Abstract
Specific small deletions within the rpoC gene encoding the β′-subunit of RNA polymerase (RNAP) are found repeatedly after adaptation of Escherichia coli K-12 MG1655 to growth in minimal media. Here we present a multiscale analysis of these mutations. At the physiological level, the mutants grow 60% faster than the parent strain and convert the carbon source 15–35% more efficiently to biomass, but grow about 30% slower than the parent strain in rich medium. At the molecular level, the kinetic parameters of the mutated RNAP were found to be altered, resulting in a 4- to 30-fold decrease in open complex longevity at an rRNA promoter and a ∼10-fold decrease in transcriptional pausing, with consequent increase in transcript elongation rate. At a genome-scale, systems biology level, gene expression changes between the parent strain and adapted RNAP mutants reveal large-scale systematic transcriptional changes that influence specific cellular processes, including strong down-regulation of motility, acid resistance, fimbria, and curlin genes. RNAP genome-binding maps reveal redistribution of RNAP that may facilitate relief of a metabolic bottleneck to growth. These findings suggest that reprogramming the kinetic parameters of RNAP through specific mutations allows regulatory adaptation for optimal growth in new environments.
Keywords: kinetics, stringent response, transcription
Mutations to the primary RNAP genes in bacteria (rpoABCZ encoding the core α, β, β′, and ω subunits and rpoD encoding the housekeeping σ70 subunit) exhibit a wide range of pleiotropic effects on bacterial phenotypes (1–5), and it has been said that mutations to RNAP genes can satisfy virtually any selection (6). We recently described the discovery of mutations in rpoB and rpoC following adaptive evolution of Escherichia coli K-12 MG1655 in glycerol M9 minimal medium (GMM) over a period of several weeks (7). Competition experiments using mutants harboring individual mutations acquired during adaptive evolution in glycerol showed that changes to the RNAP were the most dominant driving force of adaptation to this condition (8).
Some RNAP mutants exhibit changes in initiation kinetics that resemble the effects of elevated (p)ppGpp on RNAP that occur during the stringent response (3). Further, (p)ppGpp is required for growth of E. coli in minimal media (9), which raised the possibility that the adaptive changes to RNAP might permanently change the enzyme in ways similar to that achieved transiently by (p)ppGpp binding. Thus, we suggested that these mutations could be adaptive through effects on transcription that might be related to those exerted by (p)ppGpp (7).
Regulation of RNAP by (p)ppGpp, reviewed in refs. 10 and 11, is modulated by the protein DksA, which binds in the RNAP secondary channel. Binding of DksA or (p)ppGpp to the RNAP alone or together deceases the kinetic stability (i.e., lifetime or longevity) of open complexes and causes decreased transcription from promoters that form short-lived open complexes (e.g., promoters for ribosomal RNA synthesis) and increased transcription from promoters that form long-lived open complexes but bind RNAP weakly (e.g., promoters for some amino acid biosynthetic operons) (11). The reduction in open complex lifetime caused by (p)ppGpp and DksA are thought to redistribute RNAP away from rRNA transcription units to other genes, such as those required for amino acid prototrophy. (p)ppGpp also is thought to increase transcriptional pausing and decrease transcript elongation rate (12, 13). Interestingly, the (p)ppGpp regulon has previously been observed to be affected during adaptive evolution of E. coli in minimal medium (14). Therefore, we sought to understand whether the mutations to the RNAP genes were adaptive through effects related to those previously reported for so-called “stringent” RNAPs (3), or by some other means.
Here, we describe the effects of three adaptive small deletions in RNAP at multiple levels: (i) at the physiological level through changes in growth performance, (ii) at the molecular level through changes in RNAP kinetics as measured in vitro, and (iii) at a systemic level through redistribution of the polymerase and changes in gene expression.
Results
Mutations to RNAP Genes Occurring in MG1655 During Adaptation to Glycerol Minimal Medium.
Following the discovery of mutations in rpoB or rpoC in three of five fully resequenced strains of E. coli K-12 MG1655 adaptively evolved in GMM (7), an additional 45 adaptive evolution experiments of 25 d were carried out under the same condition. Targeted sequencing of selected portions of rpoB and rpoC was performed to determine the frequency and locations of mutations in RNAP genes (15). Mutations were found in the resequenced regions of rpoB or rpoC in 37 of 45 day-25 strains. The most frequent mutation was a previously unobserved 9-bp deletion rpoC(Δ3611..3619) encoding β′Δ(V1204-R1206) (eBOP43) that occurred in 31 of 45 endpoints and that is located in the so-called jaw domain of RNAP (16). Two endpoint strains had single-nucleotide changes in rpoB (encoding βH526Y or βE641K), and the remaining four strains had other small, in-frame rpoC deletions that all occurred in the so-called sequence insertion 3 (SI3) of E. coli RNAP (17). β′Δ(T1045-L1053) (del27) was found in two day-25 strains; β′Δ(M1040-R1048)::I (eBOP42) and β′Δ(G1043-N1051) were each found once. The remarkable frequency of the rpoC(Δ3611..3619) mutation may be attributed in part to a 7-bp sequence that repeats with a 9-bp interval at the location of the deletion. It has been previously observed that deletions between direct sequence repeats in other parts of the rpoC gene repeatedly arise, for example, in selection for restoration of prototrophy in ΔrelAΔspoT mutants (6).
The mutant RNAP sequences of eBOP43, del27, and eBOP42 were cloned into RNAP overexpression plasmids from which purified mutant RNAPs were prepared to study the kinetic properties of the mutant RNAPs in vitro. The mutant sequences were also recombined back into the wild-type MG1655 genome (Table 1) to study the biological effects of the mutations in isolation in vivo.
Table 1.
Strains used in this study
Physiology: Mutations to rpoC Increase Growth Rate in Minimal Media.
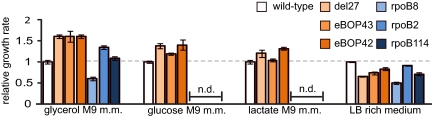
Growth rates of wild-type MG1655 and the three studied RNAP mutants were determined in M9 minimal media with glycerol, glucose, or lactate carbon source, as well as in LB rich medium (Fig. 1 and Table S1). The three RNAP mutants were found to grow 60% faster than wild type in GMM. Also, growth in both glucose and lactate M9 minimal media was significantly faster for all three mutants, indicating that the adaptive effect is not specific to only the glycerol carbon source, but extends to growth in minimal media with other carbon sources. In contrast, the growth rates of all three RNAP mutants decreased 17–34% relative to the wild-type strain in LB rich medium. These findings show that the RNAP mutants reflect a general adaptation to minimal media, with the tradeoff of slower growth in rich media.
Fig. 1.
Growth rate measurements of RNAP mutants in minimal and rich media. The growth rates of three RNAP mutants derived from E. coli adapted to growth in glycerol M9 minimal medium (del27, eBOP43, and eBOP42) and three RNAP mutants derived from rifampin-resistant E. coli were determined in triplicate in M9 minimal media with various carbon sources and in LB-rich medium. Average growth rates of mutants relative to the wild type are shown and error bars represent the SD of measurements. The glycerol-adaptive mutants grew significantly faster than wild-type in minimal media with all three tested carbon sources, but grew slower than wild-type in LB rich medium. Numerical results (absolute values) are listed in Table S1.
We next investigated possible links between the adaptive RNAP mutants, regulation by (p)ppGpp, and an improved growth rate in GMM. A ppGpp-null derivative of the del27 mutant was constructed by introducing deletions of the relA and spoT genes. The del27ΔrelAΔspoT strain was unable to grow without amino acid supplementation, showing that del27 is not phenotypically equivalent to so-called stringent RNAP mutants that allow growth of relA−spoT− strains in minimal medium (6). Furthermore, a previously characterized rifampin-resistant, stringent RNAP mutant [rpoB114(S531F)] (1) exhibited an unchanged growth rate in GMM vs. an isogenic MG1655 strain. In contrast, rifampin-resistant mutants not known to allow growth of relA−spoT− strains but that affect transcript elongation rate in vitro either decreased (rpoB8, which decreases elongation rate) or increased (rpoB2, which increases elongation rate) the growth rate in GMM (Fig. 1 and Table S1). Therefore, the improved growth rate of del27 RNAP mutant in GMM results from changes in properties of RNAP that differ from the classically defined stringent RNAP mutants. Conversely, nonstringent mutants (rpoB2 and rpoB8) affected the growth rate in GMM proportional to previously reported effects of these mutants on transcript elongation rate (18, 19).
The increased growth rates of del27, eBOP43, and eBOP42 mutants in GMM may be explained either by increased overall metabolic flux or by an increase in the fraction of carbon source being converted to biomass (biomass yield). Assuming a steady state, the uptake rate of glycerol (GUR) is an indicator of the overall rate of metabolism. We found that both biomass yield and GUR increased in all three rpoC mutants (by 14–35% and by 19–40%, respectively; Table S1).
We made two observations that could explain the increased biomass yield in the mutants. It was previously shown that a MG1655-derived strain with an flhD mutation that causes the strain to become nonmotile has a significant growth advantage in glycerol M9 minimal medium relative to the parent strain (20). Loss of motility in MG1655 adapted to growth in GMM has previously been observed (21). Interestingly, we found complete loss of motility in the eBOP43 and eBOP42 mutants and slightly decreased motility in the del27 mutant (Fig. S1). Second, when uptake of a carbon source exceeds the capability of E. coli to produce biomass, excess carbon may be excreted as acetate (22). We found that acetate appeared at a slower rate in del27 and eBOP42 growth media, and no acetate was detected in the eBOP43 growth media. Therefore, the adaptive RNAP mutations result in both a higher metabolic rate and more efficient use of the carbon source.
Molecular Biology: Adaptive RNAP Mutants Exhibit Altered Kinetics in Vitro.
Alteration of transcription kinetics through an RNAP mutation may have extensive effects on the cell state, potentially resulting in faster growth. The RNAP substitutions and deletions that we identified in E. coli adapted to GMM were located mostly in the jaw and SI3 domains of the β′ subunit of RNAP (Fig. S2). In vitro assays have previously shown that deletion of the SI3 domain or a large portion of the jaw domain destabilizes open complexes during transcription initiation and decreases pausing during elongation (16, 17). Thus, the location of the mutations suggested that they could affect the kinetics of transcription by RNAP.
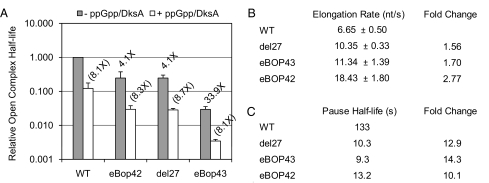
To investigate possible effects on RNAP kinetics, we used established methods to measure open complex longevity at the rrnB P1 (rRNA) promoter (23), elongation rate (16) during transcription of rpoB, and pause half-life (24) at a well-established pause site. The mutant RNAPs exhibited a decrease in open complex longevity at the rRNA promoter (Fig. 2A and Fig. S3A). Relative to wild-type RNAP, the deletions in the SI3 domain decreased the open complex half-life by a factor of 4, and the deletion in the jaw domain decreased longevity by a factor of greater than 30. Despite the decrease in inherent open complex longevity, the mutant RNAPs maintained the ability to respond to the effects of the global regulators ppGpp and DksA, similar to wild type (Fig. 2A). Thus, relative to wild type, the mutant RNAPs would be expected to decrease initiation in vivo at promoters like rrnB P1 that are sensitive to open complex longevity regardless of the physiological activity of (p)ppGpp and DksA (i.e., at both low and high (p)ppGpp and DksA levels or activities).
Fig. 2.
The small deletions in rpoC alter RNAP kinetics in vitro. (A) Comparison of open complex longevity at the rrnB P1 promoter in the absence (gray bars) and presence (white bars) of ppGpp/DksA. All open complex half-life measurements were normalized to wild type. The text above the gray bars is the relative decrease in half-life for each mutant compared with wild-type RNAP. The text in parentheses above the white bars is the relative decrease in open complex half-life in response to the presence of ppGpp/DksA for each RNAP. (B) Comparison of in vitro transcription elongation rate. Elongation was assayed by synchronized transcription on a linear T7 A1 promoter template containing a fragment of the rpoB gene. (C) Comparison of his pause half-lives. Pause half-lives were assayed using reconstituted transcription elongation complexes paused at the his pause site. Pause half-lives were measured at least twice, with the individual values within 10% error of each other. The text above each column denotes the fold decrease relative to wild type. Detailed graphs showing measurements of open complex lifetimes and elongation rates can be found in Fig. S3.
To assess how the small deletions affect transcript elongation, we first tested the effects of the deletions on the overall rate of transcript elongation using a synchronized transcription assay on a linear template containing a fragment of the rpoB gene. The mutant RNAPs exhibited a modest (1.6- to 2.8-fold) increase in elongation rate relative to wild type (Fig. 2B and Fig. S3B). The increase in elongation rate was accompanied by decreases in prominent pauses found in the rpoB gene (Fig. S4). The increase in apparent elongation rate could be caused by an increase in the rate of nucleotide addition, a decrease in pausing over the length of the transcript, or both (25).
We next investigated the observed decrease in pausing by assaying the kinetics of EC escape from a well-characterized pause site in the his operon leader region. Each mutant RNAP decreased the his pause half-life by greater than a factor of 10 (Fig. 2C). Thus, the adaptive RNAP mutants exhibited a drastic decrease in pausing at a specific model pause site as well as generally decreased pausing in the rpoB transcription unit. Although we have not ascertained whether all possible classes of pauses are affected, our findings strongly suggest that the adaptive RNAP mutants exhibit an increased elongation rate resulting at least in part from significant decreases in transcriptional pausing. Changes to pausing and elongation rate may have compound consequences, including effects on RNA folding, transcriptional–translational coupling, transcription termination, recruitment of regulatory factors, and the general rate of transcription (26).
Kinetic changes in the mutants were similar to the previously studied SI3 and jaw deletion mutants (16, 17). The consistency of changes to the kinetics of the mutant RNAPs suggests that these kinetic changes may be important to the adaptive properties of the mutants.
Systems Biology: Small Deletions in RNAP Exert Functional Changes on Gene Expression.
If the decreased open complex stability observed in vitro is significant in vivo, then the mutations to RNAP are expected to cause a redistribution of the polymerase from transcription of rRNA transcription units (TUs) to other regions of the genome (11). Redistribution would be detectable by observing the dynamic RNAP binding map in an RNAP mutant vs. MG1655 and by global measurements of gene expression.
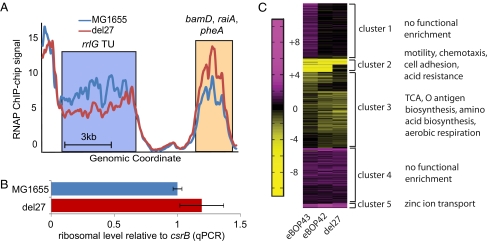
A dynamic binding map of RNAP to the genome was determined for MG1655 and the del27 mutant by chromatin immunoprecipitation using mouse antibody specific to the RNAP β-subunit followed by hybridization of immunoprecipitated DNA to a whole-genome tiling array (ChIP-chip). Signal from probes covering the seven ribosomal TUs, normalized by total positive signal across the genome, is on average 15% lower in the del27 mutant vs. MG1655 (P = 0.0003; Fig. 3A). The finding is indicative of a redistribution of the del27 RNAP from rRNA TUs to other genes.
Fig. 3.
RNAP redistribution and global expression changes due to mutations. (A) Dynamic binding map of RNAP determined by ChIP-chip in the region of the TU containing rrlG (coding 23S rRNA). On average, the binding signal of mutant RNAP across the seven E. coli ribosomal TUs decreased significantly versus wild-type signal (P = 0.0003). Binding maps provided evidence of increased binding of del27 RNAP to some TUs coding for amino acid biosynthetic genes, such as pheA. However, the binding map results do not necessarily correlate with transcriptomic measurements. (B) qPCR using primers for rRNA and csrB revealed no decrease in 23S rRNA content in the del27 mutant. (C) k-means clustering (k = 5) of log2 ratios of mutant expression relative to wild-type expression is shown. Functional enrichment was determined by using the hypergeometric test (false discovery rate: 0.05) for Gene Ontology categories.
The decreased binding of RNAP to ribosomal TUs is consistent with in vitro measurements of del27 kinetics, but is surprising given that this mutant grows 60% faster than MG1655. Previous work has estimated, for example, that the number of ribosomes per cell is approximately doubled when the growth rate of E. coli is increased 66% from 0.6 to 1.0 doublings per hour (27). Therefore, to gauge rRNA levels we used comparative PCR with primers targeting 23S rRNA and csrB (a highly expressed gene with constant expression in the mutant strains and MG1655; Fig. 3B). We observed no decrease, and if anything an increase in 23S rRNA in the del27 mutant. This is consistent with the increased elongation rate of the mutants at least partially compensating for the decreased binding of RNAP to ribosomal TUs.
Gene expression of the RNAP mutants in the midlog phase of growth in GMM was compared with that of wild-type using Affymetrix E. coli 2.0 GeneChips. The mutations were accompanied by up-regulation of 491, 728, and 439 transcripts and down-regulation of 492, 432, and 430 transcripts in the del27, eBOP43, and eBOP42 mutants, respectively. Of the 486 genes differentially expressed in all three datasets, 456 changes occur in a consistent direction for all three mutants. Consistent with the hypothesized redistribution resulting from transcription kinetic changes, eBOP43, which was seen in vitro to have a greater disruption of rrnB P1 promoter open complex longevity versus the other mutants, has more than 50% more up-regulated genes than the other mutants.
To find changes common between the mutants, we performed k-means (k = 5) clustering of the log2 ratios of mutant expression relative to wild type averaged over the three replicates (Fig. 3C and Table S2). Only genes that showed twofold or greater differential expression in at least one mutant were clustered. Although lack of knowledge about the kinetic properties of most promoters precludes linking most specific expression changes to the hypothesized redistribution of RNAP, it is of interest to speculate how the RNAP mutations increase fitness based on the functional significance of genes in these clusters. Three clusters exhibited likely functional significance, due to the presence of strongly enriched functional properties. Cluster 5 contained the most highly up-regulated genes in the mutants and was enriched for genes involved in transport of zinc ions. The moderately down-regulated genes in cluster 3 were enriched for genes involved in several metabolic functions, including TCA, aerobic respiration, and amino acid biosynthesis. Cluster 2, representing strongly down-regulated genes, was enriched for motility, chemotaxis, cell adhesion (fimbria and curli), and acid resistance genes. Down-regulation of these genes should provide a selective advantage in the growth environment, because their functions are costly in terms of energy but confer little or no benefit (20).
The cell adhesion and acid resistance genes, associated with the σS sigma factor, are strongly down-regulated in all three mutants, whereas motility and chemotaxis genes, associated with the σ28 sigma factor, are down-regulated in only two of three mutants, consistent with the results of motility assays. Down-regulation of the σ28 regulon is associated with a ∼50-fold reduction of fliA transcript encoding σ28, in the eBOP43 and eBOP42 mutants (Fig. S1). Decreased open complex longevity reduces transcription of flhDC (28), a positive regulator of genes required for flagella synthesis, providing a plausible link between changes to transcription kinetics and decreased motility in the mutants. However, the del27 RNAP has similar kinetic properties to the other two mutants, but its σ28 regulon is not strongly down-regulated, indicating that down-regulation of the σ28 regulon is not determined solely by the intrinsic kinetic changes in the mutant RNAPs.
Discussion
Mutations to genes encoding transcription regulatory hubs have been found previously in long-term adaptive evolutions of E. coli (29); the mutations to RNAP genes arising in E. coli during adaption to GMM in our experiment are additional examples of an adaptive strategy that targets a regulatory hub, in this case RNAP itself. We found (i) mutations to rpoB and rpoC genes corresponding to the RNAP jaw and SI3 regions were consistently selected for during adaptive evolution of MG1655 in GMM; (ii) these mutations are adaptive for growth of MG1655 in minimal media, at the cost of slower growth in rich media; (iii) these mutations reprogrammed the kinetic parameters of RNAP by decreasing the longevity of open complexes at a promoter, by decreasing pausing, and by increasing elongation rate; (iv) gene expression profiling and the genome-wide RNAP dynamic binding map are consistent with a redistribution of the polymerase from stringently controlled TUs to other parts of the genome; and (v) the mutations caused profound gene expression changes, including strong down-regulation of acid resistance, curlin, and fimbria genes, and sometimes motility genes.
The stringent response effector ppGpp is necessary for growth in minimal media without amino acids, and mutations in genes encoding RNAP can partially mimic the effect of (p)ppGpp (so-called stringent mutants). Thus, we initially hypothesized that the adaptive mutations might be equivalent to such stringent mutants. However, we observed that a ppGpp-null derivative of del27 was unable to grow in GMM without adding amino acids, establishing that the del27 RNAP mutation differs from stringent mutants. Additionally, a previously studied Rifr stringent mutation [rpoB114(S531F)] conferred no growth advantage in GMM. Finally, neither the kinetic changes nor the expression changes (Fig. 3C and Table S3) were fully consistent with the known effects of ppGpp, which changes gene expression in patterns different from those we observe for the adaptive mutations (30, 31) and which is generally observed to increase pausing and decrease elongation rate (12, 13) (opposite to that observed for the adaptive mutant RNAPs). Therefore we have ruled out the possibility that the mutations are adaptive simply because they mimic the effects of ppGpp.
We propose that the similar changes in open complex longevity and transcriptional pausing observed for all three adaptive RNAP mutants reprogram RNAP for better growth in minimal media. Decreased open complex longevity is expected to redistribute RNAP away from promoters with short-lived open complexes (e.g., rRNA promoters) and increase the use of promoters that are rate limited for RNAP engagement (e.g., amino acid biosynthetic operon promoters). This could explain the observed drastic changes to phenotype and to gene expression patterns. Decreased growth rate of the mutants in rich media and decreased binding of RNAP to ribosomal transcription units observed in a genome-wide dynamic binding map of RNAP support this interpretation. However, we emphasize that this redistribution of RNAP is paradoxical, given that the mutations increase growth rate, and that the rate of rRNA synthesis and consequent production of ribosomes exhibits a near-universal positive correlation with growth rate (27). The decrease in transcriptional pausing and increase in elongation rate, properties that distinguish the adaptive RNAP mutations from stringent mutations, may help compensate for the redistribution of RNAP away from ribosomal TUs and explain why no decrease in rRNA level was observed despite the reduced open complex longevity that classically decreases rrn transcription. Although antitermination proteins loaded on RNAP just downstream of rrn promoters increase the rate of transcript elongation in rrn genes to ∼80 nt/s in minimal glucose media (32), these antitermination proteins can be gradually lost as evidenced by existence of a second “boxA” loading site midway through rrn transcription units (33). Even a few pause-susceptible RNAPs are proposed to slow rRNA synthesis significantly by “roadblocking” other RNAPs (34). The adaptive mutations may reduce pausing by these nonantiterminated RNAPs and thereby increase rRNA synthesis by reducing the roadblocking effect on overall rrn gene transcription. By reprogramming RNAP with specific kinetic properties, the adaptive mutations may increase the amount of RNAP available for transcription of key genes that correspond to bottlenecks for biomass production in minimal medium. Identifying such genes will be experimentally challenging, but could be done by using genome-scale fitness profiling (35).
Although speculative, we suggest two possible bottlenecks. Despite the down-regulation of several amino acid biosynthetic genes, cysE, cysM, ilvD, and ilvM genes are up-regulated in the three studied adaptive mutants; this could indicate that cysteine or isoleucine synthesis is limiting for the growth rate in GMM. Pyrimidine biosynthesis may represent another important bottleneck to be overcome in GMM. Interestingly, the rpoB2 mutant found to have a growth advantage in GMM was previously found to relieve the pyrimidine defect of MG1655 by decreasing attenuation within the rph-pyrE operon (36). However, expression of the pyrE gene was not observed to change in the three glycerol-adaptive mutants, so it is not immediately clear that these RNAP mutations are adaptive through effects on the pyrimidine biosynthetic pathway. The possible consequences of a redistribution of RNAP resulting in adaptation are intriguing, and require further investigation.
These reproducible RNAP gene mutations are striking because they exert complex effects on the cellular phenotype, resulting in large fitness gains in GMM. Our recent work in which we resequenced wild-type E. coli adapted to lactate minimal medium (37) and a phosphoglucoisomerase mutant adapted to glucose minimal medium has continued to show the importance of mutations to the RNAP genes in adaptive evolution of E. coli, with mutations observed in rpoA, rpoB, rpoC, and rpoS. Future efforts will continue to build on understanding of the ability of mutations to the RNAP genes to adaptively rewire the transcription network of bacteria.
Materials and Methods
Adaptive Evolutions.
Forty-five parallel adaptive evolutions of E. coli K-12 MG1655 were performed over 25 d in 2 g/L GMM at 30 °C as previously described (7).
Targeted Resequencing.
Primers were designed to amplify gene positions 904–2,085 and 2,680–3,974 of rpoB and rpoC, respectively. DNA was isolated from day-25 adaptive evolution endpoints, and selected regions were amplified using ExTaq DNA polymerase (Takara). Sanger sequencing was performed by EtonBio (San Diego).
Cloning of rpoC Mutants.
The rpoC mutant alleles were cloned into the pVS7 vector. See SI Materials and Methods for details.
Kinetic Assays.
The in vitro elongation rate, open complex half-life, and pause half-life were determined as described previously (16, 23, 24, 38, 39). Details are included in SI Materials and Methods.
Site-Directed Mutagenesis and Knockouts.
A scarless form of site-directed mutagenesis, “gene gorging” (7, 40), was used to introduce the rpoC mutations from three glycerol M9 adaptive evolution endpoints into a wild-type E. coli K-12 MG1655 background. Knockouts of the relA and spoT genes were made and confirmed using the method of Datsenko and Wanner (41).
Gene Expression.
Twelve microarrays were analyzed in total. Samples from triplicate flasks of E. coli K-12 MG1655 and the three mutant knockins were collected by diluting into a 2× volume of RNA Protect when the cells reached midlog phase of growth (OD A600 ≈ 0.3) in GMM at 30 °C. RNA was extracted, synthesized into cDNA, fragmented using DNase I, labeled with biotin using terminal transferase and DNA Labeling Reagent (Affymetrix), and hybridized to Affymetrix E. coli 2.0 GeneChips using Affymetrix’s suggested protocol. Raw data have been deposited in NCBI's Gene Expression Omnibus database (42) and are accessible through GEO Series accession no. GSE15500. Microarrays were normalized using the method of Wu et al. (43). For each RNAP mutant, all active transcripts were determined by comparing against a set of 20 negative controls on the arrays (two-sample t test; false discovery rate: 0.05). Genes that were significantly expressed in neither the mutant nor wild-type strain arrays were removed from the dataset and not considered in further analyses.
Computation of Differential Expression.
For each mutant, the grand mean was subtracted from all genes within the union of active transcripts between the mutant and the wild-type arrays. Differential expression was then computed with a permuted two-sample t test. The significance cutoff was set at 1.5-fold changes and a false discovery rate of 0.03, determined using the method of Storey et al. (44).
ChIP-chip.
Detailed methods are included in SI Materials and Methods. Briefly, triplicate GMM MG1655 and del27 mutant batch cultures in GMM were harvested during midlog growth phase (OD A600 ≈ 0.3) and cross-linked using formaldehyde. Cell pellets were washed with TBS and treated with lysozyme. The lysate was sonicated and centrifuged. The lysate was incubated with either anti-RpoB mouse antibody or mock (normal) mouse antibody. Immunoprecipitated complexes were purified using Dynabeads Pan Mouse IgG (Invitrogen). After reverse cross-linking, samples were treated with RNaseA (Qiagen) and proteinase K solution (Invitrogen), and then purified with a PCR purification kit. Before shipment to NimbleGen (Reykjavík, Iceland) where sample labeling, hybridization to genomic tiling arrays, and visualization steps were carried out, gene-specific quantitative PCR was carried out for promoter regions of gapA, tpi, fbp, pgi, and dmsA using the isolated DNA samples as template to confirm immunoprecipitation quality.
Data files in .gff format were received from NimbleGen containing biweight mean-normalized log2 ratio of anti-RpoB signal relative to mock signal. To quantify relative binding of RNAP to ribosomal transcription units, the sum of average binding signal across probes representing seven ribosomal transcription units was normalized by the total binding signal across the genome.
qPCR.
RNA was extracted from triplicate cultures of MG1655 and del27 as described for gene expression, except cells were harvested at A600 ≈ 0.10 for additional confidence that the cultures were not near stationary phase. cDNA synthesized from the RNA (∼50 ng/μL) was diluted 10,000-fold. Primers were designed to amplify ≈100-bp segments of 23S rRNA and csrB (Table S4) and were verified by ensuring stable ΔCT of a cDNA sample diluted between 1,000-fold and 100,000-fold. To determine ribosomal content, triplicate technical replicates were used for each of the 12 biological replicates. The qPCR reaction was carried out using Power SYBR Green qPCR Master Mix (Applied Biosystems) in an iCycler thermocycler and optical module (Bio-Rad) using the following: 1 cycle: 95 °C, 10 min; 40 cycles: 95 °C, 15 s, 52 °C, 15 s, 72 °C, 30 s; 1 cycle: 72 °C, 10 min.
Growth Rates, Substrate Uptake Rates, and Biomass Yields.
Precultures of the strains were inoculated into GMM and at various time points during the growth curve samples were taken to measure biomass and glycerol content. Biomass was determined by measuring the absorbance of the culture at 600 nm. For all strains measured, we have empirically determined a conversion ratio of 0.54 gDW/(L · OD unit) (SI Materials and Methods). Glycerol and acetate concentrations were determined by HPLC with detection at 410 nm. GUR was calculated by measuring the depletion of glycerol over time, taking into account the amount of biomass present. Biomass yield was determined by dividing biomass produced by glycerol that had been depleted from the media.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 GM062791 and R01 GM057089 (to B.Ø.P.) and R37 GM38660 (to R.L.), and National Science Foundation Integrative Graduate Education Research Traineeship (IGERT) Plant Systems Biology Training Grant DGE-0504645 (to N.E.L.).
Footnotes
Conflict of interest statement: B.Ø.P. serves on the scientific advisory board of Genomatica, Inc.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE15500).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911253107/-/DCSupplemental.
References
- 1.Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 2.Jin DJ, Gross CA. Characterization of the pleiotropic phenotypes of rifampin-resistant rpoB mutants of Escherichia coli. J Bacteriol. 1989;171:5229–5231. doi: 10.1128/jb.171.9.5229-5231.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trinh V, Langelier MF, Archambault J, Coulombe B. Structural perspective on mutations affecting the function of multisubunit RNA polymerases. Microbiol Mol Biol Rev. 2006;70:12–36. doi: 10.1128/MMBR.70.1.12-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein-Marcuschamer D, Santos CN, Yu H, Stephanopoulos G. Mutagenesis of the bacterial RNA polymerase alpha subunit for improvement of complex phenotypes. Appl Environ Microbiol. 2009;75:2705–2711. doi: 10.1128/AEM.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy H, Cashel M. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol. 2003;371:596–601. doi: 10.1016/S0076-6879(03)71044-1. [DOI] [PubMed] [Google Scholar]
- 7.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 8.Applebee MK, Herrgård MJ, Palsson BO. Impact of individual mutations on increased fitness in adaptively evolved strains of Escherichia coli. J Bacteriol. 2008;190:5087–5094. doi: 10.1128/JB.01976-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao H, et al. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 10.Magnusson LU, Farewell A, Nyström T. ppGpp: A global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kingston RE, Nierman WC, Chamberlin MJ. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J Biol Chem. 1981;256:2787–2797. [PubMed] [Google Scholar]
- 13.Sørensen MA, Jensen KF, Pedersen S. High concentrations of ppGpp decrease the RNA chain growth rate. Implications for protein synthesis and translational fidelity during amino acid starvation in Escherichia coli. J Mol Biol. 1994;236:441–454. doi: 10.1006/jmbi.1994.1156. [DOI] [PubMed] [Google Scholar]
- 14.Cooper TF, Rozen DE, Lenski RE. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:1072–1077. doi: 10.1073/pnas.0334340100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joyce AR. PhD dissertation. La Jolla, CA: University of California at San Diego; 2007. Modeling and analysis of the E. coli transcriptional regulatory network: An assessment of its properties, plasticity, and role in adaptive evolution. [Google Scholar]
- 16.Ederth J, Artsimovitch I, Isaksson LA, Landick R. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem. 2002;277:37456–37463. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- 17.Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 18.Fisher RF, Yanofsky C. Mutations of the beta subunit of RNA polymerase alter both transcription pausing and transcription termination in the trp operon leader region in vitro. J Biol Chem. 1983;258:8146–8150. [PubMed] [Google Scholar]
- 19.Jin DJ, Gross CA. RpoB8, a rifampicin-resistant termination-proficient RNA polymerase, has an increased Km for purine nucleotides during transcription elongation. J Biol Chem. 1991;266:14478–14485. [PubMed] [Google Scholar]
- 20.Leatham MP, et al. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect Immun. 2005;73:8039–8049. doi: 10.1128/IAI.73.12.8039-8049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fong SS, Joyce AR, Palsson BO. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 2005;15:1365–1372. doi: 10.1101/gr.3832305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.el-Mansi EM, Holms WH. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J Gen Microbiol. 1989;135:2875–2883. doi: 10.1099/00221287-135-11-2875. [DOI] [PubMed] [Google Scholar]
- 23.Paul BJ, et al. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Kyzer S, Ha KS, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J Biol Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- 25.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 26.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 27.Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, editor. Escherichia coli and Salmonella. 2nd Ed. Washington, DC: American Society for Microbiology; 1996. pp. 1553–1569. [Google Scholar]
- 28.Lemke JJ, Durfee T, Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philippe N, Crozat E, Lenski RE, Schneider D. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays. 2007;29:846–860. doi: 10.1002/bies.20629. [DOI] [PubMed] [Google Scholar]
- 30.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traxler MF, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel U, Jensen KF. Effects of the antiterminator BoxA on transcription elongation kinetics and ppGpp inhibition of transcription elongation in Escherichia coli. J Biol Chem. 1995;270:18335–18340. doi: 10.1074/jbc.270.31.18335. [DOI] [PubMed] [Google Scholar]
- 33.Pfeiffer T, Hartmann RK. Role of the spacer boxA of Escherichia coli ribosomal RNA operons in efficient 23 S rRNA synthesis in vivo. J Mol Biol. 1997;265:385–393. doi: 10.1006/jmbi.1996.0744. [DOI] [PubMed] [Google Scholar]
- 34.Klumpp S, Hwa T. Stochasticity and traffic jams in the transcription of ribosomal RNA: Intriguing role of termination and antitermination. Proc Natl Acad Sci USA. 2008;105:18159–18164. doi: 10.1073/pnas.0806084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodarzi H, et al. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol. 2010;6:378. doi: 10.1038/msb.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conrad TM, et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Haugen SP, et al. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: An additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 40.Herring CD, Glasner JD, Blattner FR. Gene replacement without selection: Regulated suppression of amber mutations in Escherichia coli. Gene. 2003;311:153–163. doi: 10.1016/s0378-1119(03)00585-7. [DOI] [PubMed] [Google Scholar]
- 41.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotechnol. 2004;22:656–658. doi: 10.1038/nbt0604-656b. author reply 658. [DOI] [PubMed] [Google Scholar]
- 44.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.