Abstract
Newborn neurons in the dentate gyrus of the adult hippocampus rely upon cAMP response element binding protein (CREB) signaling for their differentiation into mature granule cells and their integration into the dentate network. Among its many targets, the transcription factor CREB activates expression of a gene locus that produces two microRNAs, miR-132 and miR-212. In cultured cortical and hippocampal neurons, miR-132 functions downstream from CREB to mediate activity-dependent dendritic growth and spine formation in response to a variety of signaling pathways. To investigate whether miR-132 and/or miR-212 contribute to the maturation of dendrites in newborn neurons in the adult hippocampus, we inserted LoxP sites surrounding the miR-212/132 locus and specifically targeted its deletion by stereotactically injecting a retrovirus expressing Cre recombinase. Deletion of the miR-212/132 locus caused a dramatic decrease in dendrite length, arborization, and spine density. The miR-212/132 locus may express up to four distinct microRNAs, miR-132 and -212 and their reverse strands miR-132* and -212*. Using ratiometric microRNA sensors, we determined that miR-132 is the predominantly active product in hippocampal neurons. We conclude that miR-132 is required for normal dendrite maturation in newborn neurons in the adult hippocampus and suggest that this microRNA also may participate in other examples of CREB-mediated signaling.
Keywords: microRNA-212, neurogenesis, plasticity, learning
The dentate gyrus of the hippocampus supports neurogenesis throughout life, and the newborn neurons generated at this site form functional synapses, integrate into the dentate network, and contribute to complex spatial learning (1, 2). Adult hippocampal neurogenesis is stimulated by spatial learning, physical exercise, and treatment with antidepressant drugs (3–5). Conversely, aging, psychosocial stress, and excess adrenal glucocorticoids are linked to decreases in this type of neurogenesis (6–8). Interestingly, these decreases may be reversible, and many of the stimuli that promote neurogenesis also promote dendritic growth and increased arbor complexity (9). This response suggests that these newborn neurons have a high degree of plasticity that is strongly influenced by external stimuli.
One potential mechanism underlying the dendritic plasticity of newborn hippocampal neurons involves the transcription factor cAMP response element binding protein (CREB), which mediates signals from a variety of signal-transduction cascades. Several studies have shown that blocking CREB leads to a dramatic decrease in hippocampal neurogenesis and dendritic arborization in newborn neurons (10, 11), although disparate results also have been reported (12). Because CREB can activate thousands of genes, it is difficult to associate specific targets with particular CREB functions. In an effort to characterize these targets, we developed a method termed “Serial Analysis of Chromatin Occupancy” to identify CREB-binding sites on a genome-wide level (13). Many noncoding RNAs, including one that encoded two microRNAs, miR-212 and miR-132, were among the CREB targets. Introduction of miR-132 into primary cortical neurons stimulated robust neurite outgrowth, whereas inhibition of miR-132 blunted neurite outgrowth under basal conditions and blocked the response to BDNF (14). We demonstrated that these effects occurred, at least in part, through the actions of miR-132 on the GTPase-activating protein, p250GAP, which inhibits the activities of Rac and Cdc42. Subsequent studies using more mature cultures showed that miR-132 and p250GAP mediated the dendritic growth and spine formation induced by depolarization and synaptic activity (15, 16). Thus, we proposed that miR-132 was both necessary and sufficient for activity-stimulated changes in dendritic growth and spine formation in neuronal cultures. Other microRNAs mediate similar effects in culture (17–19); however, it has not been determined whether miR-132 or any of these other microRNAs have these actions in vivo.
Knockout mice have proven to be an excellent tool for analyzing gene function in vivo. However, few brain microRNAs have been studied using this method, largely for technical reasons. Many brain microRNAs, such as miR-124, are transcribed from multiple locations in the genome, complicating the generation of effective knockouts. Some brain microRNAs, such as miR-134, are located in clusters, making it difficult to delete one microRNA without affecting the expression of others. In other situations, as exemplified by miR-9 and -9*, biological functions have been ascribed to both strands of the microRNA, complicating interpretation of which microRNA is responsible for a particular outcome (20). miR-132, however, is transcribed from only a single locus and is generated from a precursor that encodes only a limited number of other microRNAs. To determine which of the four potential microRNAs were generated from the miR-212/132 locus, we developed a set of bidirectional ratiometric sensors that were capable of distinguishing their activities at single-cell resolution. Ablation of the miR-212/132 locus dramatically reduced dendritic length, branching, and spine density in newborn hippocampal neurons in young adult mice. Furthermore, we show that miR-132 is the primary functional product of the miR-212/132 locus in these cells, suggesting that it is a key mediator of the dendritic phenotype.
Results
Generation of a miR-212/132 Floxed Mouse.
miR-132 is located 200 bases downstream from miR-212 in Ensembl gene ID ENSMUSG00000065537. Canonical CREB- and RE1-silencing transcription factor (REST)-binding sites lie between the two microRNA sequences, and two additional CREB-binding sites are located ~100 and 150 bases upstream (Fig. 1A). We generated a targeting vector with parallel LoxP sites 0.4 kb upstream and downstream from the miR-132 gene, creating a LoxP arm spanning 0.8 kb of the first intron. A PGK-Neo-pA cassette flanked by flippase recognition target (FRT) sites was included for clonal selection. Following selection with neomycin and confirmation of the correct integration site by Southern blot, clonal ES cells were injected into Balb/C blastocysts. Chimeric mice were crossed into a C57Bl6 mouse, and germline transmission of the floxed locus was confirmed by PCR. Expression of mature miR-132 and miR-212 in the cortex of the floxed mice was reduced by 46% and 69%, respectively (Fig. 1C), and a similar reduction occurred in the hippocampus (Fig. S1). Presumably, this hypomorphic phenotype is caused by the retention of the neomycin cassette. The homozygous floxed miR-212/132 mice were found to be viable, fertile, and grossly normal. The expression of mature miR-132 was considerably higher than the expression of miR-212 in these mice as well as in the control C57Bl6 strain (Fig. 1C). Crossing the floxed mouse with a ubiquitous Deleter-Cre mouse caused genomic deletion of the miR-212/132 locus, as assessed by PCR, and completely eliminated expression of mature miR-132 and miR-212 (Fig. 1 B and C).
Fig. 1.
Generation and validation of floxed miR-212/132 mice. (A) Schematic of miR-212/132 targeting vector before and after Cre-mediated excision. The floxed mice have a PGK-Neo cassette upstream of the miR-212/132 locus. A REST binding site is located between CRE1 and CRE2 and between miR-212 and CRE3. (B) PCR analysis showing that the floxed allele is excised only when Cre is present (lane 1) in heterozygous floxed mice. (C) Mature miR-132 and miR-212 are decreased in the cortex of floxed mice compared with C57Bl6/J mice and are absent in the knockout mice generated by crossing the floxed mouse with a Deleter-Cre mouse (miR-132: f2,15 = 20.3, P < 0.001; Tukey's post hoc test, P < 0.01 for all comparisons; miR-212: f2,15 = 98.8, P < 0.001; Tukey's post hoc test, P < 0.01 for all comparisons; error bars show SEM). There was no change in the level of miR-16, an unrelated microRNA.
Knockout of miR-212/132 in Newborn Hippocampal Neurons Decreases Dendritic Length and Arborization.
To determine whether miR-132 influences dendritic length and arborization in vivo, we stereotactically injected the dentate gyrus of homozygous floxed miR-212/132 mice with a retrovirus expressing a GFP-Cre fusion protein (21). The GFP-Cre retrovirus was active when injected into the dentate gyrus, as monitored by its effect on mice containing a floxed stop allele upstream of YFP in the ROSA26 locus (Fig. S2) (22). Along with the GFP-Cre retrovirus, we coinjected a retrovirus expressing mCherry, which allowed visualization and quantification of dendritic length, branching, and spines. C57Bl6/J mice also were injected with the mCherry virus alone as a control. To promote hippocampal neurogenesis (4), mice were housed with a running wheel from 1 d before injection until they were killed 21 d postinjection. Dendritic length and arborization were quantified in newborn neurons from floxed mice coexpressing GFP-Cre and mCherry (Flox-Cre) and were compared with neighboring newborn neurons in the same image expressing mCherry alone (Flox-Ctrl) as well as with newborn neurons in C57Bl6/J mice that had been injected with the mCherry virus alone (WT) (Fig. 2). The GFP-Cre virus had no effect on WT newborn neurons (Fig. S3). The Flox-Cre newborn neurons had dramatically decreased dendritic arbors compared with neighboring Flox-Ctrl neurons. Furthermore, the Flox-Ctrl neurons (which express about half the levels of miR-132 and miR-212 as WT neurons) had decreased length and arbors compared with WT mice, suggesting that the degree of dendritic growth depends upon the level of miR-212/132 products. Total dendritic length was decreased by 33% in the Flox-Ctrl and 76% in the Flox-Cre newborn neurons relative to WT neurons (Fig. 2G; n = 27 or 28 cells from three or four mice; one-way ANOVA, f2,79 = 83.9, P < 0.001; Tukey's post hoc test, P < 0.001 for all comparisons). Total dendritic branching decreased by 31% in the Flox-Ctrl and by 66% in the Flox-Cre newborn neurons relative to WT neurons (Fig. 2 A–F and H, one-way ANOVA, f2,79 = 68.0, P < 0.001, Tukey's post hoc test, P < 0.001 for all comparisons). To ensure that Flox-Cre newborn neurons, which often had only one dendrite, were not radial glial cells, we immunostained for glial fibrillary acidic protein (GFAP), Doublecortin (DCX), and the neuron-specific protein NeuN. We found that 96% of Flox-Cre newborn neurons costained with NeuN, and 90% costained with DCX, whereas no newborn neurons costained with GFAP, suggesting that the Flox-Cre neurons are indeed immature granule neurons and not radial glial cells (Fig. S4). Overall, these results indicate that microRNAs encoded by the miR-212/132 locus regulate dendritic length and arborization in newborn hippocampal neurons in vivo.
Fig. 2.
Loss of miR-212/132 decreases dendritic length and branching in newborn neurons in the adult hippocampus. Representative neurons are shown 21 d postinjection. (A) C57Bl6/J mouse injected with mCherry virus. (C and E) Floxed miR-212/132 mouse injected with mCherry and GFP-Cre. (C shows a neuron from a floxed mouse that did not coexpress GFP-Cre.) (B, D, F, and H) Dendritic branching is decreased in Flox-Ctrl and Flox-Cre mice (f2,79 = 68.0, P < 0.001; Tukey's post hoc test, P < 0.001 for all comparisons). (G) Total dendritic length is decreased in Flox-Ctrl and Flox-Cre neurons (f2,79 = 83.9, P < 0.001; Tukey's post hoc test, P < 0.001 for all comparisons; error bars show SEM).
We next examined whether products from the miR-212/132 locus affected dendritic spine formation. The severity of the dendritic phenotype prevented us from assessing spine density in many Flox-Cre newborn neurons because they lacked secondary dendrites and their dendritic arbor barely extended beyond the granule cell layer (Fig. 2F). Consequently, we quantified spine density on second- and third-order dendrites in the inner half of the molecular layer. Flox-Cre neurons that had secondary dendrites had a 24% decrease in spine density compared with WT neurons and a 20% decrease compared with Flox-Ctrl newborn neurons (Fig. S5; n = 30 dendrites from three or four mice; one-way ANOVA, f2,87 = 11.1, P < 0.001; Tukey’s post hoc test, P < 0.01 WT or Flox-Ctrl compared with Flox-Cre).
miR-132 Is the Predominant microRNA Generated from the miR-212/132 Cluster.
Deletion of the miR-212/132 locus ablated four potential microRNAs, miR-132, -212, -132* and -212*. To determine which of these microRNAs are functionally important in the newborn hippocampal neurons, we developed a set of bidirectional ratiometric sensors capable of distinguishing each potential microRNA. These sensors feature a bidirectional promoter to express simultaneously distinct transcripts encoding a green fluorescent protein from Aequorea coerulscens (AcGFP) and a red fluorescent protein, Discosoma sp. Express-1 (DsRedEx1) (Fig. 3A). In the 3′ UTR of the AcGFP transcript, we incorporated three perfectly complementary microRNA recognition elements (MRE) that corresponded to the mature microRNA of interest (Fig. 3A). Thus, the level of AcGFP expression reflects the activity of the microRNA in a particular cell. We minimized the effects of variable expression levels and nonspecific transcriptional regulation by normalizing the AcGFP level to the internally expressed DsRedEx1 that lacked MREs. This approach provided a ratiometric measure that represents the activity of the specific microRNA with single-cell resolution.
Fig. 3.
Ratiometric sensors for microRNA activity. (A) A bidirectional promoter (Bi-CMV) simultaneously drives expression of AcGFP whose 3′ UTR contains three MREs. DsRedEx1 is expressed in the antisense direction as an internal control. Lentiviral packaging elements include psi and self-inactivating (SIN) LTRs. (B) SH-SY5Y neuroblastoma cells expressing the 132-MRE sensor were transfected with increasing amounts (up to 10 nM total) of miR-132 mimic and a miR-Scrm mimic (pink = 0 nM; cyan = 0.1 nM; orange = 1.0 nM; green = 10 nM). The G/R ratio in 1 × 104 individual cells per condition was assessed using flow cytometry and graphed as a cumulative frequency distribution. Black and blue lines represent the response of the negative control No-MRE sensor to the same concentrations of miR-132 mimic. (C) The 132-MRE sensor responded to the exogenous (0.1 nM) miR-132 mimic (blue) but not to the highly related miR-212 mimic (red). (D) 132-MRE and 212-MRE sensors were expressed in SH-SY5Y cells under growth (white bars) and differentiation (black bar) conditions (1% serum + 10 ng/mL retinoic acid for 2 d). The G/R ratios were normalized to that of the Scrm-MRE negative control. (E) Addition of 2′O-methyl inhibitors specific to miR-132 confirmed that the 132-MRE sensor detected endogenous miR-132 in SH-SY5Y cells. (F) The same concentrations of 2′O-methyl inhibitors did not affect expression of the No-MRE control vector.
To evaluate its dynamic range, we expressed the sensor for miR-132 (132-MRE) with increasing amounts of miR-132 mimic in SH-SY5Y neuroblastoma cells and used flow cytometry to obtain ratiometric green/red (G/R) measurements in 1 × 104 individual red-positive cells per condition (Fig. 3B). A decreased G/R ratio indicated microRNA activity. Notably, we detected endogenous miR-132 in these cells (Fig. 3B, pink lines compared with black lines) and observed a dose-dependent decrease in the G/R measurement of 132-MRE that corresponded to increasing amounts of expressed miR-132 mimic (Fig. 3B, cyan, orange, and green lines, respectively). Importantly, the miR-132 mimic had no effect on a control sensor lacking MREs (designated “No-MRE”; Fig. 3B, dark blue lines compared with black lines). Sensors for miR-212 and miR-212* had similar responses, indicating that both could be used to detect their cognate microRNAs (Fig. S6A). MREs containing mismatches at positions 9–11 responded in the same manner but with a narrower dynamic range.
To assess their specificity, we tested various combinations of sensors and microRNA mimics. Importantly, expression of scrambled microRNA mimics (miR-Scrm) did not change the ratios, and the mimics were highly specific for particular sensors (Fig. S6B). Notably, 132-MRE could even distinguish between ectopically expressed miR-132 and miR-212 mimics (Fig. 3C), which have identical seed sequences and whose mature sequences differ by only four nucleotides (Fig. S6C). Using Taqman assays, we confirmed that endogenous expression of miR-132 and miR-212 fell well within the range of the ability of the sensors to distinguish the ectopically expressed mimics (Fig. S7A).
We next tested whether the sensors could detect induced microRNA activity. Using Taqman assays, we had detected miR-132 and a much lower level of miR-212 in SH-SY5Y cells under basal conditions (Fig. S7B). Treatment with retinoic acid produced equivalent fold-increases in both miR-132 and miR-212; however, the absolute level of miR-132 was considerably higher (Fig. S7B). We applied our sensors to the same paradigm and, under basal conditions, saw a decreased G/R ratio between 132-MRE and a sensor with a scrambled MRE (Scrm-MRE) and no significant difference between Scrm-MRE and 212-MRE (Fig. 3D). Upon treatment with retinoic acid, the G/R ratio of 132-MRE decreased further, and we observed a small but significant decrease of the G/R ratio in 212-MRE (Fig. 3D). This observation suggests that the sensors were capable of detecting endogenous and inducible microRNA activity. To confirm that the decrease in the observed G/R ratio was the result of miR-132 activity, we used increasing concentrations of a 2′O-methyl miR-132 inhibitor and observed a corresponding return of the ratio back to the No-MRE sensor control (Fig. 3E). Importantly, the inhibitor had no significant effects on the No-MRE sensor control (Fig. 3F).
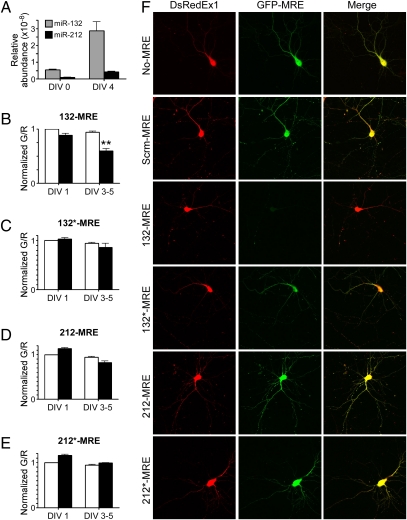
To establish the specificity and sensitivity of the sensors in primary neurons, we performed Taqman assays for miR-132 and miR-212 on a population of dissociated hippocampal neurons that had been freshly dissected or cultured for 4 d. In both cases, we observed a greater abundance of miR-132 than miR-212 (Fig. 4A). To monitor the activity of each microRNA, we introduced the sensors 132-MRE, 132*-MRE, 212-MRE, and 212*-MRE into dissociated hippocampal cultures and used flow cytometry to assay the G/R ratio either the next day or after culturing for 3–5 d in vitro (DIV 3–5) (Fig. 4 B–E). Approximately 1–5 × 103 individual red-positive live cells were evaluated per condition for each experiment, and experiments were repeated with seven different cultures. At DIV 1, we did not observe significant differences in the G/R ratio for any of the microRNAs. In contrast, at DIV 3–5 the sensors reported a significant level of miR-132 activity but did not detect significant miR-132*, -212, or -212* activity (Fig. 4 B–E). This result was confirmed using quantitative confocal microscopy of individual neurons at DIV 1 (Fig. S8) and DIV 4 (Fig. 4F) in which imaging conditions were calibrated based on green-only and red-only controls (Fig. S9). Moreover, the predominance of miR-132 activity in these neurons extended to DIV 17 (Fig. S8).
Fig. 4.
miR-132 is the predominant functional product of the miR-212/132 locus in primary hippocampal neurons. (A) Taqman assays from DIV 0 and 4 dissociated hippocampal cultures measuring the relative abundance of mature miR-132 and miR-212. (B–E) Sensors (black bars) for each of the putative microRNAs from the miR-212/132 locus were expressed in dissociated hippocampal cultures and assessed with flow cytometry after DIV 1 or DIV 3–5. The median G/R ratios of 5 × 103 neurons were normalized to that of control sensors (white bars) in the same experiment, and the SEM from seven cultures is shown. **P < 0.01; ANOVA P < 0.001. (F) Representative confocal images of the sensors in DIV 4 neurons. Scrm-MRE control corresponds to C. elegans miR-239b.
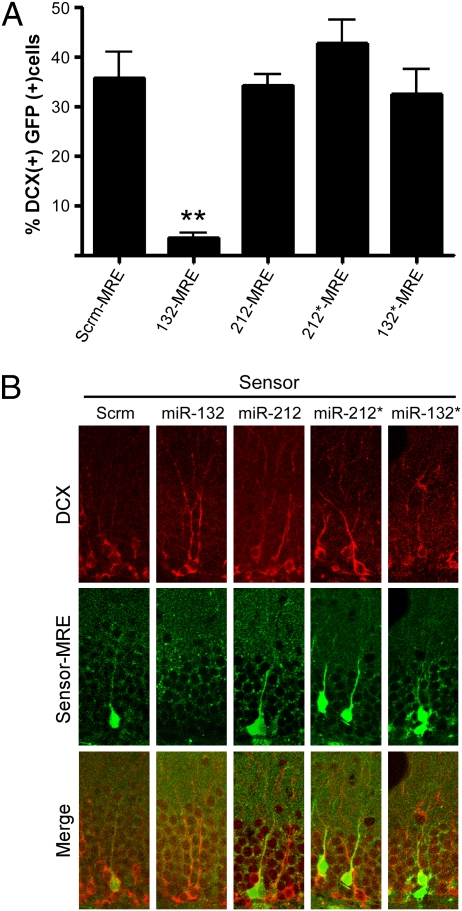
To determine whether miR-132 was responsible for the observed dendritic growth, arborization, and spine phenotypes in newborn neurons, we asked which microRNAs were active in DCX-positive neurons in the dentate gyrus of young adult C57Bl6/J mice. The expression of the red fluorescent protein from the (−) strand was insufficient to monitor transduced cells after lentiviral infection, so we compared the percentage of DCX-positive neurons that expressed GFP (indicating low microRNA activity) after infection with equal titers of lentiviral sensors. With the control Scrm-MRE, 36% of the DCX-positive cells showed GFP expression (Fig. 5), and a similar percentage was observed for the 212-MRE, 132*-MRE, and 212*-MRE sensors. However, only 4% of the DCX-positive cells in mice injected with the 132-MRE sensor expressed GFP, indicating that miR-132, but not miR-132*, -212, or -212*, is active in immature granule neurons. Thus, we propose that the lack of miR-132 is responsible for the phenotypic changes in the adult newborn neurons.
Fig. 5.
miR-132 is the primary functional product of the miR-212/132 locus in immature granule neurons in vivo. The dentate gyrus of WT mice was stereotactically injected with lentiviral microRNA sensors, and immature granule neurons were detected with DCX staining (Alexa Fluor 647). (A) The mean percentage of DCX-positive neurons that also express GFP (f4,106 = 23.4; Dunnett's multiple comparison post hoc test **P < 0.01; Error bars show SEM). (B) Representative confocal images (n = 15 sections from two or three mice per condition).
Discussion
MicroRNAs are essential for normal brain development and for establishing the functional connectivity of the brain, as evidenced by the decreased dendritic complexity, altered spine morphology, and cognitive defects caused by disruption of the microRNA processing machinery (23, 24). Linking individual microRNAs to specific brain functions has been difficult, especially in the context of intact animals. As a result, most studies have relied on cell-culture models and have used microRNA mimics, inhibitors, or sponges (25), all of which have the potential for off-target effects and can produce partial phenotypes. Furthermore, these approaches have a mixed record in recapitulating the effects of genetic knockouts or even can have opposite effects (26, 27). Generating knockout models has been challenging because brain microRNAs often are transcribed from multiple genetic loci or are located within clusters.
Ablation of miR-212/132 is comparatively simple because a single deletion affects, at most, four potential microRNAs. Still, understanding such a deletion requires establishing which of the four microRNAs from the miR-212/132 locus are functional. To address this question, we developed a series of ratiometric fluorescent sensors specific for each possible microRNA. Surprisingly, the sensors revealed that only one of the four potential microRNAs miR-132, is functional in hippocampal neurons. Thus, we could use the conditional knockout mouse strain that we developed to determine the function of miR-132 alone.
To determine the consequences of deleting the miR-212/132 locus in vivo, we examined newborn neurons within the young adult hippocampus. Hippocampal neurogenesis is exquisitely sensitive to spatial learning tasks, exposure to an enriched environment, and physical exercise. Studies have shown extensive changes in dendritic arborization and spine formation in newborn neurons that persist for several months after training for just 6 d in a Morris water maze (3). This morphological plasticity is much more evident in newborn neurons than in mature neurons, providing us with a sensitized system for examining the contribution of miR-132.
To test whether miR-132 regulates dendritic maturation in vivo, we injected Cre-expressing retroviruses into the dentate gyrus of floxed miR-132 mice and examined the morphology of the newborn neurons. By using retroviral Cre infection, we could prevent microRNA expression acutely, eliminating the possibility of compensating effects during brain development that can occur with a germline knockout. Newborn neurons expressing Cre had a profound dendritic phenotype characterized by a reduction in dendritic length, branching, and spine density. Notably, our sensors confirmed that miR-132 activity predominated in these specific neurons. Together with our previous findings, these results suggest that miR-132 plays an important role in neuronal development and maturation in vivo.
Loss of CREB signaling has been shown to impair dendritic growth and arborization of newborn hippocampal neurons (11). Our findings demonstrate that ablating a single CREB target, miR-212/132, causes a similar dendritic phenotype in newborn hippocampal neurons, suggesting that microRNAs from this locus mediate some of these CREB effects. This notion is consistent with our previous experiments using neuronal cultures in which we found that miR-132 inhibitors largely blocked the effects of CREB on dendrite maturation. Previously, we had identified p250GAP as a target mediating miR-132 effects on dendritic outgrowth in cultured neurons; whether p250GAP is sufficient for these effects in vivo is under investigation. Other miR-132 targets have been identified, including methyl CpG-binding protein 2 (MeCP2), p120RasGAP, and p300 (28–30), and it will be interesting to determine whether any of these contribute to the dendritic phenotype.
In addition to its role in dendrite maturation, CREB is necessary for the survival of newborn granule neurons (10). Thus, it will be important to determine whether newborn neuron survival depends on miR-132. The effects of miR-132 on dendritic growth and neuronal survival could be interdependent; a large proportion of the newborn hippocampal neurons undergo apoptosis (31), and survival may depend on whether immature neurons become functionally integrated into circuits. A severe dendritic phenotype caused by loss of CREB signaling or miR-132 expression could prevent integration, rendering these cells more susceptible to elimination.
Two recent papers implicated striatal miR-212 as an essential component of the mechanism underlying cocaine addiction (32, 33). Our data indicated that mature miR-132 is expressed at a significantly higher level than mature miR-212 in hippocampus and cortex. Although low levels of mature miR-212 expression could be detected, functional activity, as measured using the sensors, is minimal under basal conditions, suggesting a potential threshold (34). Without the information provided by the microRNA sensors, it would have been difficult to rule out the possibility that the low level of miR-212 expression results from different PCR primer efficiencies. However, the expression and sensor data combined indicate that miR-212 levels are considerably lower than those of miR-132. Whether the levels of miR-212 are sufficient to mediate a functional response is uncertain. An advantage of the sensors is that they allow monitoring of specific microRNA activity in individual cells, as we demonstrated using the immature neuron marker, DCX. It will be interesting to determine whether miR-212 is functionally active in dorsal striatal neurons.
In conclusion, we have demonstrated that miR-132 mediates the dendritic outgrowth of newborn neurons in the adult hippocampus in vivo. Furthermore, we have shown that miR-132 is the primary functional product of the miR-212/132 locus in both hippocampal cultures and newborn granule neurons. Thus, we have identified a mechanism whereby CREB signaling through microRNAs can promote the growth and maturation of newborn neurons in the hippocampus. Future studies will elucidate the generality of this pathway and how it may underlie activity-dependent changes in other parts of the brain.
Materials and Methods
Floxed miR-132/212 mice were generated by Ozgene. Stereotactic injections were performed as previously described (35). The CAG-GFP-Cre retrovirus (21) was a generous gift from F. Gage (Salk Institute, La Jolla, CA). Full experimental details for mice generation, viruses, cell culture, immunohistochemistry, imaging, and flow cytometry can be found in SI Materials and Methods. All experiments were approved by the Oregon Health and Science University Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Brian Druker and the Oregon Health and Science University Knight Cancer Institute for access to the Aria II cell sorter. Confocal microscopy and analysis were carried out at the Advanced Light Microscopy Core in the Oregon Health and Science University Jungers Center. Retroviruses were packaged in the Vollum Viral Core Laboratory. We thank Rukayat Taiwo for experimental assistance with in vivo injections, immunohistochemistry, and image analysis. This work was supported by National Institutes of Health Grants NS067811 and P30NS061800, the Howard Hughes Medical Institute, and a NARSAD Young Investigator Award (to B.W.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015691107/-/DCSupplemental.
References
- 1.van Praag H, et al. Functional neurogenesis in the adult hippocampus. Nature. 2002;415:1030–1034. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupret D, et al. Spatial relational memory requires hippocampal adult neurogenesis. PloS One. 2008;3:e1959. doi: 10.1371/journal.pone.0001959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tronel S, et al. Spatial learning sculpts the dendritic arbor of adult-born hippocampal neurons. Proc Natl Acad Sci U S A. 2010;107(17):7963–7968. doi: 10.1073/pnas.0914613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- 5.Santarelli L, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- 6.Gould E, McEwen BS, Tanapat P, Galea LA, Fuchs E. Neurogenesis in the dentate gyrus of the adult tree shrew is regulated by psychosocial stress and NMDA receptor activation. J Neurosci. 1997;17:2492–2498. doi: 10.1523/JNEUROSCI.17-07-02492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: Age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron HA, Gould E. Adult neurogenesis is regulated by adrenal steroids in the dentate gyrus. Neuroscience. 1994;61:203–209. doi: 10.1016/0306-4522(94)90224-0. [DOI] [PubMed] [Google Scholar]
- 9.Jin K, et al. Neurogenesis and aging: FGF-2 and HB-EGF restore neurogenesis in hippocampus and subventricular zone of aged mice. Aging Cell. 2003;2:175–183. doi: 10.1046/j.1474-9728.2003.00046.x. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa S, et al. Regulation of neurogenesis in adult mouse hippocampus by cAMP and the cAMP response element-binding protein. J Neurosci. 2002;22:3673–3682. doi: 10.1523/JNEUROSCI.22-09-03673.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jagasia R, et al. GABA-cAMP response element-binding protein signaling regulates maturation and survival of newly generated neurons in the adult hippocampus. J Neurosci. 2009;29:7966–7977. doi: 10.1523/JNEUROSCI.1054-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gur TL, et al. cAMP response element-binding protein deficiency allows for increased neurogenesis and a rapid onset of antidepressant response. J Neurosci. 2007;27:7860–7868. doi: 10.1523/JNEUROSCI.2051-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Impey S, et al. Defining the CREB regulon: A genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 14.Vo N, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wayman G, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Impey S, et al. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43:146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 18.Siegel G, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tashiro A, Sandler VM, Toni N, Zhao C, Gage FH. NMDA-receptor-mediated, cell-specific integration of new neurons in adult dentate gyrus. Nature. 2006;442:929–933. doi: 10.1038/nature05028. [DOI] [PubMed] [Google Scholar]
- 22.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark KL, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 25.Ebert MS, Sharp PA. MicroRNA sponges: Progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carè A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 27.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anand S, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein ME, et al. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 30.Lagos D, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 31.Biebl M, Cooper CM, Winkler J, Kuhn HG. Analysis of neurogenesis and programmed cell death reveals a self-renewing capacity in the adult rat brain. Neurosci Lett. 2000;291:17–20. doi: 10.1016/s0304-3940(00)01368-9. [DOI] [PubMed] [Google Scholar]
- 32.Hollander J, et al. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Im H-I, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 35.Cetin A, Komai S, Eliava M, Seeburg PH, Osten P. Stereotaxic gene delivery in the rodent brain. Nat Protoc. 2006;1:3166–3173. doi: 10.1038/nprot.2006.450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.