We show here that the relative expression levels of the Myc-driven miR-371–3 and miR-100/let-7a-2/miR-125b clusters have strong clinical significance. The ability of the miR signature to stratify patients with HB relies on the robustness of the defined molecular subtypes and their strong correlation with biological parameters that impinge on survival, such as cell differentiation and invasion. These data are in line with our previous report showing that HB prognostic subtypes defined by a 16-gene signature reflect liver developmental stage (11). Recent studies have evidenced high-grade HCC subtypes with stem/progenitor features that share with aggressive HBs expression of the early hepatic markers AFP, KRT19, and EpCAM (20, 21, 31, 32).
Although heterogeneous miR profiles have been detected in HCC, reflecting multiple etiological factors and genetic lesions (16, 33, 34), molecular matches between poorly differentiated HBs and HCCs support the view that a miR signature deduced from HB subtypes might be relevant for HCC classification. In particular, the effects of several miRs of the HB signature on cell migration, invasiveness, and metastatic spread probably account for its ability to distinguish invasive and metastatic HCCs. Our data support the view that differential expression of Myc-regulated miRs might contribute to liver fate specification and provide a characteristic signature of cancer progenitor cells. Targeting this regulatory circuitry might be beneficial for adapted therapy of Myc-related liver cancer.
Materials and Methods
Patients and Tissue Samples.
Frozen tumor specimens from 65 patients (Table S1) and seven nontumor liver samples were collected from different hospitals in France. Most patients were enrolled in clinical trials of the Childhood Liver Tumour Strategy Group (SIOPEL) (35). Informed patient consent was obtained at each medical center, and this study was approved by the Institutional Review Board of the Institut Pasteur. Human fetal liver RNAs were purchased from the BioChain Institute. We used the χ2 test for comparisons between groups. Kaplan–Meier estimates of overall survival time in the various groups were compared using the log-rank test.
Cell Lines, Constructs, and Transfection Experiments.
The HB cells Huh6 and HepG2 were maintained as described (36). A retroviral vector allowing simultaneous expression of the three miRs of the miR-371–3 cluster was constructed using pMSCV-PIG-IRES-GFP (a kind gift of Scott W. Lowe, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), as described in SI Materials and Methods. For siRNA transfection, we used Interferin (Ozyme). miR expression was inhibited by transfection of 5–50 nM specific miRIDIAN micro-RNA Inhibitor (Dharmacon) with Lipofectamine 2000 (Invitrogen).
Quantitative ChIP.
ChIP analysis in hepatoma cells was performed as described (37). For ChIP on HB specimens, the step preceding immunoprecipitation was adapted from Farnham's laboratory protocol (http://farnham.genomecenter.ucdavis.edu/protocols/tissues.html). In qPCR assays, we used 1 or 2 μL of ChIP or Input sample and Sybr-Green MasterMix (Applied Biosystems). Primers can be found in SI Materials and Methods.
Cell Proliferation, Soft Agar Assays, and in Vivo Tumorigenic Assays.
Proliferation and soft agar assays were performed as described previously (36). For in vivo tumorigenic assays, experimental protocols were conducted in accordance with the Institutional Guidelines for the Care and Use of Laboratory Animals. Huh6 cells were injected into the flanks of athymic nu/nu BALB/c mice (Charles River Laboratories), and tumor volume was measured as described (36).
ISH and Western Blot Analysis.
ISH was performed on formaldehyde-fixed tissue microarray or single tumor sections using 5′-digoxigenin–labeled LNA-modified probes for miR-122 and miR-371, following the instructions of the manufacturer (Exiqon). For Western blotting, protein extracts resolved by SDS/PAGE were analyzed as described (36) using Lats2 monoclonal antibody (clone ST-3D10; Abnova) and c-Myc and actin antibodies from Santa Cruz and Sigma.
RNA Extraction and qPCR.
RNA was prepared from liver tissues and cell lines with TRIzol (Invitrogen) or mirVana (Ambion). cDNA was generated with SuperScript II reverse transcriptase (Invitrogen), followed by qPCR using the Sybr-Green MasterMix as described (11). We used TaqMan microRNA assays (Applied Biosystems) to quantify mature miR expression relative to RNU43.
Microarrays and Statistical Evaluation.
miRs were profiled using the miRNA microarray V 2.0 (Microarray Shared Resource, Comprehensive Cancer Center, Ohio State University Medical Center), and raw data were normalized by the print-tip loess method. For supervised analysis and tumor classification, we used the Class prediction tool of BRB Array Tools (BRB ArrayTools software, version 3.6.0a; http://linus.nci.nih.gov/BRB-ArrayTools.html), keeping the classification provided by the majority of algorithms. Comparison of the mRNA/miR datasets is detailed in SI Materials and Methods. The HCC miR database has been previously published (22). We used dChip software (http://biosun1.harvard.edu/complab/dchip/) for hierarchical clustering with four miRs. GSEA and pathway analysis were carried out as described (11).
Supplementary Material
Acknowledgments
We thank C. Trépo (Department of Hepato-gastroenterology, Hotel-Dieu Hospital, and Institut National de la Santé et de la Recherche Médicale U871, Lyon, France) for providing the HepaRG cell line, S. W. Lowe (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for miR retroviral vector, A. Goga for helpful discussions, and pathologists involved in SIOPEL studies. Research was supported by grants from the Association pour la Recherche contre le Cancer and the Nausicaa Association. S.C. was supported by the Institut Pasteur. X.W.W. was supported by the Intramural Research Program Grant Z01-BC 010313 of the Center for Cancer Research of the National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE20971). For the miR database of X.W.W. for HCC, sample data and annotation can be found in the GEO database (accession no. GSE6857).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009009107/-/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DH, Saetrom P, Snøve OJ, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann A. The emerging family of hepatoblastoma tumours: From ontogenesis to oncogenesis. Eur J Cancer. 2005;41:1503–1514. doi: 10.1016/j.ejca.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 9.Koch A, et al. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the β-catenin gene. Cancer Res. 1999;59:269–273. [PubMed] [Google Scholar]
- 10.Hirschman BA, Pollock BH, Tomlinson GE. The spectrum of APC mutations in children with hepatoblastoma from familial adenomatous polyposis kindreds. J Pediatr. 2005;147:263–266. doi: 10.1016/j.jpeds.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 11.Cairo S, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 13.Chang TC, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.le Sage C, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji J, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Ozsolak F, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JS, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita T, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budhu A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong DJ, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suh MR, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Stadler BM, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 30.Huang Q, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 31.Boyault S, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 32.Hoshida Y, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladeiro Y, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 34.Pineau P, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perilongo G, Shafford E, Plaschkes J, Liver Tumour Study Group of the International Society of Paediatric Oncology SIOPEL trials using preoperative chemotherapy in hepatoblastoma. Lancet Oncol. 2000;1:94–100. doi: 10.1016/s1470-2045(00)00018-8. [DOI] [PubMed] [Google Scholar]
- 36.Renard CA, et al. Tbx3 is a downstream target of the Wnt/β-catenin pathway and a critical mediator of β-catenin survival functions in liver cancer. Cancer Res. 2007;67:901–910. doi: 10.1158/0008-5472.CAN-06-2344. [DOI] [PubMed] [Google Scholar]
- 37.Cairo S, et al. PML interacts with Myc, and Myc target gene expression is altered in PML-null fibroblasts. Oncogene. 2005;24:2195–2203. doi: 10.1038/sj.onc.1208338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.