Abstract
The important family of G protein-coupled receptors has so far not been targeted very successfully with conventional monoclonal antibodies. Here we report the isolation and characterization of functional VHH-based immunoglobulin single variable domains (or nanobodies) against the chemokine receptor CXCR4. Two highly selective monovalent nanobodies, 238D2 and 238D4, were obtained using a time-efficient whole cell immunization, phage display, and counterselection method. The highly selective VHH-based immunoglobulin single variable domains competitively inhibited the CXCR4-mediated signaling and antagonized the chemoattractant effect of the CXCR4 ligand CXCL12. Epitope mapping showed that the two nanobodies bind to distinct but partially overlapping sites in the extracellular loops. Short peptide linkage of 238D2 with 238D4 resulted in significantly increased affinity for CXCR4 and picomolar activity in antichemotactic assays. Interestingly, the monovalent nanobodies behaved as neutral antagonists, whereas the biparatopic nanobodies acted as inverse agonists at the constitutively active CXCR4-N3.35A. The CXCR4 nanobodies displayed strong antiretroviral activity against T cell-tropic and dual-tropic HIV-1 strains. Moreover, the biparatopic nanobody effectively mobilized CD34-positive stem cells in cynomolgus monkeys. Thus, the nanobody platform may be highly effective at generating extremely potent and selective G protein-coupled receptor modulators.
Keywords: nanobody, CXCR4 antagonist, chemotaxis, HIV, stem cell mobilization
The G protein-coupled receptors (GPCRs) family, composed of more than 800 members, represents the largest family of drug target proteins to date. GPCRs are targeted therapeutically mostly by small molecules (1), whereas they appear to be difficult targets for the growing field of antibody-based therapeutic agents. This may be explained by the limited availability of highly purified GPCR preparations (2) and poor accessibility to the receptor's cryptic potentially antigenic sites, such as the ligand binding pocket located within the transmembrane regions. A new means to target GPCRs is by using nanobodies. These are based on VHHs, which are immunoglobulin single variable domains (12–15 kDa) of heavy-chain antibodies that occur naturally in the Camelidae family, including camels and llamas (Fig. S1A). Nanobodies have several favorable biological and physicochemical characteristics, including low immunogenicity, increased tissue penetration, high solubility, and physical stability (3). Most important, VHH-based immunoglobulin single variable domains are able to bind to cryptic antigenic sites like enzyme active sites and conserved epitopes of infectious agents that are normally not recognized by conventional antibodies and engineered Fab and scFv fragments (4–7). In addition, nanobody constructs with improved potencies or multiple valencies or specificities are easy to generate using short peptide linkers (8).
To investigate the potential of nanobodies to target and modulate GPCR function, we selected the CXCR4 chemokine receptor. CXCR4 and CXCL12 play a central role in stem cell homing, organogenesis, tissue repair, and inflammation (9–11), but also in the metastatic spread of cancer cells (12). In addition, CXCR4 serves a coentry receptor for HIV (13, 14). The quest for CXCR4 ligands, inhibiting activation by CXCL12 and/or binding of HIV, led to the discovery of various synthetic small molecule ligands and short peptides. Recently, the prototype CXCR4 antagonist AMD3100 (plerixafor) has proven its clinical efficiency in inhibiting CXCR4-using viruses in HIV-infected individuals (15), and in enhancing granulocyte colony-stimulating factor (G-CSF)-induced stem cell mobilization in patients with non-Hodgkin lymphoma and multiple myeloma (16–18). The therapeutic half-life of AMD3100 is limited (1.5 h), which is favorable for stem cell mobilization but undesirable for other therapeutic indications targeting CXCR4. This might be circumvented by the use of antibody-based therapeutic agents. Conventional CXCR4-blocking antibodies and antibody fragments showing anti-HIV, anti-inflammatory, and/or anticancer activities have also been described and are still under preclinical evaluation and development (9, 19–24). Yet, the development of antibody-based therapeutic agents targeting CXCR4 has mostly been hampered by incomplete CXCR4 blockage (25, 26).
In this study we describe inhibitory VHH-based immunoglobulin single variable domains, nanobodies and constructs thereof, directed against a GPCR generated by using a combined whole cell immunization, phage display, and counterselection method. Within a few months, we generated highly potent CXCR4 nanobodies and constructs thereof which inhibited chemotaxis and HIV-1 entry, and induced the mobilization of CD34+ stem cells, demonstrating the power of this technique for the development of nanobody (VHH-based immunoglobulin single variable domain)-based GPCR drug candidates.
Results
Generation of Potent and Selective CXCR4 Binding Nanobodies.
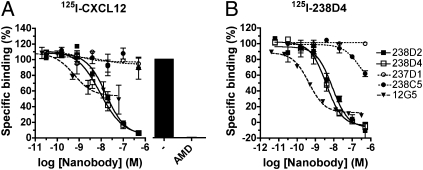
To generate VHH-based immunoglobulin single variable domains against a functional properly folded CXCR4, llamas were immunized with intact CXCR4-expressing HEK293T cells. A phage nanobody library was subsequently generated from peripheral blood mononuclear cells (PBMCs) collected from these animals. After two rounds of counterselection (Fig. S1B), nanobodies present in the periplasma fractions of 180 isolated clones were screened for competition of 125I-CXCL12 binding to CXCR4 (Fig. S1C). Approximately 13% of the clones were found to inhibit specific 125I-CXCL12 binding by at least 30%. Among these clones, approximately 25% show an inhibition of more than 70%. Following sequencing and purification of identified hits, CXCR4 binding characteristics of several nanobodies were investigated. 238D2 and 238D4 displaced specifically bound 125I-CXCL12 on cell membranes from HEK293T cells transiently expressing CXCR4, showing affinities for CXCR4 in the low nanomolar range (Fig. 1A and Table S1). Other VHH-based immunoglobulin single variable domains were unable to displace 125I-CXCL12 at the highest tested concentration (0.5 μM; 237D1, 238C5). The monoclonal antibody 12G5, previously reported to label a subpopulation of CXCR4 (25, 26), potently but partially displaced specifically bound 125I-CXCL12 from CXCR4 (Fig. 1A).
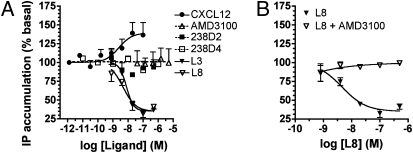
Fig. 1.
Characterization of CXCR4-specific nanobodies. (A) Dose–response curve of potential specific CXCR4 nanobodies and the CXCR4-specific monoclonal antibody 12G5 in a 125I-CXCL12 displacement assay performed with membranes of transiently transfected HEK293T-CXCR4. (B) Displacement assay of 125I-238D4 with CXCR4 nanobodies and the CXCR4-specific monoclonal antibody 12G5.
Subsequently, 238D2 and 238D4 were radiolabeled, and both 125I-238D2 and 125I-238D4 selectively bound to membranes from HEK293T cells transiently expressing CXCR4, but not to cells expressing human CXCR3 (Fig. S2A). Both nanobodies specifically competed each other for binding to CXCR4 (Fig. 1B and Fig. S2B). Furthermore, AMD3100 displaces 125I-238D2 and 125I-238D4 with affinities comparable to those obtained against 125I-CXCL12 (Table 1), indicating that also AMD3100 competes with 238D2 and 238D4 for the same receptor. Also 12G5 potently inhibited 125I-238D2 (Fig. S2B) and 125I-238D4 (Fig. 1B). Other nanobodies (237D1, 238C5, and 116B2) failed to inhibit binding of 125I-238D2 (Fig. S2B) or 125I-238D4 (Fig. 1B) to the receptor at concentrations lower than 100 nM (Table S1).
Table 1.
Receptor affinity, relative potency and maximal displacement of 125I-CXCL12 by bivalent nanobodies in comparison with their monovalent counterparts
| Nanobody | Displacement, % | Ki, nM | Relative potency |
| 238D2-15GS-238D4 (L3) | 92 | 0.35 | 28*/17† |
| 238D2-20GS-238D4 (L8) | 103 | 0.36 | 27*/17† |
| 238D2-20GS-238C5 (L6) | 110 | 44.7 | 0.18* |
| 238D2-20GS-238B10 (L10) | 43‡ | >100 | <0.10* |
| 238D2 + 238D4 (1:1) | 92 | 10.0 | 1.0*/0.6† |
*Potency relative to those of monovalent 238D2.
†Potency relative to those of monovalent 238D4.
‡Maximum not reached at the highest test concentration of 0.5 μM, displacement at 0.5 μM.
Nanobodies Inhibit CXCR4-Mediated Signaling and Chemotaxis.
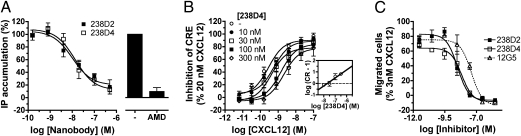
In an effort to functionally characterize the 238D2 and 238D4 nanobodies, we measured their ability to activate or inhibit G protein signaling in HEK293T cells transiently cotransfected with cDNAs encoding for CXCR4 and Gαqi5. Upon stimulation of the Gαi-coupled CXCR4 by CXCL12, the chimeric Gαqi5 protein transduces signals to the Gαq signaling pathway (27), resulting in an accumulation of inositol phosphates (EC50, 12.9 nM). No agonist activity was observed for the nanobodies 238D2 or 238D4 up to a concentration of 100 nM (Fig. S3A). However, 238D2 and 238D4 fully inhibited the CXCL12-induced accumulation of inositol phosphates with potencies (i.e., Ki values) of 3.1 and 4.7 nM, respectively (Fig. 2B). In cAMP response element (CRE) reporter gene analysis, stimulation of CXCR4 by CXCL12 inhibited the forskolin-induced activation of CRE (EC50, 0.17 nM) as a result of Gαi protein activation. The addition of increasing concentrations of CXCR4 nanobodies antagonized the CXCL12-induced response. Schild analysis showed linearity between log (CR − 1) and −log (nanobody) (M) with slopes not significantly different from unity (0.91 ± 0.20 and 0.71 ± 0.17 for 238D2 and 238D4, respectively; Fig. 2C and Fig. S3B), indicating competitive antagonism for both nanobodies. Based on the Schild plot data, Kb values of 23 and 20 nM were calculated for 238D2 and 238D4, respectively.
Fig. 2.
The monovalent nanobodies 238D2 and 238D4 are potent antagonists of CXCR4. (A) CXCL12-induced inositol phosphate formation in CXCR4 and Gαqi5-expressing HEK293T cells preincubated with 238D2 or 238D4 nanobodies. (B) HEK293T cells transfected with CXCR4 together with the pCRE/β-gal reporter gene display CRE activation upon CXCL12 stimulation. The 238D4 nanobody shows competitive antagonism as shown by Schild–Plot analysis (Inset). (C) CXCL12-induced chemotaxis of Jurkat cells is inhibited in the presence of the nanobodies 238D2 and 238D4, as well as with the CXCR4 monoclonal antibody 12G5 in a dose-dependent manner.
In CXCR4-expressing Jurkat leukemia T cells, CXCL12 induced chemotaxis with a typical bell-shaped profile and a EC50 of 0.39 nM for the first phase of the concentration response curve (Fig. S3C). 238D2 and 238D4 did not induce significant migration of Jurkat cells by themselves (Fig. S3C) but concentration-dependently inhibited the migration of Jurkat cells toward 3 nM CXCL12 with potencies of 3.2 and 6.8 nM, respectively (Fig. 2C). The antibody 12G5 also fully inhibited CXCL12-induced chemotaxis, albeit with a reduced potency of 40.2 nM (Fig. 2C).
Selectivity Profile CXCR4 Nanobodies.
At concentrations up to 2.5 μM, 238D2 and 238D4 did not alter the agonist-induced inhibition of the forskolin (3 μM)-induced activation of CRE in HEK293T cells transiently transfected with CXCR1, CXCR2, CXCR3, CXCR6, CCR5, or CCR7 chemokine receptors or histamine H4 receptors or in cells endogenously expressing β2-adrenoreceptors (Fig. S4A). The selectivity of the nanobodies for CXCR4 versus CXCR7, another CXCL12-recognizing chemokine receptor that does not activate Gαi proteins (28), was tested in a 125I-CXCL12 competition binding assay. 238D2 and 238D4 (2.5 μM) were unable to displace radiolabeled CXCL12 from CXCR7-expressing HEK293T cells. In addition, no binding of the CXCR4 nanobodies 238D4 and 238D2 could be detected on cells expressing hCXCR3 (Fig. S4B) and hCXCR7 (Fig. S4C) by using flow cytometry, whereas the monoclonal antibodies 498801 and 11G8 detected specific expression of CXCR3 and CXCR7, respectively. These results demonstrate a more than 1,000-fold selectivity of 238D2 and 238D4 for CXCR4 versus all other GPCRs tested.
Epitope Mapping of Nanobody Binding Sites.
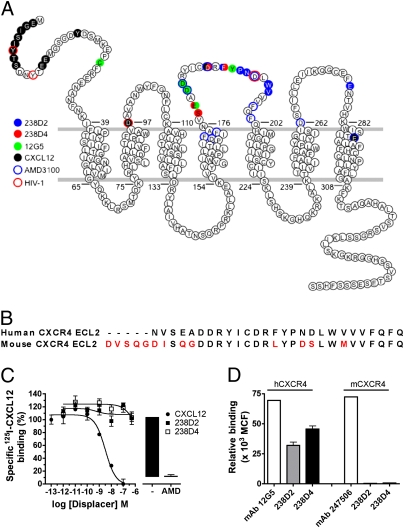
CXCL12 is known to bind to the N-terminal part and the extracellular loops (ECLs) of CXCR4 (29–31). To define the binding sites of the 238D2 and 238D4 nanobodies, we applied the shotgun mutagenesis technology platform from Integral Molecular (32). 238D2 and 238D4 reactivities were tested for binding to a CXCR4 mutation library, created by random mutagenesis. The monoclonal antibody 12G5 used as a positive control appeared affected by mutagenesis of E179, confirming the previously reported binding of 12G5 to ECL2 of CXCR4 (26). The analysis indicated that 238D2 and 238D4 both bind to ECL2 amino acid residues, but displaying distinct binding modes (Fig. 3A and Table S2). Loss of 238D4 binding was apparent after mutagenesis of amino acids D187, F189, E179, and S178 in ECL2. In contrast, 238D2 seemed to interact with ECL2 amino acid residues F189, N192, W195, P191, V196 and also with E277 located in ECL3. Importantly, F189, positioned in ECL2, appears critical for binding of both nanobodies (Table S2). Because ECL2 is critical for binding of the VHH-based immunoglobulin single variable domains, and differs considerably between human and murine CXCR4 (Fig. 3B), we hypothesized that both nanobodies would not interact with mCXCR4. Both 238D2 and 238D4 indeed do not bind mCXCR4 as demonstrated by their inability to displace 125I-CXCL12 from mCXCR4 in a radioligand binding assay (Fig. 3C). In addition, 238D2 and 238D4 detected expression of hCXCR4 but not mCXCR4 on transiently transfected cells in a FACS-based assay (Fig. 3D).
Fig. 3.
Epitope mapping of CXCR4-specific nanobodies unmasks species-specific activity. (A) Snake plot representation of human CXCR4 with highlighted amino acids involved in the binding of 238D2 (blue filled circles), 238D4 (red filled circles), 12G5 (green filled circles), CXCL12 (black filled circles), AMD3100 (blue open circles), and HIV-1 (red open circles). Multiple color codes show differential involvement of specific amino acids with various ligands or HIV-1. (B) Sequence alignment of the ECLs of human and mouse CXCR4 (red indicates differences). (C) Membranes of HEK293T transiently transfected with mouse CXCR4 specifically bind to human 125I-CXCL12, which can be displaced by AMD3100 and cold CXCL12, but not by the nanobodies 238D2 or 238D4. (D) HEK293T cells transiently transfected with human or mouse CXCR4 were assayed for the binding of specific monoclonal antibodies (12G5 and 247506 in the case of human and mouse CXCR4, respectively) and the nanobodies 238D2 and 238D4. Bound antibodies or nanobodies were detected by flow cytometry using the appropriate conjugated secondary antibodies. MCF, mean channel fluorescence.
Biparatopic Nanobodies Display Enhanced Potency and Different Mode of Action.
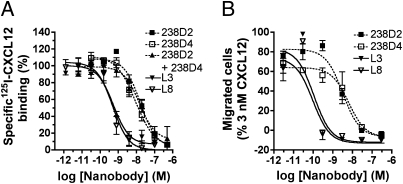
For other target proteins, generation of bi- or multivalent nanobody constructs resulted in a significant increase in avidity (33–35). Here, we engineered a series of biparatopic nanobody constructs on the basis of 238D2 and 238D4, with repetitive GGGGS sequences altering in the linker size between 15 aa and 20 aa. As seen in Fig. 4A, this strategy resulted in as much as a 27-fold increase in apparent affinity to CXCR4 for both biparatopic nanobody constructs [238D2-15GS-238D4 (L3) and 238D2-20GS-238D4 (L8)]. Linking of 238D2 to the inactive nanobody 238B10 or to the low affinity nanobody 238C5 resulted in a decreased receptor affinity (Table 1). This may be a result of increased ligand size and steric hindrance and excludes the possibility that the linker increases CXCR4 affinity on its own. Furthermore, competition in binding between 238D2 and 238D4 (Fig. 1C and Fig. S2B) and the lack of increased potency for displacing 125I-CXCL12 binding by equimolar mixing of 238D2 and 238D4 (Table 1) argue against a positive cooperative effect resulting from allosteric binding on the same receptor molecule. Importantly, both 238D2-15GS-238D4 (L3) and 238D2-20GS-238D4 (L8) showed enhanced inhibitory potency as they fully antagonized the chemoattractant effects of CXCL12 with a potency of 96 pM and 109 pM, respectively (Fig. 4B).
Fig. 4.
Bivalent nanobodies show increased affinity and inhibitory potency compared with their monovalent counterparts. (A) Competition binding experiments of 125I-CXCL12 on membranes of HEK293T cells transfected with CXCR4 in the presence of monovalent nanobodies 238D2, 238D4, a mixture of 238D2 with 238D4, or the biparatopic nanobodies L3 and L8. (B) CXCL12-induced chemotaxis experiment of Jurkat cells in the presence of monovalent nanobodies 238D2, 238D4, or the biparatopic nanobodies L3 and L8.
In view of the differential binding epitopes of 238D2 and 238D4 and the increased affinity of the biparatopic nanobodies, we defined their mode of action using a constitutively active CXCR4 mutant (CAM) N3.35A (Ballesteros–Weinstein numbering of class A GPCRs) equivalent to CXCR4-N119A (36). Both 238D2 and 238D4 bind to CXCR4-N3.35A but could not fully displace 125I-CXCL12 at concentrations up to 1 μM (Fig. S5). HEK293T cells transiently expressing CXCR4-N3.35A show a three to eight times higher basal rate of inositol phosphate accumulation compared with WT CXCR4 or mock-transfected cells, and constitutive activity can be further stimulated by CXCL12 (Fig. 5A). No significant agonistic or inverse agonistic activity was apparent for 238D2, 238D4, or AMD3100 in cells expressing the constitutively active CXCR4 mutant (N3.35A). Interestingly, the most potent biparatopic nanobody constructs, 238D2-15GS-238D4 (L3) and 238D2-20GS-238D4 (L8), behaved as inverse agonists at CXCR4-N3.35A (reduction of 59 ± 3%; Fig. 5B). The 238D2-20GS-238D4 (L8)-induced reduction of basal inositol phosphate accumulation was antagonized by AMD3100, confirming that the observed inverse agonistic effect is mediated via CXCR4 (Fig. 5C).
Fig. 5.
CXCR4-specific nanobodies behave as neutral antagonists or inverse agonists on the constitutively active mutant of CXCR4. (A) Modulation of the constitutive formation of inositol phosphates in HEK293T cells transfected with CXCR4-N3.35A and Gαqi5 by CXCL12, AMD3100, 238D2, 238D4, L3, or L8. (B) Dose-dependent inhibition of the constitutive production of inositol phosphates in HEK293T cells transfected with CXCR4-N3.35A and Gαqi5 by L8. Treatment of the cells with AMD3100 blocks the inverse agonistic activity of L8.
CXCR4 Nanobodies Display Anti-HIV-1 Activity.
The ability of 238D2 and 238D4 to block the entry of the CXCR4-using (X4) HIV-1 strain NL4.3, the CCR5-using (R5) HIV-1 strain BaL, and the dual-tropic (R5/X4) HIV-1 strain HE was investigated on human MT-4 cells (endogenously expressing CXCR4 but not CCR5), human PBMCs (endogenously expressing CXCR4 and CCR5), or U87 cells stably transfected with human CD4 and CCR5 (U87.CD4.CCR5) or human CD4 and CXCR4 (U87.CD4.CXCR4), respectively. Nanomolar concentrations of 238D2 and 238D4 equipotently inhibited virus replication of the X4 NL4.3 and R5/X4 HE strains in both MT-4 cells and PBMCs (Table 2). Furthermore, both VHH-based immunoglobulin single variable domains were highly active against HIV-1 NL4.3 in U87.CD4.CXCR4 cells. The biparatopic nanobody constructs 238D2-15GS-238D4 (L3) and 238D2-20GS-238D4 (L8) showed a significantly increased inhibitory potency for the replication of the X4 HIV-1 strain NL4.3 in MT-4, PBMCs, and U87.CD4.CXCR4 cells at picomolar concentrations (Table 2). Both nanobody constructs also potently inhibited the entry of the dual-tropic R5/X4 HIV-1 strain HE into MT-4 cells, endogenously expressing CXCR4 but not CCR5. The biparatopic nanobodies were approximately one order less potent in the inhibition of HIV-1 HE replication in PBMCs, which endogenously express CCR5 in addition to CXCR4 and thus can partially circumvent a blocking of CXCR4. As expected, 238D2 and 238D4 and the biparatopic nanobody constructs failed to inhibit the infectivity of the R5 BaL strain into PBMCs and U87.CD4.CCR5 cells. For comparison, the small molecule CCR5 antagonist maraviroc effectively inhibited BaL replication in PBMCs (IC50, 3.13 nM) and U87.CD4.CCR5 cells (IC50, 0.72 nM). The nanobodies did not induce cellular toxicity or morphological changes in the different cell types evaluated at their highest concentration (500 nM). These results confirm that the observed activity of the described nanobodies depends solely on the interaction with CXCR4.
Table 2.
Anti-HIV-1 activity profile (IC50 in nM) of the monovalent and biparatopic nanobodies
| Monovalent nanobodies |
Bivalent nanobodies |
|||
| Cell/HIV-1 strain (receptor tropism) | 238D2 | 238D4 | L3 | L8 |
| MT-4/NL4.3 (CXCR4) | 30.7 | 40.3 | 0.25 | 0.25 |
| MT-4/HE (CCR5/CXCR4) | 26.7 | 82.0 | 0.16 | 0.10 |
| PBMC/NL4.3 (CXCR4) | 30.9 | 13.7 | 0.5 | 0.2 |
| PBMC/HE (CCR5/CXCR4) | 44.7 | 13.6 | 1.5 | 2.3 |
| PBMC/BaL (CCR5) | >500 | >500 | >500 | >500 |
| U87.CD4.CXCR4/NL4.3 (CXCR4) | 44.5 | 107.0 | 0.36 | 0.35 |
| U87.CD4.CCR5/BaL (CCR5) | >500 | >500 | >500 | >500 |
CXCR4 Nanobody Induces Stem Cell Mobilization.
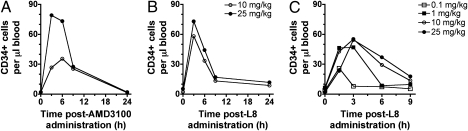
The CXCR4/CXCL12 axis contributes to the residence of hematopoietic cells in the bone marrow. In view of the recent approval of plerixafor (AMD3100) for the mobilization and transplantation of hematopoietic stem cells (16), we investigated if the CXCR4 nanobodies can also be used for this therapeutic indication. Cynomolgus monkeys injected with AMD3100 effectively displayed increased numbers of WBCs and CD34+ stem cells in the circulation, with both cell types being mobilized with similar kinetics (Fig. 6A and Fig. S6A). The maximum WBC and stem cell release was observed at 3 to 6 h after the start of infusion and the number of circulating cells returned to basal values after 24 h. Importantly, single i.v. administration (10 and 25 mg/kg) of biparatopic nanobody 238D2-20GS-238D4 (L8) induced mobilization of WBCs and stem cells to a similar extent and with similar kinetics compared with AMD3100, with a maximum CD34+ stem cell release 3 h after treatment (Fig. 6B and Fig. S6B). To gain insight in the early release of WBCs and stem cells following 238D2-20GS-238D4 (L8) administration, monkeys received different amounts of L8 (0.1, 1, 10, and 25 mg/kg) with a 30-min i.v. infusion and blood samples were taken 0, 1.5, 3, 6, and 9 h after administration (Fig. 6C and Fig. S6C). The biparatopic nanobody 238D2-20GS-238D4 (L8) was well tolerated at all doses tested and no treatment-related clinical signs were observed. In monkeys treated at 10 and 25 mg/kg, maximum CD34+ cell release was recorded at 3 h after the start of infusion. Decrease in circulating stem cells was appreciable starting from 6 h after treatment. Treatment with nanobody at 0.1 and 1 mg/kg still produced an increase in circulating stem cells at 1.5 h after the start of infusion. The return to basal number of CD34+ cells was observed at 3 and 6 h after treatment of animals dosed with 0.1 mg/kg and 1 mg/kg, respectively. Thus, the biparatopic nanobody L8 induced stem cell mobilization in a dose-dependent manner.
Fig. 6.
CXCR4-specific biparatopic nanobody L8 induces stem cell mobilization in vivo to a similar extent as AMD3100. (A) Two independent monkeys were injected s.c. with 1 mg/kg of the CXCR4-specific antagonist AMD3100 and blood samples were taken over a period of 24 h after administration. (B) Two independent monkeys were injected i.v. with the biparatopic CXCR4-specific nanobody L8 at 10 mg/kg or 25 mg/kg. Blood samples were taken over a period of 24 h after administration. (C) Four independent monkeys were injected i.v. with various amounts of L8 nanobody, namely 0.1 mg/kg, 1 mg/kg, 10 mg/kg, or 25 mg/kg. Blood samples were withdrawn up to 9 h after administration. In all experiments, the presence of CD34+ stem cells in the blood was determined by flow cytometry analysis.
Discussion
Highly potent and selective functional llama-derived immunoglobulin single variable domains (i.e., nanobodies) were generated against the chemokine receptor CXCR4, known to be involved in stem cell physiology, inflammation, HIV entry, and promotion of tumor growth and metastasis. The nanobodies 238D2 and 238D4 act as competitive CXCR4 antagonists, binding to the same binding site as CXCL12, as shown in Schild analysis of CRE reporter gene inhibition data. The full and competitive blocking of CXCR4 by these VHH-based immunoglobulin single variable domains, to our knowledge, has not been described for conventional antibodies like 12G5, possibly as a result of allosteric and/or receptor state-dependent binding of conventional surface-binding antibodies to CXCR4 (25, 26). The small-molecule ligand AMD3100—also fully inhibiting CXCL12 binding—binds to well characterized sites within this cavernous binding pocket of CXCR4 within the transmembrane helices and the ECL2 (37). Cavity binding was previously observed for enzyme-inhibiting nanobodies and nanobodies directed against cryptic epitopes of infectious agents (4–6). This feature seems to be unique for VHH-based immunoglobulin single variable domains and can be attributed to the penetration of their extremely long and flexible CDR loops into cavity structures (6). Epitope mapping showed that the identified nanobodies in this study bind to distinct but partially overlapping sites in ECL2, with 238D2 having an additional anchoring point in ECL3. As 238D2 and 238D4 are highly selective for human but not murine CXCR4 and because not all amino acids from the nanobodies epitopes differ between both species, we suggest that minor changes in ECL2 are sufficient to affect the nanobodies’ affinity for CXCR4.
To further optimize the pharmacological profile of the VHH-based immunoglobulin single variable domain, we engineered biparatopic nanobody constructs. The strategy of linking active nanobodies to dimeric, trimeric, or even pentameric complexes has previously successfully been applied to target proteins that exist in a constitutive or inducible dimeric or oligomeric complex such as the Escherichia coli verotoxin (35), TNF-α (33), and the EGF receptor (34). In this study we show that biparatopic CXCR4 nanobodies present antagonistic properties against CXCL12-induced chemotaxis and CXCR4-mediated HIV-1 entry in picomolar concentrations, which, to our knowledge, represents the most potent CXCR4 inhibitors described so far. Although it was generally considered that rhodopsin-like GPCRs such as CXCR4 act as monomers, there is now accumulating evidence that GPCRs exist as dimers or oligomers. Dimerization or oligomerization of GPCRs is thought to have important implications for GPCR maturation, GPCR trafficking, ligand binding, and GPCR signaling (38, 39). Constitutive homodimerization of CXCR4 has previously been shown by using resonance energy transfer, coimmunoprecipitation, and protein crosslinking (40–42). As both 238D2 and 238D4 have distinct but also overlapping epitopes, the increased potency of the biparatopic nanobody constructs may indicate binding to two CXCR4 molecules in close proximity and support the hypothesis of CXCR4 homodimers (or oligomers) as functional units at the cell membrane. Interestingly, the monovalent and biparatopic nanobodies display a different mode of action, as the monovalent nanobodies behave as neutral antagonists, and the linked nanobodies behave as inverse agonists at the constitutively active CXCR4-N3.35A. AMD3100, like the monovalent nanobodies, acts as neutral antagonist. A significant number of top-selling GPCR drugs behave as inverse agonists rather than neutral antagonists (43), and it has been claimed that inverse agonists may have specific therapeutic benefits compared with neutral antagonists for several diseases, including cancer (44). CXCR4 is overexpressed in the majority of tumors, which is often associated with increased levels of basal activity (12, 45). As such, the use of inverse agonistic CXCR4 biparatopic nanobodies could be beneficial. The (patho)physiological relevance of constitutive activity of CXCR4 and benefit of inverse agonism of CXCR4 ligands awaits further research.
Besides their anti-HIV-1 entry activity, CXCR4 nanobodies also act as rapid and efficient stem cell-mobilizing agents in cynomolgus monkeys. Targeted interference of the CXCL12–CXCR4 axis by AMD3100 has previously been shown to effectively and rapidly mobilize hematopoietic stem cells in patients with multiple myeloma or non-Hodgkin lymphoma (46, 47). In this study we show that the biparatopic nanobody induces CD34+ stem cell mobilization as rapidly and nearly as effectively as AMD3100. Because patients with multiple myeloma and non-Hodgkin lymphoma show poor mobility in response to G-CSF and a significant number of patients, particular those undergoing chemotherapy, are resistant to G-CSF, the CXCR4 nanobodies present a new class of effective stem cell mobilizers.
In conclusion, our findings show the successful generation of inhibitory nanobodies directed against a GPCR. The method used here resulted, within a few months, in highly specific and potent (picomolar) nanobodies inhibiting CXCR4-mediated signaling and inducing stem cell mobilization in vivo. Half-life extension methods used for conventional antibody fragments (Fabs and scFvs), such as PEGylation or fusion to serum albumin (48, 49), could be used for tailoring the half-life of nanobodies (50) and increase their therapeutic window depending on the therapeutic indication. Thus, the CXCR4 nanobodies may open up a new avenue for the development of novel therapeutic agents for CXCR4-related diseases.
Materials and Methods
Detailed materials and methods can be found in SI Materials and Methods.
Immunization, Library Construction, and Selection of Nanobodies.
Nanobody libraries were generated using PBMCs isolated from two different llamas immunized with HEK293T cells transfected with CXCR4. The nanobody phage library was generated by RT-PCR with at least 107 transformants. Counterselections with membranes of CHO cells overexpressing CXCR4 versus nontransfected cells in a first round, and membranes of COS-7 cells overexpressing CXCR4 versus nontransfected cells in the second round, identified potential CXCR4-specific nanobodies. The presence of CXCR4-specific phages was tested using phage ELISA. Details are described in SI Materials and Methods.
Epitope Mapping of CXCR4 Nanobodies Binding Sites.
CXCR4 mutation library was created by random mutagenesis using the shotgun mutagenesis platform from Integral Molecular. A total of 731 clones were generated with all residues mutated at least once (100%), at least twice (98.8%), and 76.2% or 19.9% of the clones presented one or two mutated amino acids, respectively. All mutant CXCR4 were tagged at their N- and C-termini with V5 and Flag epitopes, respectively, allowing detection of cell surface and full-length expression using anti-V5 and anti-Flag antibodies. Nanobodies bound to CXCR4 mutants were detected by using an anti-Myc antibody.
Stem Cell Mobilization.
All experiments were conducted in strict compliance with European Economic Community and Italian guidelines for laboratory animal welfare. Cynomolgus monkeys were given AMD3100 (1 mg/kg) s.c. at the doses of 1 mg/kg whereas the nanobody (0.1, 1, 10, or 25 mg/kg) was administered in a single i.v. infusion over a period of 30 min. Mortality, clinical signs, and food consumption were monitored daily. Body weight was recorded once in the pretest period, the day of treatments, and then once weekly.
Supplementary Material
Acknowledgments
We thank Claesand and Rebecca Provinciael for excellent technical assistance with the anti-HIV experiments. We also thank Guus van Dongen and Dennis Verzijl for the technical feedback on labeling and handling procedures of nanobodies. This work was funded by Ablynx and supported by Institute for the Promotion of Innovation by Science and Technology in Flanders Grant IWT70050 from the Flemish government, “Fonds voor Wetenschappelijk Onderzoek” Grant G-485-08, and Centers of Excellence of Katholieke Universiteit Leuven Grant EF/05/015.
Footnotes
Conflict of interest statement: M.G.-P., S.D.V., B.S., H.U., W.V., M.S., H.J.D.H., and P.V. are/were paid employees of Ablynx.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012865107/-/DCSupplemental.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Gupta A, Heimann AS, Gomes I, Devi LA. Antibodies against G-protein coupled receptors: Novel uses in screening and drug development. Comb Chem High Throughput Screen. 2008;11:463–467. doi: 10.2174/138620708784911465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol. 2007;77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Genst E, et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci USA. 2006;103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson KA, et al. Structure of an IgNAR-AMA1 complex: Targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure. 2007;15:1452–1466. doi: 10.1016/j.str.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Lauwereys M, et al. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998;17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stijlemans B, et al. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J Biol Chem. 2004;279:1256–1261. doi: 10.1074/jbc.M307341200. [DOI] [PubMed] [Google Scholar]
- 8.Van Bockstaele F, Holz JB, Revets H. The development of nanobodies for therapeutic applications. Curr Opin Investig Drugs. 2009;10:1212–1224. [PubMed] [Google Scholar]
- 9.Gonzalo JA, et al. Critical involvement of the chemotactic axis CXCR4/stromal cell-derived factor-1 alpha in the inflammatory component of allergic airway disease. J Immunol. 2000;165:499–508. doi: 10.4049/jimmunol.165.1.499. [DOI] [PubMed] [Google Scholar]
- 10.Matthys P, et al. AMD3100, a potent and specific antagonist of the stromal cell-derived factor-1 chemokine receptor CXCR4, inhibits autoimmune joint inflammation in IFN-gamma receptor-deficient mice. J Immunol. 2001;167:4686–4692. doi: 10.4049/jimmunol.167.8.4686. [DOI] [PubMed] [Google Scholar]
- 11.Ratajczak MZ, et al. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia. 2006;20:1915–1924. doi: 10.1038/sj.leu.2404357. [DOI] [PubMed] [Google Scholar]
- 12.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 14.Liang X. CXCR4, inhibitors and mechanisms of action. Chem Biol Drug Des. 2008;72:97–110. doi: 10.1111/j.1747-0285.2008.00681.x. [DOI] [PubMed] [Google Scholar]
- 15.Hendrix CW, et al. AMD3100 HIV Study Group Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. J Acquir Immune Defic Syndr. 2004;37:1253–1262. doi: 10.1097/01.qai.0000137371.80695.ef. [DOI] [PubMed] [Google Scholar]
- 16.Cashen AF. Plerixafor hydrochloride: A novel agent for the mobilization of peripheral blood stem cells. Drugs Today (Barc) 2009;45:497–505. doi: 10.1358/dot.2009.45.7.1395292. [DOI] [PubMed] [Google Scholar]
- 17.DiPersio JF, et al. 3101 Investigators Phase III prospective randomized double-blind placebo-controlled trial of plerixafor plus granulocyte colony-stimulating factor compared with placebo plus granulocyte colony-stimulating factor for autologous stem-cell mobilization and transplantation for patients with non-Hodgkin's lymphoma. J Clin Oncol. 2009;27:4767–4773. doi: 10.1200/JCO.2008.20.7209. [DOI] [PubMed] [Google Scholar]
- 18.DiPersio JF, et al. 3102 Investigators Plerixafor and G-CSF versus placebo and G-CSF to mobilize hematopoietic stem cells for autologous stem cell transplantation in patients with multiple myeloma. Blood. 2009;113:5720–5726. doi: 10.1182/blood-2008-08-174946. [DOI] [PubMed] [Google Scholar]
- 19.McKnight A, et al. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 21.Ottaiano A, et al. Inhibitory effects of anti-CXCR4 antibodies on human colon cancer cells. Cancer Immunol Immunother. 2005;54:781–791. doi: 10.1007/s00262-004-0636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strizki JM, Turner JD, Collman RG, Hoxie J, González-Scarano F. A monoclonal antibody (12G5) directed against CXCR-4 inhibits infection with the dual-tropic human immunodeficiency virus type 1 isolate HIV-1(89.6) but not the T-tropic isolate HIV-1(HxB) J Virol. 1997;71:5678–5683. doi: 10.1128/jvi.71.7.5678-5683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaday GG, et al. CXCR4 and CXCL12 (SDF-1) in prostate cancer: inhibitory effects of human single chain Fv antibodies. Clin Cancer Res. 2004;10:5630–5639. doi: 10.1158/1078-0432.CCR-03-0633. [DOI] [PubMed] [Google Scholar]
- 24.Xu C, Sui J, Tao H, Zhu Q, Marasco WA. Human anti-CXCR4 antibodies undergo VH replacement, exhibit functional V-region sulfation, and define CXCR4 antigenic heterogeneity. J Immunol. 2007;179:2408–2418. doi: 10.4049/jimmunol.179.4.2408. [DOI] [PubMed] [Google Scholar]
- 25.Baribaud F, et al. Antigenically distinct conformations of CXCR4. J Virol. 2001;75:8957–8967. doi: 10.1128/JVI.75.19.8957-8967.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carnec X, Quan L, Olson WC, Hazan U, Dragic T. Anti-CXCR4 monoclonal antibodies recognizing overlapping epitopes differ significantly in their ability to inhibit entry of human immunodeficiency virus type 1. J Virol. 2005;79:1930–1933. doi: 10.1128/JVI.79.3.1930-1933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coward P, Chan SD, Wada HG, Humphries GM, Conklin BR. Chimeric G proteins allow a high-throughput signaling assay of Gi-coupled receptors. Anal Biochem. 1999;270:242–248. doi: 10.1006/abio.1999.4061. [DOI] [PubMed] [Google Scholar]
- 28.Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood. 2009;113:6085–6093. doi: 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- 29.Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem. 2000;275:23736–23744. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- 30.Kofuku Y, et al. Structural basis of the interaction between chemokine stromal cell-derived factor-1/CXCL12 and its G-protein-coupled receptor CXCR4. J Biol Chem. 2009;284:35240–35250. doi: 10.1074/jbc.M109.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou N, et al. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J Biol Chem. 2001;276:42826–42833. doi: 10.1074/jbc.M106582200. [DOI] [PubMed] [Google Scholar]
- 32.Paes C, et al. Atomic-level mapping of antibody epitopes on a GPCR. J Am Chem Soc. 2009;131:6952–6954. doi: 10.1021/ja900186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coppieters K, et al. Formatted anti-tumor necrosis factor alpha VHH proteins derived from camelids show superior potency and targeting to inflamed joints in a murine model of collagen-induced arthritis. Arthritis Rheum. 2006;54:1856–1866. doi: 10.1002/art.21827. [DOI] [PubMed] [Google Scholar]
- 34.Roovers RC, et al. Efficient inhibition of EGFR signaling and of tumour growth by antagonistic anti-EFGR nanobodies. Cancer Immunol Immunother. 2007;56:303–317. doi: 10.1007/s00262-006-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, et al. Pentamerization of single-domain antibodies from phage libraries: a novel strategy for the rapid generation of high-avidity antibody reagents. J Mol Biol. 2004;335:49–56. doi: 10.1016/j.jmb.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 36.Zhang WB, et al. A point mutation that confers constitutive activity to CXCR4 reveals that T140 is an inverse agonist and that AMD3100 and ALX40-4C are weak partial agonists. J Biol Chem. 2002;277:24515–24521. doi: 10.1074/jbc.M200889200. [DOI] [PubMed] [Google Scholar]
- 37.Wong RS, et al. Comparison of the potential multiple binding modes of bicyclam, monocylam, and noncyclam small-molecule CXC chemokine receptor 4 inhibitors. Mol Pharmacol. 2008;74:1485–1495. doi: 10.1124/mol.108.049775. [DOI] [PubMed] [Google Scholar]
- 38.Dalrymple MB, Pfleger KD, Eidne KA. G protein-coupled receptor dimers: functional consequences, disease states and drug targets. Pharmacol Ther. 2008;118:359–371. doi: 10.1016/j.pharmthera.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Prinster SC, Hague C, Hall RA. Heterodimerization of g protein-coupled receptors: specificity and functional significance. Pharmacol Rev. 2005;57:289–298. doi: 10.1124/pr.57.3.1. [DOI] [PubMed] [Google Scholar]
- 40.Babcock GJ, Farzan M, Sodroski J. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. J Biol Chem. 2003;278:3378–3385. doi: 10.1074/jbc.M210140200. [DOI] [PubMed] [Google Scholar]
- 41.Lagane B, et al. CXCR4 dimerization and beta-arrestin-mediated signaling account for the enhanced chemotaxis to CXCL12 in WHIM syndrome. Blood. 2008;112:34–44. doi: 10.1182/blood-2007-07-102103. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, He L, Combs CA, Roderiquez G, Norcross MA. Dimerization of CXCR4 in living malignant cells: Control of cell migration by a synthetic peptide that reduces homologous CXCR4 interactions. Mol Cancer Ther. 2006;5:2474–2483. doi: 10.1158/1535-7163.MCT-05-0261. [DOI] [PubMed] [Google Scholar]
- 43.Milligan G. Constitutive activity and inverse agonists of G protein-coupled receptors: A current perspective. Mol Pharmacol. 2003;64:1271–1276. doi: 10.1124/mol.64.6.1271. [DOI] [PubMed] [Google Scholar]
- 44.Kenakin T. Inverse, protean, and ligand-selective agonism: Matters of receptor conformation. FASEB J. 2001;15:598–611. doi: 10.1096/fj.00-0438rev. [DOI] [PubMed] [Google Scholar]
- 45.Beider K, et al. Interaction between CXCR4 and CCL20 pathways regulates tumor growth. PLoS ONE. 2009;4:e5125. doi: 10.1371/journal.pone.0005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Clercq E. The AMD3100 story: The path to the discovery of a stem cell mobilizer (Mozobil) Biochem Pharmacol. 2009;77:1655–1664. doi: 10.1016/j.bcp.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 47.Flomenberg N, et al. The use of AMD3100 plus G-CSF for autologous hematopoietic progenitor cell mobilization is superior to G-CSF alone. Blood. 2005;106:1867–1874. doi: 10.1182/blood-2005-02-0468. [DOI] [PubMed] [Google Scholar]
- 48.Chapman AP, et al. Therapeutic antibody fragments with prolonged in vivo half-lives. Nat Biotechnol. 1999;17:780–783. doi: 10.1038/11717. [DOI] [PubMed] [Google Scholar]
- 49.Holt LJ, Herring C, Jespers LS, Woolven BP, Tomlinson IM. Domain antibodies: Proteins for therapy. Trends Biotechnol. 2003;21:484–490. doi: 10.1016/j.tibtech.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 50.Harmsen MM, et al. Passive immunization of guinea pigs with llama single-domain antibody fragments against foot-and-mouth disease. Vet Microbiol. 2007;120:193–206. doi: 10.1016/j.vetmic.2006.10.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.