Abstract
Mitosis ensures equal genome segregation in the eukaryotic lineage. This process is facilitated by microtubule attachment to each chromosome via its centromere. In centromeres, canonical histone H3 is replaced in nucleosomes by a centromere-specific histone H3 variant (CENH3), providing the unique epigenetic signature required for microtubule binding. Due to recent findings of alternative CENH3 nucleosomal forms in invertebrate centromeres, it has been debated whether the classical octameric nucleosomal arrangement of two copies of CENH3, H4, H2A, and H2B forms the basis of the vertebrate centromere. To address this question directly, we examined CENH3 [centromere protein A (CENP-A)] nucleosomal organization in human cells, using a combination of nucleosome component analysis, atomic force microscopy (AFM), and immunoelectron microscopy (immuno-EM). We report that native CENP-A nucleosomes contain centromeric alpha satellite DNA, have equimolar amounts of H2A, H2B, CENP-A, and H4, and bind kinetochore proteins. These nucleosomes, when measured by AFM, yield one-half the dimensions of canonical octameric nucleosomes. Using immuno-EM, we find that one copy of CENP-A, H2A, H2B, and H4 coexist in CENP-A nucleosomes, in which internal C-terminal domains are accessible. Our observations indicate that CENP-A nucleosomes are organized as asymmetric heterotypic tetramers, rather than canonical octamers. Such altered nucleosomes form a chromatin fiber with distinct folding characteristics, which we utilize to discriminate tetramers directly within bulk chromatin. We discuss implications of our observations in the context of universal epigenetic and mechanical requirements for functional centromeres.
Keywords: alternative nucleosomes, cell division, histone variant, regional centromeres
Each eukaryotic chromosome has a centromere, which serves as the sole attachment point for spindle microtubules during mitosis (1). Sister centromeres undergo a distinct phase change from independent discrete spots in the interphase nucleus to a paired constriction on mitotic chromosomes. At the base of the constriction, centromeric DNA is packaged within variant nucleosomes composed of a centromere-specific histone H3 (CENH3). In metazoans, centromeric DNA alone is insufficient to dictate centromere location, as evidenced by the formation of satellite-free neocentromeres (2, 3). That a common epigenetic identity prevails can be deduced from a number of studies spanning evolutionary, biochemical, cytological, genetic, and genomic methods (4–22), all of which pinpoint CENH3 as the key epigenetic marker for active centromeres in eukaryotes.
Another general feature of CENH3 nucleosomes is that they must provide a stable foundation for kinetochore proteins (23), while still allowing displacement during DNA replication. Studies investigating structural features of centromeric chromatin that contribute to its function have reported that it is refractory to nuclease digestion (24–26) and topologically distinctive (27, 28). Direct examinations of Drosophila and yeast CENH3 chromatin indicate that native centromeric nucleosomes can adopt tetrameric (29) or hexameric states (30), respectively, in contrast to the octameric nucleosomal configuration reported for ectopically expressed CENH3 (31). In vitro, centromere-like AT-rich DNA excludes octameric nucleosomes, suggesting that the geometry of native centromeric DNA can adapt to noncanonical conformations (32). Indeed, recent analysis of yeast and Drosophila CENH3 nucleosomes show that they wrap DNA into a right-handed configuration, which is structurally incompatible with canonical octameric organization (33, 34). The right-handed direction of the DNA wrap dictated by tetrameric nucleosomes would be the same as the direction of the twist of double helix, providing a plausible solution for how centromeres are able to provide a solid structural basis for the kinetochore (35).
In vitro, in contrast to the flexible structures described for invertebrate CENH3s (29,30, 32), the loop-1/alpha-2 [centromere protein A (CENP-A) targeting domain (CATD)] domain of human CENH3 (CENP-A) reduces flexibility in alpha-2 of H4 (18). In the context of nucleosome structure, CENP-ACATD induced stiffness in H4 is proposed to create a compacted, left-handed octameric nucleosome. This model is attractive, because it predicts an inflexible centromeric chromatin fiber, which should be resistant to disassembly (18, 36). However, subsequent dissections reveal that the CATD domain also drives instability in CENP-A octameric nucleosomes in vitro (37). Consequently, these seemingly incompatible in vitro observations present a mechanistic hurdle for understanding how CENP-A nucleosomes function without destabilizing the mitotic kinetochore in vivo. Therefore, it is of interest to dissect properties of CENP-A chromatin in vivo.
We report here an investigation of CENP-A chromatin in interphase human cells. Our nucleosome component analysis shows that CENP-A nucleosomes are composed of equimolar amounts of CENP-A, H2A, H2B, and H4, contain centromeric alpha satellite DNA, and are bound to kinetochore proteins CENP-B and CENP-C. Such native CENP-A nucleosomes form chromatin fibers with distinctive folding characteristics, and when measured by atomic force microscopy (AFM) are consistently one-half the height and volume of canonical octameric nucleosomes under a range of ionic conditions. Furthermore, immunoelectron microscopy (immuno-EM) analysis detects stable DNA organization and single copies of CENP-A and H2B in each CENP-A nucleosome. These observations offer direct evidence in support of tetrameric organization of CENP-A nucleosomes in human cells. Tetrameric nucleosomes can also be discriminated directly from bulk octamers within human chromatin. We discuss mechanisms by which tetramer nucleosomes may arise, propose an epigenetic model by which they contribute to centromere recognition, and suggest a universal role for CENP-A tetramers in withstanding mitotic torque.
Results
Native Human CENP-A Nucleosomes Have a Heterotypic Composition.
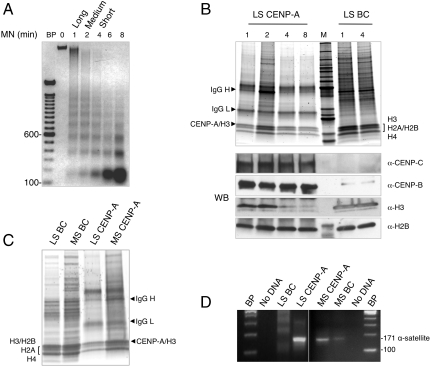
We purified native CENP-A nucleosomes from interphase HeLa nuclear extracts to examine their composition and physical dimensions. Taking into consideration that CENP-A chromatin may interconvert between folded and unfolded fibers, we used low salt (LS, 50 mM NaCl) and moderate salt (MS, 350 mM NaCl) buffers to extract chromatin fibers made soluble by sequential micrococcal nuclease digestion (38). Analysis of bulk chromatin fibers (BC) reveals a classical nucleosomal DNA ladder (Fig. 1A), and protein analysis detects the full complement of histones (Fig. 1B, BC). To avoid shearing the chromatin by centrifugal forces, we coupled anti-CENP-A antibodies to magnetic beads and performed gentle native CENP-A chromatin immunopurification (NChIP) (39).
Fig. 1.
Native human CENP-A nucleosomes have a heterotypic composition. (A) DNA gel shows that bulk chromatin (BC) obtained from 0-, 1-, 2-, 4-, and 8-min micrococcal nuclease (MN) digests of HeLa nuclei have typical nucleosome protection ladders. (B) Krypton-X stained protein gels show that BC has canonical histones H3, H2A, H2B, and H4 in 1- and 4-min MN extracts, and CENP-A IP has all four histones in 1-, 2-, 4-, and 8-min MN extracts. The intense marker band is 50 kDa. Western blot (WB) panel demonstrates that CENP-C, CENP-B, and H2B are present in all time points of LS CENP-A IP, whereas H3 is depleted in shorter CENP-A arrays. (C) Krypton-X stained protein gels show that MS and LS CENP-A IP have H2A, H2B, and H4. (D) PCR analysis shows that the D17Z1 171-bp centromeric alpha satellite repeat is enriched in LS and MS CENP-A IP.
CENP-A nucleosomal components eluted from the immune beads were analyzed by ultrasensitive protein gels (Fig. 1B), and by Western blot analysis (Fig. 1B, WB). Regardless of chromatin fiber length, CENP-A, H2A, H2B, and H4 are equimolar components of native CENP-A chromatin [Fig. 1B, LS CENP-A immunopurification (IP), WB α-H2B; Fig. 1C, MS CENP-A IP; and Fig. S1A, WB α-H2B]. Consistent with previous reports of CENP-A and H3 domains alternating within metazoan centromeres (10, 11), histone H3 is present in long arrays of CENP-A, but rapidly depleted in shorter arrays (Fig. 1B, α-H3, compare lanes 2 and 4). In LS and MS, native and tagged CENP-As associate stably with H2A, H2B and H4 (Fig. 1 B and C, and Fig. S1B), whereas transiently overexpressed CENP-A does not (Fig. S1B). Kinetochore proteins CENP-B and CENP-C are enriched within LS and MS CENP-A chromatin, suggesting that native CENP-A nucleosomes are representative of active centromeres (Fig. 1B, WB α-CENP-C and α-CENP-B, and Fig. S1A). To confirm that native CENP-A IPs derive from centromeric locations, we used the D17Z1 171 bp α-satellite repeats as a marker for human centromeres (40). Indeed, this centromeric DNA is enriched in both LS and MS CENP-A chromatin compared to the equivalent BC input (Fig. 1D). LS and MS chromatin fractions of native CENP-A contain similar histone components, bind CENP-B and CENP-C, and contain centromeric α-satellite DNA. From these data, we infer that LS and MS CENP-A IPs derive from active centromeric regions in human cells.
Native CENP-A Chromatin Fibers Have Distinctive Folding Motifs.
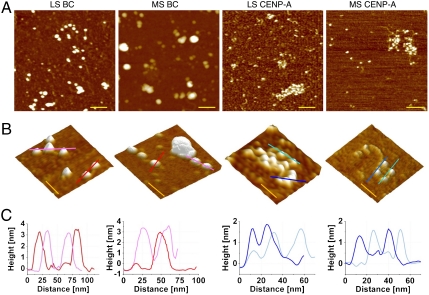
We next sought to determine the characteristics of native CENP-A chromatin fibers by AFM. Regardless of whether extracted from LS or MS, BC and CENP-A nucleosomes present the beads-on-a-string motif, consistent with stable nucleosomal configuration (Fig. 2 A and B, and Fig. S2). CENP-A arrays are also close packed, but generally shorter (5–25 nucleosomes) than BC arrays (20–50 nucleosomes), suggesting that nuclease release is more effective for AT-rich centromeric chromatin than for bulk (24, 25, 29, 39). As reported previously (38), BC chromatin is generally open in LS and more compacted in MS (Fig. 2 A and B, and Fig. S2A), whereas over 60% of CENP-A chromatin arrays of equivalent length are compacted in LS and resist ionic condensation in MS (Fig. 2 A and B, and Fig. S2). Therefore, CENP-A chromatin manifests unique folding behavior. Line section profiles of individual CENP-A nucleosomes indicate that they have similar widths, but shorter heights relative to BC nucleosomes (Fig. 2C).
Fig. 2.
Native CENP-A chromatin has distinctive folding motifs. Representative AFM images showing (A) LS BC, MS BC, LS CENP-A, and MS CENP-A chromatin have classical nucleosomal organization but differ in compaction. LS BC has open beads-on-a-string structure, whereas LS CENP-A chromatin remains compacted. Conversely, MS BC is heavily compacted, whereas MS CENP-A resists condensation. (Scale bar: 100 nm.) (B) Representative 3D topography images showing folding differences between BC and CENP-A, with (C) single-molecule line scans below showing that CENP-A nucleosomes are one-half the height of BC nucleosomes. (Scale bar: 50 nm.)
Native CENP-A Nucleosomes Have Tetrameric Dimensions.
During our analysis of compaction above, we noted that individual CENP-A nucleosomes were shorter than BC nucleosomes (Fig. 2C, line section profiles). AFM serves as a reliable tool for quantitative assessment of nucleosomal configuration because it can distinguish octamers from tetramers (41). Previous AFM measurements of native Drosophila CENH3 nucleosomes yield one-half the height of canonical octameric nucleosomes, indicating that they are tetrameric at interphase (29, 42).
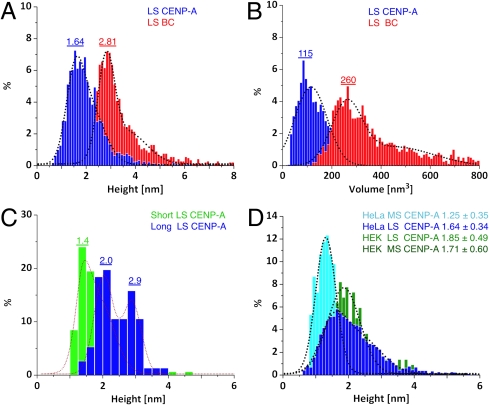
Consequently, we automated our analysis protocols (SI Text), in order to obtain unbiased dimensions for large numbers of CENP-A and BC nucleosomes (Table S1). Consistent with previously published AFM height estimates of classical octameric organization (29, 41, 42), LS BC nucleosomes range in height from 2.5–4.5 nm (Fig. 3A, red). In contrast, 75% of LS CENP-A nucleosomes yield one-half octameric heights, ranging from 1.4–2.0 nm, with a distinct 25% subpopulation ranging from 1.7–3.0 nm (Fig. 3A, dark blue). We also assessed volumes, which are an independent measure of nucleosomal organization. BC nucleosomes yield a mean volume of 260 nm3 in good accordance with octameric volumes (AFM methods), whereas CENP-A nucleosomes have ∼1/2 the octameric volume, at 115 nm3 (Fig. 3B, red vs. dark blue).
Fig. 3.
Native CENP-A nucleosomes have tetramer dimensions. (A) LS CENP-A nucleosomes are lower in height compared to LS BC nucleosomes. (B) LS CENP-A nucleosomes occupy smaller volumes compared to LS BC nucleosomes. (C) LS CENP-A nucleosomes have a bimodal distribution of CENP-A tetramers and H3 octamers in long chromatin and enrich exclusively in tetramers in short chromatin. (D) LS and MS HeLa CENP-A and HEK LS CENP-A nucleosomes show overlap in tetrameric dimensions. BC nucleosomes (red), LS and MS CENP-A nucleosomes (dark and light blue), short and long CENP-A (light green and dark blue), and HEK CENP-A nucleosomes (green). Means are shown above peaks. Data are summarized in Table S1.
In long centromeric fibers, H3 domains alternate with CENP-A domains (10, 11). We reasoned it should be possible to purify both CENP-A tetramers and bulk octamers using long fibers as input. Indeed, from lightly digested extracts, LS CENP-A IP yields equal proportions of tetramers and classical octamers (Fig. 3C, dark blue, and Fig. S3A), whereas, from short chromatin fibers, CENP-A IP yields tetramers exclusively (Fig. 3C, green, and Fig. S3A). These data are consistent with Western blot analysis, which show that H3 is present in long CENP-A arrays, but is depleted in shorter arrays (Fig. 1C, WB α-H3). This experiment demonstrates that H3 octamers and CENP-A tetramers coexist as discrete populations. Thus, the far end of the tetrameric range (2.0–2.3 nm) seen in LS CENP-A IP above (Fig. 3A) most likely derives from H3 nucleosomes, whereas the 75% fraction ranging from 1.7–2.3 nm reflects CENP-A nucleosomes alone. This interpretation is supported by an examination of MS samples, in which MS BC nucleosomes have a broad distribution ranging from 3–6 nm in height, (Table S1), whereas MS CENP-A nucleosomes have tetrameric heights tightly distributed from 1.3–1.8 nm (Fig. 3D, light blue). MS BC volumes are broadly distributed from 250–550 nm3 (Fig. S3B, red), whereas MS CENP-A chromatin yields volumes clustering around 81 nm3 (Fig. S3B, light blue).
We extended our analysis to noncarcinoma HEK cell lines, and observed that LS and MS HEK CENP-A nucleosomes yield heights corresponding closely to HeLa LS CENP-A nucleosomes (Fig. 3D, green vs. dark blue, and Fig. S3C). Similarly, HEK LS and MS CENP-A volumes cluster from 80–100 nm3 (Fig. S3D). To exclude the possibility that antibody binding and elution protocols might disrupt CENP-A chromatin obtained from native extracts, we expressed a fusion CENP-A protein (SI Materials and Methods), expressed at a lower level than endogenous CENP-A, and localized correctly to human centromeres (Fig. S4 A and B). Long CENP-A chromatin fibers were enriched by binding a strep column in LS, eluted with biotin in physiological salt, and visualized by AFM as nucleosomal beads (Fig. S4C). These fusion CENP-A nucleosomes yield a bimodal distribution with ∼60% at 1.3 nm and ∼40% at 2.3 nm in height (Fig. S4D), consistent with the bimodal distribution obtained from LS CENP-A above (Table S1).
Overall, when comparing the range of heights and volumes of ∼4,000 CENP-A nucleosomes (1.4–2.3 nm, 81–160 nm3, Table S1), to those of ∼4,900 BC nucleosomes (2.5–4.5 nm, 240–360 nm3, Table S1), it is apparent that BC nucleosomes yield dimensions consistent with octameric organization, whereas CENP-A nucleosomes do not. Therefore, tetrameric dimensions are a particular feature of native and tagged CENP-A chromatin.
CENP-A Nucleosomes Contain One Copy of CENP-A and H2B, and Have Accessible C Termini.
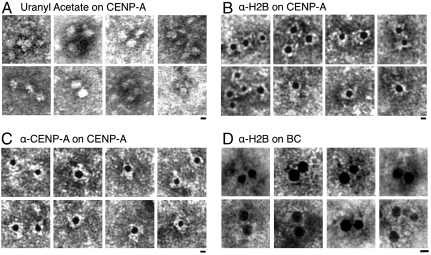
One explanation for the tetrameric structures we report for CENP-A above (Table S1) might be that DNA is dissociated and what remains is a protein tetramer. A priori, this interpretation would seem unlikely, because α-satellite DNA is enriched in CENP-A IPs (Fig. 1D), and linker DNA can be visualized by AFM (Fig. 2 A and D). Nevertheless, we used the intercalating drug uranyl acetate (UA) (38) to detect DNA within the same samples of LS CENP-A chromatin for which tetrameric heights were observed by AFM (Table S1). We observed that UA-stained DNA forms an electron-dense region around CENP-A nucleosomes (Fig. 4A), yielding widths of CENP-A nucleosomes at 10 ± 1 nm (N = 31), consistent with stable heterotypic particles.
Fig. 4.
CENP-A nucleosomes have single copies of CENP-A and H2B, and exposed C termini electron micrographs showing (A) uranyl-acetate-stained CENP-A nucleosomes are 10-nm beads on a string. (B) Anti-CENP-A 5-nm gold bead (black bead) detects single epitope per LS CENP-A nucleosome. (C) Anti-H2B 5-nm gold bead staining shows that a single C-terminal epitope of H2B is present and accessible in LS CENP-A nucleosomes. (D) Anti-H2B 5-nm gold bead staining of bulk chromatin is highly inefficient, but detects examples of dual labels. Particles larger than 25 nm in diameter were observed and excluded from this analysis. (Scale bar: 5 nm.) Images were resized for comparison.
Immuno-EM permits mapping histones on chromatin at single molecule resolution (43). Therefore, we chose an anti-CENP-A antibody that recognizes the N terminus of CENP-A, which should be accessible regardless of whether CENP-A nucleosomes are tetrameric or octameric. Secondary-antibody-coupled 5-nm gold beads subsequently bind the primary antibody. Of 576 CENP-A nucleosomes detected by the CENP-A antibody, no examples of dual labels could be found, which is consistent with tetrameric organization (Fig. 4B and Table 1). In contrast, bulk nucleosomes stained with anti-H3 yield both single and double labels (Table 1 and Fig. S5).
Table 1.
CENP-A nucleosomes have one copy of CENP-A and H2B
| Chromatin | Antibody | Nucleosomes observed, N | Single 5-nm bead | Two 5-nm beads | Nm between beads* |
| CENP-A | CENP-A | 576 | 576 | 0 | > 12 |
| CENP-A | H2B | 588 | 588 | 0 | > 12 |
| Bulk | H3 | 632 | 589 | 43 | < 9 |
| Bulk† | H2B | 114† | 109 | 5 | < 9 |
*Center-to-center distances were plotted for gold beads.
†Efficiency of detection of H2B’s C-terminal epitope was 30-fold less for bulk nucleosomes compared to CENP-A nucleosomes.
Even though H2A and H2B copurify with CENP-A from long, medium, and short chromatin (Fig. 1 B and C, and Fig. S1A), one intriguing possibility is that CENP-A nucleosomes have weaker associations with H2A/H2B (30). Thus, the tetrameric nucleosomes measured in AFM experiments above could reflect CENP-A2/H42 instead of CENP-A/H2AB/H4 tetramers. In order to distinguish between these two possibilities, we tested if H2B was present within individual CENP-A nucleosomes. We chose an antibody that detects the C terminus of H2B, reasoning that this epitope is exposed in heterotypic tetramers, whereas it is relatively inaccessible in octamers, and absent in CENP-A2/H42 tetramers (34). We first tested bulk chromatin with anti-H2B, detecting 30-fold less H2B staining in bulk nucleosomes compared to CENP-A nucleosomes (Table 1), consistent with H2B C termini being buried within the bulk octameric core (34). In five cases, dual H2B labels were observed for bulk nucleosomes within 10 nm of each other, consistent with octameric configuration (Fig. 4D and Table 1). However, in 588 CENP-A nucleosomes readily detected by H2B labeling, only a single epitope is present in each CENP-A nucleosome (Fig. 4C), with no examples of dual labels. These data indicate that the C terminus of H2B is present and easily accessed in CENP-A nucleosomes (Table 1). Therefore, immuno-EM data indicate that tetramer height CENP-A nucleosomes are present in stable heterotypic nucleosomal arrays containing DNA, single copies of CENP-A, H2A, H2B, and H4, and that the C-terminal interface of the CENP-A nucleosome is accessible.
Tetrameric Nucleosomes Can Be Directly Discerned in Human Chromatin.
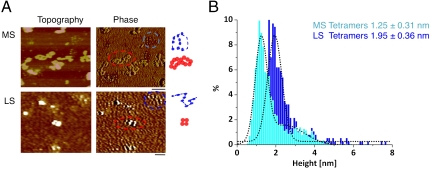
Our data suggested a distinct nucleosomal population possessing tetramer features exists in human chromatin. We reasoned we might be able to discriminate such domains directly in chromatin by scanning nuclear extracts for three properties: open chromatin structure, shorter heights, and differential surface properties. In this experiment, chromatin fibers are very long and generally appear compacted in physiological salt (38). Indeed, from MS, the vast majority of bulk chromatin forms highly compacted fibers (Fig. 5A, MS, red circles, graphic to the right). In cases where nucleosomes can be seen as well-separated beads, these heights are in correspondence to octameric heights and volumes (Fig. S6 B and C). However, a distinct subset of chromatin resists compaction and has smaller nucleosomes (Fig. 5A, MS, blue circles, graphic to the right). These nucleosomes tend to have a darker phase image than bulk chromatin, which is indicative of differential surface properties (Fig. 5A, phase image, blue vs. red circle). Similarly, in LS, a small population of such diminutive nucleosomes can be easily identified amongst octameric nucleosomes (Fig. 5A, LS, blue vs. red circle).
Fig. 5.
Tetrameric nucleosomes can be directly discriminated in human chromatin. (A) Topography (height) and phase image of MS and LS HeLa bulk chromatin, with red dotted line circling an example of highly folded bulk chromatin with light phase and taller nucleosomes, and blue dotted line circling an example of extended chromatin with smaller nucleosomes and darker phase (B) Height and (C) volume measurements of LS and MS nucleosomes such as those highlighted in blue above yields values consistent with tetramers.
The phase and folding signatures observed are a qualitative measure of differential material properties. To obtain a quantitative measure, we used topography to measure the heights of the differential nucleosomes. We measured 345 such smaller nucleosomes from LS, whose mean heights and volumes are 1.95 nm and 100 nm3, respectively, and 6,980 such nucleosomes from MS, whose mean heights and volumes are 1.25 nm and 97 nm3, respectively (Fig. 5B and Fig. S6A, light blue and dark blue), in contrast to the vast majority of bulk octamers with classical dimensions (Fig. S6 B and C). These tetrameric dimensions, folding motifs, and the close packing of tetrameric nucleosomes are in excellent accordance with our observations of LS and MS CENP-A chromatin above (Table S1), indicating that tetrameric organization is the signature of an exclusive class of nucleosomes.
Discussion
Native IP experiments show that purified CENP-A nucleosomes contain equimolar amounts of H2A, H2B, and H4 (Fig. 1 B and C, and Fig. S1A) and are enriched in centromeric α-satellite DNA (Fig. 1D) regardless of ionic conditions. Transiently overexpressed CENP-A does not incorporate into chromatin, but can stably associate with centromeres (Fig. S1 B and C). Inner kinetochore proteins CENP-B and CENP-C copurify with native CENP-A chromatin preparations, whereas histone H3 is depleted (Fig. 1B, WB). Single molecule imaging shows that CENP-A chromatin forms beads-on-a-string structures, which are compacted in LS, and resist condensation in MS, relative to equivalent length BC arrays (Fig. 2 A–C). AFM height measurements show that the majority of LS and MS CENP-A IP nucleosomes are one-half the dimensions of canonical octameric nucleosomes (Fig. 3 A–D). Such tetramer-height nucleosomes predominate across three different experimental conditions and two human cell lines (Table S1) and can be clearly distinguished in mixtures with bulk octamers (Fig. 3C). Electron microscopy analysis of CENP-A chromatin shows individual nucleosomes connected to each other by linker DNA, with four to five nucleosomes forming an array (Fig. 4A), in which single epitopes of CENP-A are recognized efficiently (Fig. 4B). The C terminus of H2B is relatively inaccessible in bulk chromatin, whereas, in CENP-A nucleosomes, a single C terminus of H2B is readily detected (Fig. 4C and Table 1). These traits are indicative of stable organization, wherein each CENP-A nucleosome contains one copy of CENP-A, H2A, H2B, and H4, allowing access to the C-terminal interface. Nucleosomes with folding motifs and dimensions identical to those of CENP-A nucleosomes can be discriminated from bulk chromatin directly within nuclear spreads (Fig. 5A and Fig. S6 A). Put together, these data are compatible with a heterotypic tetrameric nucleosomal organization. Because CENP-A nucleosomes contain centromere-specific DNA repeats and are associated with CENP-C/CENP-B, we favor the view that they derive from active centromeres (44).
In salt dialysis experiments in vitro, CENH3s reconstitute into octamers robustly (18, 29–31, 36, 39). However, in this report, we detect primarily tetrameric CENP-A species in vivo, suggesting that they originate from biological processes. One simple possibility is that specific chaperones constrain CENH3 deposition into tetramers (6–9, 30, 33). An alternative possibility is that CENH3 tetramers are generated by dynamic forces such as mitotic torque (1), transcription, or emerge as an epigenetic mark through replication (45). Either proposal is consistent with observations that CENP-A octamers are far less stable than H3 octamers in vitro and dissociate when challenged by polyacidic molecules (37). In vivo, noncoding centromeric RNA is transcribed upstream of CENP-A assembly (46–48). Thus, RNA could serve as a polyacidic competitor directly (46), or recruit CENP-C, which interacts with the C terminus of CENP-A (49), potentially blocking octamer formation. Concurrently, whereas CENP-A nucleosomes comigrate with bulk nucleosomes on sucrose gradients, high-resolution mapping of α-satellite chromatin in vivo reveals that the CENP-A footprint spans only 100 bp of DNA, rather than the canonical 150 bp (19, 39). The balance could be driven to favor other CENP-A forms prior to replication, when α-satellite transcription is repressed (47, 48), and CENP-A chromatin is remodeled (50). Because our experiments were performed in asynchronous cells, it remains to be determined if CENH3 tetramers originate from dynamic processes and whether alternate forms exist at other points of the cell cycle (45).
Could tetrameric chromatin confer a functional advantage during mitosis? A recent study has proposed that alternating CENP-A/H3 domains fold into a planar amphipathic arrangement, stabilized by cross-array CENP-C binding (51). This notion is supported by previous reports that CENP-C interacts with both centromeric DNA and the C terminus of CENP-A in vivo (52). Amphipathic sheets can also self-assemble by burying hydrophobic residues between planes. Therefore, a speculative proposal is that the CENP-A chromatin fiber folds into a planar motif to avoid exposing hydrophobic C-terminal interfaces, whereupon CENP-C binds exposed C termini across planes, yielding an accordion-like tensility to the centromere. This proposal is consistent with superresolution live microscopy and biochemical data indicating that the CENP-A/CENP-C chromatin fiber is highly compliant (53, 54), and provides a mechanistic basis for how spring-like behavior can be generated. Future experiments will test whether unconventional centromeric nucleosomes can provide a mechanical counterpoint to mitotic torque (35). However, insofar CENH3s are evolutionarily divergent in their N-, loop-1, loop-2, and C- terminal domains (15), it is remarkable that such diversity could result in similar organization and folding properties, suggesting that centromere function is universally rooted in a conserved chromatin fiber.
Materials and Methods
Detailed methods regarding chromatin preparation, immunopurification, Tag-CENP-A expression and purification, PCR, antibodies used for ChIP, and Western blot analysis are described in SI Materials and Methods
AFM Image Collection.
Sample preparation and imaging methods were as stated (SI Materials and Methods). MS and LS data were obtained from 10 experimental repetitions. Bulk chromatin and CENP-A-IP chromatin were imaged in parallel, two technical replicates were performed for each experimental pair, and all the particles in each image were collected. Approximately 4,000 CENP-A-IP and 4,900 bulk nucleosomes were measured (see Table S1 for detailed analysis).
Array Folding Measurements.
Chromatin arrays were defined as > 5 nucleosomes attached by visible linker DNA. Typical CENP-A arrays range from 5–25 nucleosomes, but BC arrays were longer. We manually scored CENP-A and BC arrays (each containing 5–15 nucleosomes, Fig. S2), qualitatively assessing the ratio of “open” beads-on-a-string vs. “closed” compacted arrays (38).
Height and Volume Measurements.
Standard height and volume measurements were performed (29, 42) using National Institutes of Health Image J using automated counting (see SI Text for detailed description). The volume of an octameric nucleosome is ∼400 nm3, half of which is the water volume in the histone core (SI Text), thereby loss of hydration results in ∼200–300 nm3 (29, 41, 42). Experimentally observed values for LS BC (V = 260 nm3, H = 2.8–4 nm) are in good accordance with those predicted (34) and observed previously for octameric nucleosomes (41, 42). Tetramer nucleosome heights and volumes are predicted to be one-half of LS octamer values, specifically, V = 100–150 nm3 and H = 1.4–2 nm.
Supplementary Material
Acknowledgments.
We thank Carl Wu for use of the Typhoon scanner, Gary Karpen for inspiring us to test LS conditions, Malte Bussiek and Martin Westermann for advice on AFM and immuno-EM experiments, respectively. We thank Gordon Hager, Tom Misteli, Steve Henikoff, Sam John, Minh Bui, and Delphine Quènet for critical comments on the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009563107/-/DCSupplemental.
References
- 1.Bloom K, Joglekar A. Towards building a chromosome segregation machine. Nature. 2010;463:446–456. doi: 10.1038/nature08912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 3.Amor DJ, et al. Human centromere repositioning “in progress”. Proc Natl Acad Sci USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker RE, Rogers K. Genetic and genomic analysis of the AT-rich centromere DNA element II of Saccharomyces cerevisiae. Genetics. 2005;171:1463–1475. doi: 10.1534/genetics.105.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 6.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci USA. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi T, et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci USA. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hori T, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 23.Santaguida S, Musacchio A. The life and miracle of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloom KS, et al. Chromatin conformation of yeast centromeres. J Cell Biol. 1984;99:1559–1568. doi: 10.1083/jcb.99.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polizzi C, Clarke L. The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J Cell Biol. 1991;112:191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, et al. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Hooser AA, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114(Pt 19):3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 28.Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–4139. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:1798–1809. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 31.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visnapuu ML, Greene EC. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat Struct Mol Biol. 2009;16:1056–1062. doi: 10.1038/nsmb.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 35.Bouck DC, Joglekar AP, Bloom KS. Design features of a mitotic spindle: balancing tension and compression at a single microtubule kinetochore interface in budding yeast. Annu Rev Genet. 2008;42:335–359. doi: 10.1146/annurev.genet.42.110807.091620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 37.Conde e Silva N, et al. CENP-A-containing nucleosomes: Easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 38.Woodcock CL, Horowitz RA. Electron microscopy of chromatin. Methods. 1997;12:84–95. doi: 10.1006/meth.1997.0450. [DOI] [PubMed] [Google Scholar]
- 39.Yoda K, Morishita S, Hashimoto K. Histone variant CENP-A purification, nucleosome reconstitution. Methods Enzymol. 2004;375:253–269. doi: 10.1016/s0076-6879(03)75017-4. [DOI] [PubMed] [Google Scholar]
- 40.Willard HF, Waye JS. Chromosome-specific subsets of human alpha satellite DNA: Analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol. 1987;25:207–214. doi: 10.1007/BF02100014. [DOI] [PubMed] [Google Scholar]
- 41.Tomschik M, Karymov MA, Zlatanova J, Leuba SH. The archaeal histone-fold protein HMf organizes DNA into bona fide chromatin fibers. Structure. 2001;9:1201–1211. doi: 10.1016/s0969-2126(01)00682-7. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Dalal Y, Henikoff S, Lindsay S. Single-epitope recognition imaging of native chromatin. Epigenet Chromatin. 2008;1 doi: 10.1186/1756-8935-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frado LL, Mura CV, Stollar BD, Woodcock CL. Mapping of histone H5 sites on nucleosomes using immunoelectron microscopy. J Biol Chem. 1983;258:11984–11990. [PubMed] [Google Scholar]
- 44.Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalal Y, Bui M. Down the rabbit hole of centromere assembly and dynamics. Curr Opin Cell Biol. 2010;22:292–402. doi: 10.1016/j.ceb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong LH, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 48.Lu J, Gilbert DM. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J Cell Biol. 2007;179:411–421. doi: 10.1083/jcb.200706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du Y, Topp CN, Dawe KR. DNA bindingof centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ribeiro SA, et al. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trazzi S, et al. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS One. 2009;4:1–13. doi: 10.1371/journal.pone.0005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.