Abstract
Engineered glyphosate resistance is the most widely adopted genetically modified trait in agriculture, gaining widespread acceptance by providing a simple robust weed control system. However, extensive and sustained use of glyphosate as a sole weed control mechanism has led to field selection for glyphosate-resistant weeds and has induced significant population shifts to weeds with inherent tolerance to glyphosate. Additional weed control mechanisms that can complement glyphosate-resistant crops are, therefore, urgently needed. 2,4-dichlorophenoxyacetic acid (2,4-D) is an effective low-cost, broad-spectrum herbicide that controls many of the weeds developing resistance to glyphosate. We investigated the substrate preferences of bacterial aryloxyalkanoate dioxygenase enzymes (AADs) that can effectively degrade 2,4-D and have found that some members of this class can act on other widely used herbicides in addition to their activity on 2,4-D. AAD-1 cleaves the aryloxyphenoxypropionate family of grass-active herbicides, and AAD-12 acts on pyridyloxyacetate auxin herbicides such as triclopyr and fluroxypyr. Maize plants transformed with an AAD-1 gene showed robust crop resistance to aryloxyphenoxypropionate herbicides over four generations and were also not injured by 2,4-D applications at any growth stage. Arabidopsis plants expressing AAD-12 were resistant to 2,4-D as well as triclopyr and fluroxypyr, and transgenic soybean plants expressing AAD-12 maintained field resistance to 2,4-D over five generations. These results show that single AAD transgenes can provide simultaneous resistance to a broad repertoire of agronomically important classes of herbicides, including 2,4-D, with utility in both monocot and dicot crops. These transgenes can help preserve the productivity and environmental benefits of herbicide-resistant crops.
Keywords: herbicide resistance, weed management, genetically modified crops, AOPP herbicides
Since their introduction in the mid-1990s, genetically modified (GM) crops have delivered many benefits to agricultural growers by providing increased productivity, reduced pesticide use, and greater flexibility and efficiency in farm management. The area devoted to GM crop plantings has steadily increased at a rate of ∼20%/y globally for over a decade, with 134 million ha of GM crops planted worldwide in 2009. Continued growth is expected in the near future because of rapid adoption of this technology in developing countries (1). Over 90% of global GM crop plantings contain a trait conferring resistance to the broad-spectrum herbicide glyphosate (1). These include soybean, cotton, maize, and canola. Because glyphosate resistance provides a simple and convenient solution for control of a wide spectrum of broadleaf and grass weeds, farmers have rapidly adopted glyphosate-resistant (GR) crops. However, in many instances, traditional best agronomic practices for avoiding natural development of GR weeds in the field have been abandoned or reduced. These practices include crop rotation, herbicide mode of action rotation, tank mixing of multiple herbicide modes of action, and incorporation of mechanical weed control with chemical and cultural methods.
Glyphosate is commonly used in noncrop areas for total vegetation control. With the introduction of GR crops, glyphosate has been applied to some crop fields one to three times per year for more than 15 consecutive years. This increased application frequency, in combination with higher use rates and the increasing hectarage being treated, has placed heavy selection pressure on weed species to acquire naturally occurring resistance mechanisms to glyphosate (2–5). To date, 19 weeds, including both grass and broadleaf species, have developed natural resistance to glyphosate (6), most of which have been reported in the past 8 y, which is coincident with increasing use of GR crops (7, 8). Moreover, weeds that had previously not been an agronomic problem before the wide deployment of GR crops, such as Ipomoea, Amaranthus, Chenopodium, Taraxacum, and Commelina species (9–11), are now becoming more prevalent and difficult to control. With this trend, growers are rapidly losing the advantages of GR crops.
One of the most effective approaches for weed resistance management is the use of herbicides with differing modes of action, either in rotation as part of a weed management program or in combination as mixtures (12, 13). One strategy to complement and sustain the powerful weed control technology conferred by the GR trait is to deploy an additional trait that can confer resistance to herbicides with alternative modes of action. The plant hormone mimic 2,4-dichlorophenoxyacetic acid (2,4-D) was the first synthetic herbicide to be commercially developed and has been used agronomically and in noncrop applications for broad-spectrum broadleaf weed control for over 60 y (14). In addition to low cost, 2,4-D has environmentally friendly properties such as short environmental persistence and low toxicity to humans and wildlife (15, 16). Although 2,4-D remains one of the most widely used herbicides globally, its complex, plant-specific mode of action has not led to widespread natural resistance development (17), and only isolated cases of resistant weed species have been reported (6). Engineered 2,4-D resistance could offer a potentially robust, cost-effective herbicide-resistant trait that could complement glyphosate resistance.
In this work, we show that members of a class of bacterial enzymes [aryloxyalkanoate dioxygenases (AADs)], which efficiently cleave 2,4-D in vitro into nonherbicidal dichlorophenol and glyoxylate, can provide robust field resistance to 2,4-D when expressed in maize and soybean crops. In addition, we have discovered that two unique members of this class of enzymes have surprisingly broad substrate ranges for xenobiotic herbicides. AAD-1 has the additional ability to inactivate potent grass-active herbicides that act through a different mode of action (inhibition of lipid biosynthesis). AAD-12 can cleave pyridyloxyacetate auxins that are structurally diverse members of the synthetic auxin herbicide family related to 2,4-D and provide extended spectra of weed control. These attributes indicate the utility of these types of enzymes for herbicide-resistant crop applications. They may offer growers options to overcome the significant developing issues of GR weeds as well as provide the ability to use herbicides that differ in both chemical structure and mode of action through a single transgene.
Results and Discussion
Identification of 2,4-D Degrading Enzymes.
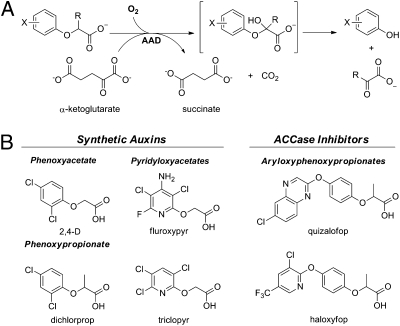
Previous studies have established that the enzyme TfdA is responsible for the first enzymatic step in the 2,4-D mineralization pathway of the soil bacterium Ralstonia eutropha (18). TfdA catalyzes the oxygenolytic cleavage of 2,4-D to nonherbicidal dichlorophenol and glyoxylate through an Fe(II)/α-keto acid-dependent dioxygenase reaction (Fig. 1A) (19). The enzyme was also shown to confer 2,4-D resistance when expressed in transgenic cotton plants (20). Sequences for a large number of bacterial genes that encode homologs of TfdA have now been deposited in genetic databases. However, most of these have not been biochemically characterized or more significantly, tested for attributes that could confer optimal 2,4-D resistance in transgenic crops. We identified sequences of candidate genes for potential 2,4-D–resistant crop utility by using a bioinformatic approach to identify homologs of TfdA in the National Center for Biotechnology Information (NCBI) genetic sequence database. Fe(II)/α-ketoglutarate–dependent dioxygenases typically contain a triad of conserved histidine residues comprising the Fe(II)-coordinating active site (21), and therefore, although overall similarity to TfdA may be relatively low, sequences with this histidine triad motif can be attributed to the Fe(II)/α-ketoglutarate–dependent dioxygenase family. Genes encoding candidate enzymes were synthesized and expressed in Escherichia coli, and the biochemical and physiological properties of the recombinant enzymes were assessed. This range-finding study enabled us to focus on two lead homologs, RdpA from Sphingobium herbicidivorans and SdpA from Delftia acidovorans, that have 28% and 31% amino acid sequence identity to TfdA, respectively.
Fig. 1.
(A) General reaction catalyzed by AAD enzyme. (B) Aryloxyalkanoate herbicidal compounds that are substrates for AADs.
To establish a uniform and unbiased nomenclature for the enzymes that we have characterized, we refer to these enzymes generically as AADs and further refer to RdpA and SdpA as AAD-1 and AAD-12, respectively. The chemical structures of herbicidal compounds included in the generic aryloxyalkanoate chemical family descriptor are shown in Fig. 1B. AAD-1 has been previously shown to cleave the herbicidally active R-enantiomer of dichlorprop and was also reported to have a low level of activity on 2,4-D (22). Screening an expanded set of structurally related herbicides, we unexpectedly found that AAD-1 has a unique ability to enantioselectively cleave members of the aryloxyphenoxypropionate (AOPP) class of potent grass-selective herbicides, including R-cyhalofop and R-quizalofop (Table 1). These herbicides act by an entirely different mechanism than that of the synthetic auxins and specifically inhibit the monomeric acetyl-CoA carboxylases of grasses (23). The efficiency of in vitro AOPP cleavage approaches that of the optimal substrate, R-dichlorprop, and is significantly higher than the activity of AAD-1 on 2,4-D (Table 1). This was surprising given the extended and bulky aryloxy substituents on the AOPPs. These data show that AAD-1 can cleave two distinct classes of herbicides covering broadleaf and grass-selective modes of action.
Table 1.
In vitro kinetic constants of AAD-1 and AAD-12 for various herbicide substrates
| Enzyme | Substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1·s−1 × 103) |
| AAD-1 | ||||
| 2,4-D | 683 ± 76 | 0.31 ± 0.01 | 0.45 | |
| MCPA | 527 ± 38 | 0.11 ± 0.002 | 0.21 | |
| R-dichlorprop | 93 ± 16 | 5.22 ± 0.36 | 56.3 | |
| R-cyhalofop | 96 ± 7 | 3.12 ± 0.06 | 32.5 | |
| R-quizalofop | 144 ± 9 | 3.34 ± 0.09 | 23.3 | |
| AAD-12 | ||||
| 2,4-D | 185 ± 25 | 3.46 ± 0.14 | 18.6 | |
| MCPA | 283 ± 67 | 4.18 ± 0.60 | 14.7 | |
| S-dichlorprop | 118 ± 35 | 3.57 ± 0.46 | 30.2 | |
| S-cyhalofop | 279 ± 22 | 1.52 ± 0.08 | 5.43 | |
Assays were performed using 0.1- to 1.5-μM enzyme. MCPA, 2-methyl-4-chlorophenoxyacetic acid.
In contrast to AAD-1, AAD-12 has been previously reported to selectively cleave the S-enantiomer of dichlorprop (24). We found in our biochemical studies that AAD-12 can also cleave the S-enantiomers of the AOPP herbicides (Table 1). Because these are the herbicidally inactive enantiomers of these compounds, this AAD-12 activity is not of utility for providing crop resistance to these chiral herbicides. However, AAD-12 has significantly greater in vitro activity on 2,4-D (an achiral substrate) than AAD-1 (Table 1) (25). Testing a series of pyridyloxyacetate compounds as substrates, we found that AAD-12 was capable of degrading the synthetic auxin herbicides triclopyr and fluroxypyr at rates of 4% and 16%, respectively, of 2,4-D (at 1 mM substrate). AADs have not previously been shown to work on substrates containing a pyridine ring. This activity gives AAD-12 potential utility for providing resistance to a wider repertoire of synthetic auxins beyond 2,4-D and thus enables expanded broadleaf weed control.
Validation of Plant Herbicide Resistance Provided by AADs.
Our initial investigations into the broad in vitro substrate specificities of the AAD-1 and AAD-12 enzymes were intriguing. Our next goal was to evaluate if expression of these enzymes in transgenic plants could provide protection against the damaging effects of 2,4-D and other herbicides. Arabidopsis thaliana was first used as a rapid and facile model system for validation of in planta activity of the AADs. An expression cassette containing either aad-1 or aad-12 genes (synthesized with plant-preferred codon usage) under control of a constitutive promoter was introduced into Arabidopsis plants through Agrobacterium tumefaciens-mediated gene delivery using phosphinothricin acetyltransferase (PAT) gene as a selectable marker (Figs. S1 and S2). T1 generation plants were selected with glufosinate herbicide, and transformants were analyzed by Southern blot and PCR for presence of the genes and by Western blot and ELISA for expression of the appropriate proteins.
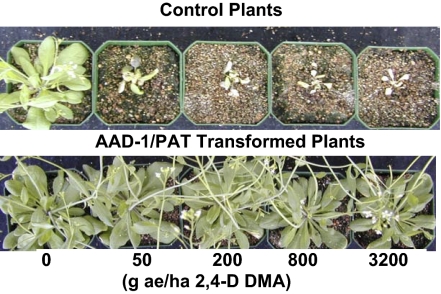
Application of 2,4-D onto untransformed Arabidopsis plants severely injured them, even at doses as low as 50 g acid equivalent (ae)/ha. In contrast, transgenic Arabidopsis plants expressing either AAD-1 and AAD-12 exhibited little or no visible signs of 2,4-D damage, even when treated with up to 3.2 kg ae/ha of the herbicide (Fig. 2 and Fig. S3). This application rate is about 5- to 10-fold higher than typical field use rates of 2,4-D and therefore, indicated that the introduced AAD enzymes could function well in plants.
Fig. 2.
Aad-1 transformed Arabidopsis response to 2,4-D in the greenhouse.
In addition, plants expressing AAD-12 survived applications of triclopyr and fluroxypyr at rates of up to 2.24 kg ae/ha. Some temporary epinastic symptoms were visible on rates exceeding 0.28 g ae/ha fluroxypyr on AAD-12–expressing plants; however, the treated plants recovered, whereas control plants were severely injured or died.
Crop Transformation with AADs.
For broad utility, a transgene must provide robust herbicide resistance in major crops where management of weeds is critical to maximize production. Maize, soybean, cotton, rice, and canola were chosen as crop targets for transformation with AAD-1 or AAD-12 expression cassettes similar to those used in Arabidopsis. Here, we describe the results obtained for maize and soybean as relevant representative monocot and dicot crops. To obtain transgenic events with adequate expression of AAD-1 and AAD-12 in these crops, various vectors with different regulatory elements were designed.
For soybean transformation, a binary vector pDAB4468 was constructed with a transfer DNA (T-DNA) insert carrying an aad-12 gene cassette under the control of an A. thaliana ubiquitin 10 promoter (AtUbi10) and a pat gene cassette driven by the Cassava Vein Mosaic Virus promoter (CsVMV) as the selectable marker for transformation (Fig. S4). Multiple transgenic soybean events were produced through Agrobacterium-mediated transformation of the public elite variety Maverick. Events with one intact copy of T-DNA insert were selected for further evaluation and characterization. T1 seed was collected from self-pollinated T0 putative transformants, T1 plants were screened for resistance to glufosinate, and seeds from multiple subsequent generations of transformants (up to T5) were collected. The heritability and stability of the gene in soybean were investigated over multiple generations. Progeny testing and molecular analysis revealed that the inheritance of the aad-12 gene was stable across all of the tested generations and followed typical single-locus Mendelian segregation for selected soybean events (Table 2). When sprayed with varying levels of 2,4-D, most of the transgenic soybean events showed 2,4-D resistance exceeding typical agricultural use rates, and the selected events were resistant to rates of ≥4.48 kg ae/ha under greenhouse and field conditions. The gene and herbicide resistance were heritable across multiple generations from T0 through T5 (Table S1 and Fig. S5). Selected events showed excellent resistance to 2,4-D at 4.48 kg ae/ha when applied at the V3 stage in field trials (eight times the recommended level of agricultural applications) (Fig. 3). Initial field studies with multiple selected soybean events at several locations in the United States showed agronomic performance equivalent to that of the nontransformed control variety, including date to flowering, lodging, phenotype, and yield. Transgenic plots were treated with 2,4-D applications comprising a seasonal total of 3.36 kg ae/ha (or 3 lb ae/acre) at both preemergence (postplant) and approximately V4 or R2 growth stages.
Table 2.
Inheritance of the AAD-1 maize and AAD-12 soybean
| AAD-1 maize event A T2 seeds | AAD-1 maize event B T2 seed | AAD-12 soybean event C F2 seed | AAD-12 soybean event D F2 seed | WT maize | WT soybean | |
| Resistant | 61 | 55 | 102 | 78 | 0 | 0 |
| Sensitive | 27 | 18 | 45 | 22 | 30 | 30 |
| Totals | 88 | 73 | 147 | 100 | 30 | 30 |
| R (%) | 69.3 | 75.3 | 69.4 | 78.0 | 0 | 0 |
| S (%) | 30.7 | 24.7 | 30.6 | 22.0 | 100 | 100 |
| χ2 | 0.2184 | 0.9461 | 2.4694 | 0.7866 | ||
| P value | >0.05 | >0.05 | >0.05 | >0.05 | ||
| Ratio | 3:1 | 3:1 | 3:1 | 3:1 |
T1 maize events were self-pollinated to produce T2 seed; T2 seeds were progeny-tested. Maize was sprayed with quizalofop and verified with either ELISA or Southern blot analysis. T4 soybean events were crossed with elite soybean variety to produce F1 seeds that were planted to produce F2 seeds. Soybeans were sprayed with 2,4-D and verified with either ELISA or Southern blot analysis. WT, wild-type.
Fig. 3.
Transgenic and nontransgenic soybean plants response to 2,4-D in the field.
For maize transformation, plasmid pDAS1740 was constructed containing an aad-1 gene under the control of the maize ubiquitin promoter (ZmUbi1) (Fig. S6). The recipient maize line Hi-II was transformed using direct insertion of the DNA fragment from plasmid pDAS1740 through silicon carbide fiber-mediated transformation (26). Putative transgenic maize plants were screened with R-haloxyfop, and multiple transgenic maize events were produced. Events with one single intact copy of aad-1 plant transcription unit (PTU) were selected for further evaluation and characterization. Seeds from multiple generations, including self-pollinated generations (up to T4), converted inbred generations (BC1, BC2, and BC3S1), and hybrid test crosses, were collected. Progeny testing and molecular analysis revealed that the inheritance of the aad-1 gene was stable across all of the generations evaluated and followed typical single-locus Mendelian segregation (Table 2).
The transgenic maize plants were resistant to applications of the grass-active AOPP herbicides (cyhalofop or quizalofop) at rates of 0.28–0.56 kg ae/ha under greenhouse and field conditions. Maize is normally highly susceptible to these herbicides. Although 2,4-D is used as a broadleaf herbicide with little or no activity on grasses, it can produce significant malformations of maize plants when applied at late seedling stages. This leads to restrictions on its use in maize to plants that are before the six-leaf stage or under 18 cm. Transgenic maize events showed resistance to 2,4-D application rates up to 4.48 kg ae/ha, with no 2,4-D–induced brace root malformations being observed. The gene and herbicide resistance was heritable over multiple generations of selfed lines (T0 through T4), converted inbred lines (BC1, BC2, and BC3S1), and test hybrid crosses (Table S2 and Fig. S7). Selected events showed excellent resistance to quizalofop at rates of up to 0.28 kg ae/ha when applied at the V6 stage in field trials. This is four to eight times the recommended field rate for control of volunteer maize, showing that the aad-1 gene can serve as an excellent selectable marker in field situations (Fig. 4). Initial field studies with multiple selected maize events across multiple locations in the United States showed equivalent agronomic performance to conventional controls, including date to silking, date to pollination, date to maturity, lodging, phenotype, and yield. Transgenic plots were treated with 2,4-D applications, with a seasonal total of 3.36 kg ae/ha at preemergence (postplant) and approximately V4 and V8 growth stages and/or a quizalofop application at a rate of 0.092 kg ae/ha at approximately V6 growth stage. These data show that a single aad-1 gene confers robust field resistance to two different herbicides with distinct modes of action. This is in contrast to other transgenic mechanisms of field-level herbicide resistance, such as glyphosate and glufosinate resistance, that confer resistance to a single herbicide chemistry.
Fig. 4.
Transgenic and nontransgenic maize plants response to quizalofop in the field.
Potential Impact of AAD-Derived Herbicide-Resistant Traits.
2,4-D is a synthetic auxin herbicide that kills the target weed by mimicking the natural plant growth hormone auxin [indole-3-acetic acid (IAA)] and binds to recently discovered IAA receptors in plants (17). It has been used effectively for over six decades to efficiently control a wide spectrum of broadleaf weeds (27). When applied to dicotyledonous plants at effective doses, 2,4-D causes uncontrolled and disorganized plant growth that leads to death (17). It can also have undesirable morphological effects on seedling grass species. Despite its widespread use, very few 2,4-D–resistant weed species have been identified (6). Of these few species, none have showed significant spread from initial sites of discovery, and none have significant economic importance (28). The lack of widespread development of 2,4-D–resistant weeds may be because of the genetic redundancy in auxin/2,4-D receptors, the essentiality of auxin perception for plant development, and/or the pleiotropic nature of the downstream auxin effects (29–31). These observations suggest that the frequency of 2,4-D–resistant weed appearance may be low.
Robust crop 2,4-D resistance can assist growers by increasing the flexibility of weed control practices at low cost. Moreover, it can help overcome or slow the development of GR weeds. A significant advantage provided by higher rates of 2,4-D is the potential for residual control of broadleaf weeds. Glyphosate is a nonresidual herbicide and does not provide control of later-germinating weeds. Resistance provided by AADs can also provide the ability to tank mix a broad-spectrum broadleaf weed control herbicide with commonly used residual weed control herbicides. These herbicides are typically applied before or at planting but often are less effective on emerged, established weeds that may exist in the field before planting. By extending the utility of aryloxyacetate auxin herbicides such as 2,4-D and fluroxypyr to include at-plant, preemergence, or preplant applications, the flexibility of residual weed control programs can be increased.
In grass crops such as maize, the AAD-1 gene confers robust resistance to grass-active AOPP herbicides, such as quizalofop and cyhalofop, that are otherwise highly injurious to these crops. AAD-1 simultaneously provides resistance to 2,4-D. The enhanced resistance of maize and other monocot crops to 2,4-D conferred by AAD-1 will enable use of these auxinic herbicides without the potential for known auxin-induced effects, such as crop leaning, inhibited leaf unfurling (rat-tailing), stalk brittleness, or deformed brace roots, that lead to the current growth-stage restrictions on 2,4-D application to grass crops.
In summary, the broad xenobiotic substrate selectivity of the AAD enzymes that we have characterized provides diverse mechanisms to incorporate additional established highly effective herbicide modes of action with the convenience of herbicide-resistant crops in an integrated weed-shift management strategy. When stacked with other herbicide resistance traits, for example, with the widely used GR traits, AAD-derived herbicide resistance will provide farmers with a practical tool to control current GR weeds, such as horseweed, as well as multiple pigweed and ragweed species and newly developing GR weeds. It also may slow weed-resistance development by diversifying herbicide selection pressures with additional modes of action and differing chemical structures. AAD-based herbicide-resistant traits will provide a tool to protect and sustain the value of current herbicides and herbicide-resistant crops.
Materials and Methods
Expression and Purification of AAD-1 and AAD-12 Enzymes.
Aad-1 and aad-12 genes were cloned using standard PCR amplification and molecular cloning techniques. The primers were designed according to published DNA sequences to amplify the mature protein coding region (24, 25). The fragment encoding the full-length gene was then cloned in frame into a pET-based vector (pET280) and transformed into E. coli BL21 (DE3) for protein expression. Typical induction conditions for the AAD-1 and AAD-12 proteins were with 75 μM isopropyl-β-d-thiogalactopyranoside (IPTG) in Luria Broth with appropriate antibodies and cells grown overnight at 25 °C for optimal soluble protein expression. Cell paste was harvested by centrifugation, and cell lysate was prepared either by using sonication or microfluidization methods. The protein was purified from the soluble fraction using standard ion exchange and hydrophobic interaction chromatography separations followed by size exclusion chromatography. Typically, after three steps of column separation, the target protein had a purity of >99%. The homogeneity and enzymatic activity of the protein were further validated by HPLC and activity assays. Protein concentration was determined by total amino acid hydrolysis.
In Vitro Enzyme Assay.
AAD enzymatic activity was measured by colorimetric detection of the product phenol using a protocol modified from that of Fukumori and Hausinger (32) in a 96-well microplate format. Assays were performed in a total volume of 0.15 mL 20 mM Mops (pH 6.75) containing 200 μM NH4FeSO4, 200 μM sodium ascorbate, 1 mM α-ketoglutarate, and the appropriate substrate and enzyme. Assays were initiated by addition of the aryloxyalkanoate substrate, enzyme, or α-ketoglutarate at time 0. After 15 min incubation at 25 °C, the reaction was terminated by addition of 30 μL of a 1:1:1 mix of 50 mM Na EDTA, pH 10 buffer, and 0.2% 4-aminoantipyrine; then, 10 μL 0.8% potassium ferricyanide were added. After 15 min, the absorbance at 510 nm was recorded in a spectrophotometer. Blanks contained all reagents except for enzyme. Assays with pyridyloxyacetate substrates were performed in the same way, except that release of the pyridinol was directly observed by the increase of absorbance at 325 nm or by a succinyl-CoA synthetase-coupled assay (33).
Gene Redesigns for Plant Expression.
Both of the aad-1 and aad-12 genes (RdpA and SdpA) were identified from the NCBI database (http://www.ncbi.nlm.nih.gov) with accession numbers of AF516752 and AY327575, respectively. To improve production of the recombinant protein in plants, both AAD DNA sequences were codon-optimized for plant expression. The resulting synthetic gene sequences encoded identical protein sequences, except for the addition of an alanine residue in the second position resulting from the creation of an NcoI site (CCATGG) across the ATG start codon to enable subsequent cloning operations. The plant-optimized aad-1 and aad-12 genes were synthesized at Picoscript and confirmed by sequencing.
Constructs for testing in Arabidopsis.
The aad-1 gene was cloned into plasmid pDAB726 as an NcoI–SacI fragment. The resulting construct was designated pDAB720, containing AtUbi10 /aad-1/Nt Osm3′UTR/AtuORF1 3′UTR. The fragment containing the described cassette in pDAB720 was then cloned into the corresponding Not I site of the binary vector pDAB3038. The resulting binary vector, pDAB721 (Fig. 2), containing the following cassette (AtUbi10 promoter/aad-1/Nt OSM 3′UTR/AtuORF1 3′UTR//CsVMV promoter/pat/AtuORF25/26 3′UTR) was confirmed through restriction enzyme digestion and DNA sequencing.
The aad-12 gene was cloned into plasmid pDAB726 as an NcoI–SacI fragment. The resulting construct was designated pDAB723, containing AtUbi10 promoter/aad-12/NtOsm3′UTR/AtuORF1 3′UTR. The fragment containing the described cassette in pDAB723 was then cloned into the Not I site of the binary vector pDAB3038. The resulting binary vector, pDAB724 (Fig. S1), containing the following cassette (AtUbi10 promoter/aad-12/NtOsm 3′UTR/AtuORF1 3′UTR//CsVMV promoter/pat/AtuORF25/26 3′UTR) was confirmed through restriction enzyme digestion and DNA sequencing.
AAD constructs for soybean and maize.
Aad-12–containing construct pDAB4468 for soybean transformation was similar to construct pDAB724 with some modification. The T-DNA insert encompassing the aad-12 and pat expression cassettes consisted of RB7 MAR/AtUbi10 promoter/aad-12/AtuORF23 3′UTR//CsVMV promoter/pat/ AtuORF1 3′UTR (Fig. S3). The resulting binary vector, pDAB4468, was confirmed by restriction enzyme digestion and DNA sequencing.
Aad-1–containing construct pDAS1740 was derived from pDAB721 by simply eliminating the PAT expression cassette. Aad-1–containing construct pDAS1740 was digested by restriction enzyme FseI flanking the aad-1 expression cassette. The released fragment containing RB7 MAR/ZmUbi1 promoter/aad-1/ZmPer5 3′ UTR//RB7 MAR was purified and used for silicon carbide fiber-mediated transformation of maize (Fig. S5).
Plant Transformation.
Agrobacterium-mediated transformation of Arabidopsis and soybean.
The genetically engineered aad gene cassettes in the binary vectors pDAB721 and pDAB724 were introduced into electrocompetent A. tumefaciens strain Z707S using a protocol from Weigel and Glazebrook (34). The presence of the gene insert in selected Agrobacterium colonies was verified with PCR analysis using vector-specific primers. Arabidopsis was transformed using the floral dip method (35).
The disarmed A. tumefaciens strain EHA101 carrying the binary vector pDAB4468 was used for soybean transformation. Transformation of soybean varieties was carried out by following the cotyledonary node Agrobacterium-mediated transformation system (36). Briefly, Maverick soybean seeds were germinated on basal media, and cotyledonary nodes were isolated and infected with Agrobacterium. Shoot initiation, shoot elongation, and rooting media were supplemented with cefotaxime, timentin, and vancomycin for removal of Agrobacterium. Glufosinate selection was used to inhibit the growth of nontransformed shoots. Selected shoots were transferred to rooting medium for root development and then, transferred to soil mix for acclimatization of plantlets. Terminal leaflets of selected plantlets were leaf-painted with glufosinate to screen for putative transformants. Surviving plantlets were deemed to be putative transformants. The screened plants were sampled and analyzed by molecular approaches to confirm the insertion of selectable marker gene and/or the gene of interest.
Silicon carbide fiber-mediated transformation of maize.
DNA fragment containing the genetically engineered aad-1 gene cassette was released from plasmid pDAS1740 by FseI digestion and introduced into recipient maize line Hi-II through silicon carbide fiber (Whiskers)-mediated transformation (26). Immature embryos of maize were aseptically removed from the developing caryopsis, callused on semisolid media, initiated in liquid suspension cultures, cryopreserved, thawed, and reestablished as embryogenic suspensions. The reestablished suspensions were agitated with isolated pDAS1740/Fsp I DNA fragment and silicon carbide fibers to introduce the DNA into the cells. After 3 d of growth on nonselective, semisolid media, the cells were transferred to a medium containing the herbicide (R)-haloxyfop to select for cells expressing the aad-1 gene. The callus that survived on the herbicide-containing medium proliferated and produced embryogenic tissue that was presumably genetically transformed. Callus samples were taken for molecular analysis to verify the presence of the transgene and the absence of the ampicillin resistance gene from the vector backbone, although they were not included in the DNA fragment used for transformation. The embryogenic tissue was then manipulated to regenerate whole transgenic plants and transferred to a greenhouse. The plants were sprayed with the AOPP herbicide quizalofop to confirm herbicide resistance. Surviving plants were crossed with proprietary inbred maize lines to obtain T1 seed from the initially transformed T0 plants.
Characterization of Herbicide Resistance in Transformed Plants.
Freshly harvested transformed Arabidopsis T1 seeds were planted, and the T1 plants at cotyledon [5 d after planting (DAP)] and two to four leaf-growth stages (10 DAP) were sprayed with glufosinate at a rate of 0.28 kg active ingredient (ai)/ha per application. Survivors (plants actively growing) were identified 5–7 d after the final spraying and transplanted individually into 3-in pots filled with potting media. Surviving T1 plants from glufosinate spray were confirmed for expression of the PAT protein using a PAT ELISA (EnviroLogix, Inc.). Plants were then treated with various rates of 2,4-D at 0.0125 kg ae/ha and elevated rates of 0.05, 0.2, 0.8, or 3.2 kg ae/ha. Similarly, AAD-1 maize plants were selected either in the greenhouse or field with quizalofop (0.14–0.56 kg ai/ha) and subsequently challenged with 2,4-D (0.56–4.48 kg ae/ha). AAD-12 transgenic soybean plants were selected with herbicide glufosinate (0.411 kg ai/ha) and subsequently challenged with 2,4-D (0.56–4.48 kg ae/ha) in the field.
Heritability of Genes in Transgenic Plants.
Herbicide resistance.
T1 and T2 Arabidopsis populations (from self-pollination of T1 plants) were screened with glufosinate. Approximately 75% of the T2 individuals survived selective rates of glufosinate. Survivors were sprayed with varying rates of 2,4-D (0.05–3.2 kg ae/ha) as previously noted. Plant survival and level of injury were rated at 7 and 14 d after treatment (DAT). The level of injury observed among T2 siblings was consistent with that observed in the individuals in T1 generation. For soybean events, up to generation T5 and the F2 generation after crossing with elite varieties were used to study transgene heritability. For maize events, self-pollinated inbred generations (up to T4) and back-cross generations (BC1, BC2, and BC3S1) were studied for the AAD-1 gene heritability. The data gathered indicated that the resistance is transferable across all of the tested generations (Table S1 and Fig. S6).
Southern blot analysis.
For DNA analysis, total genomic DNA was isolated from individual plant tissues using the cetyltrimethylammonium bromide (CTAB) method. A total of 9 μg leaf genomic DNA or 10 μg genomic callus DNA was subjected to an overnight digestion using selected restriction enzymes. DNA was then separated by gel electrophoresis and transferred to a nylon membrane as described by Memelink et al. (37). The hybridization probes were generated using a PCR-based incorporation of a digoxigenin (DIG)-labeled nucleotide, (DIG-11)-dUTP, to DNA fragments generated by primers specific to the gene elements and other regions from target plasmid. The PCR synthesis of the probes was performed using PCR DIG Probe Synthesis Kit (Roche Diagnostics). Labeled probes were hybridized to the target DNA on the nylon membranes using the DIG Easy Hyb Solution (Roche Diagnostics). DIG-labeled DNA molecular weight marker II was used to determine the hybridizing fragment size on the Southern blots.
DIG-labeled probes bound to the nylon membranes were visualized using CDP-Star Chemiluminescent Nucleic Acid Detection System (Roche Diagnostics). Blots were exposed to chemiluminescent film for one or more time points to detect the hybridizing fragments and visualize the molecular weight standards.
Western blot analysis.
For Western blot analysis, ∼50–100 mg leaf tissue were extracted with 500 μL plant extraction buffer (PBS containing 0.05% Tween 20 and 0.5% BSA) using the Geno/Grinder (Model 2000–115; Certiprep). Extracts were centrifuged, and the supernatant containing the soluble proteins was used for Western blots or ELISA. Plant extracts were incubated with Laemmli sample buffer at 95 °C for 10 min and electrophoretically separated in 8–16% Tris·Glycine Precast gel. Proteins were then electro-transferred onto nitrocellulose membrane using a standard protocol. After blocking in 4% skim milk in PBS, protein was detected by antiserum followed by goat anti-rabbit/HRP conjugates. The detected protein was visualized by chemiluminescence substrate (ECL Western Analysis Reagent; Amersham).
ELISA Analysis of Plant Tissues.
AAD-1 ELISA.
An AAD-1–specific monoclonal antibody (2 μg/mL) was immobilized on microtiter wells to capture AAD-1 protein in plant extracts (described above). Various concentrations of plant extracts were incubated in the wells. To prepare a standard curve, different concentrations of purified AAD-1 proteins (100, 50, 25, 12.5, 6.25, 3.13, 1.56, and 0 ng/mL) were also put into the microtiter wells on the same assay plate in duplicates. The plate was incubated for 1 h and washed four times with PBS with 0.05% Tween 20 (PBST). Biotinylated anti–AAD-1 monoclonal antibody (1 μg/mL) solution in PBST was added and incubated for 1 h to detect the bound AAD-1 protein. After another wash step, neutravidin–alkaline phosphatase (AP) conjugate (Pierce Biotech) was added to each well and incubated for another 1 h. pNPP (p-nitrophenyl phosphate) solution (AP substrate; 100 μL) was added, and after the 30- to 45-min incubation, the absorbance at 405 nm was measured using a microtiter plate reader. The concentration of proteins in plant extracts was extrapolated from a standard curve developed on the same plate.
AAD-12 ELISA.
Purified rabbit antibody (1 μg/mL) was immobilized on microtiter wells to capture AAD-12 protein in plant extracts as described above for AAD-1. HRP-conjugated anti–AAD-12 monoclonal antibody (1 μg/mL) solution in PBST was added and incubated for 1 h to detect the bound AAD-12 protein. After another wash step, 3,3′,5,5′ Tetramethylbenzidine (TMB) substrate (100 μL) was added, and after the 15-min incubation, 100 μL stop solution (1 N HCl) was added to stop the reaction. The absorbance at 450 nm was measured using a microtiter plate reader.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013154107/-/DCSupplemental.
References
- 1.James C. Global Status of Commercialized Biotech/GM Crops: 2009. Ithaca, NY: ISAAA; 2009. ISAAA Brief No 41. [Google Scholar]
- 2.Powles SB, Preston C. Evolved glyphosate resistance in plants: Biochemical and genetic basis of resistance. Weed Technol. 2006;20:282–289. [Google Scholar]
- 3.Simarmata M, Kaufmann JE, Penner D. Potential basis of glyphosate resistance in California rigid ryegrass (Lolium rigidum) Weed Sci. 2003;51:678–682. [Google Scholar]
- 4.Lorraine-Colwill DF, et al. Investigations into the mechanism of glyphosate resistance in Lolium rigidum. Pestic Biochem Physiol. 2003;74:62–73. [Google Scholar]
- 5.Miller SD, et al. Risks of weed spectrum shifts and herbicide resistance in glyphosate-resistant cropping systems. Proc West Soc Weed Sci. 2003;56:61–62. [Google Scholar]
- 6.Heap I. The International Survey of Herbicide Resistant Weeds. 2010. [Accessed August 14, 2010]. Available at www.weedscience.com.
- 7.Cerdeira AL, Duke SO. The current status and environmental impacts of glyphosate-resistant crops: A review. J Environ Qual. 2006;35:1633–1658. doi: 10.2134/jeq2005.0378. [DOI] [PubMed] [Google Scholar]
- 8.Heap, I The International Survey of Herbicide Resistant Weeds. [Accessed August 10, 2010]. Available at www.weedscience.com.
- 9.Coble HD, Warren LS. Weed control investigations in maize, cotton, crop rotations, soybean, small grain. Annual report of the Department of Crop Science. N C State University. 1997;28:103–113. [Google Scholar]
- 10.Shaner DL. The impact of glyphosate-tolerant crops on the use of other herbicides and on resistance management. Pest Manag Sci. 2000;56:320–326. [Google Scholar]
- 11.Gianessi LP. Economic and herbicide use impacts of glyphosate-resistant crops. Pest Manag Sci. 2005;61:241–245. doi: 10.1002/ps.1013. [DOI] [PubMed] [Google Scholar]
- 12.Service RF. Agbiotech. A growing threat down on the farm. Science. 2007;316:1114–1117. doi: 10.1126/science.316.5828.1114. [DOI] [PubMed] [Google Scholar]
- 13.Behrens MR, et al. Dicamba resistance: Enlarging and preserving biotechnology-based weed management strategies. Science. 2007;316:1185–1188. doi: 10.1126/science.1141596. [DOI] [PubMed] [Google Scholar]
- 14.Industry Task Force II on 2,4-D Research Data Issue Backgrounder. 2005. [Accessed August 10, 2010]. Available at http://www.24d.org/background/24D-Backgrounder-Benefits.pdf.
- 15.Tu M, Hurd C, Randall JM. Weed Control Methods Handbook. The Nature Conservancy, Version April 2001. 2001. [Accessed August 10, 2010]. Available at http://www.invasive.org/gist/handbook.html.
- 16.United States Environmental Protection Agency Pesticides reregistration, 2,4-D RED facts. [Accessed August 14, 2010];2006 Available at http://www.epa.gov/oppsrrd1/REDs/factsheets/24d_fs.htm. [Google Scholar]
- 17.Sterling TM, Hall JC. Mechanism of action of natural auxins and the auxinic herbicides. In: Roe MR, Kuhr RJ, Burton JD, editors. Herbicide Activity: Toxicology, Biochemistry and Molecular Biology. Amsterdam: IOS Press; 1997. pp. 111–141. [Google Scholar]
- 18.Streber WR, Timmis KN, Zenk MH. Analysis, cloning, and high-level expression of 2,4-dichlorophenixyacetic monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987;169:2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smejkal CW, et al. Substrate specificity of chlorophenoxyalkanoic acid-degrading bacteria is not dependent upon phylogenetically related tfdA gene types. Biol Fertil Soils. 2001;33:507–513. [Google Scholar]
- 20.Bayley C, et al. Engineering 2,4-D resistance into cotton. Theor Appl Genet. 1992;83:645–649. doi: 10.1007/BF00226910. [DOI] [PubMed] [Google Scholar]
- 21.Hogan DA, Smith SR, Saari EA, McCracken J, Hausinger RP. Site-directed mutagenesis of 2,4-dichlorophenoxyacetic acid/alpha-ketoglutarate dioxygenase. Identification of residues involved in metallocenter formation and substrate binding. J Biol Chem. 2000;275:12400–12409. doi: 10.1074/jbc.275.17.12400. [DOI] [PubMed] [Google Scholar]
- 22.Müller TA, Byrde SM, Werlen C, van der Meer JR, Kohler HP. Genetic analysis of phenoxyalkanoic acid degradation in Sphingomonas herbicidovorans MH. Appl Environ Microbiol. 2004;70:6066–6075. doi: 10.1128/AEM.70.10.6066-6075.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright TR, et al. International Patent WO/2005/107437. 2005 [Google Scholar]
- 24.Müller TA, Fleischmann T, van der Meer JR, Kohler HP. Purification and characterization of two enantioselective α-ketoglutarate-dependent dioxygenases, RdpA and SdpA, from Sphingomonas herbicidovorans MH. Appl Environ Microbiol. 2006;72:4853–4861. doi: 10.1128/AEM.02758-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westendorf A, Muller RH, Babel W. Purification and characterization of the enantiospecific dioxygenases from Delftia acidovorans MC1 initiating the degradation of phenoxypropionates and phenoxyacetate herbicides. Acta Biotechnol. 2003;23:3–17. [Google Scholar]
- 26.Petolino JF, Arnold NL. Whiskers-mediated maize transformation. In: Scott MP, editor. Methods in Molecular Biology: Transgenic Maize. Vol. 526. Clifton, NJ: Humana Press; 2009. pp. 59–67. [DOI] [PubMed] [Google Scholar]
- 27.Mockaitis K, Estelle M. Auxin receptors and plant development: A new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- 28.Heap IM. The occurrence of herbicide-resistant weeds worldwide. Pestic Sci. 1997;51:235–243. [Google Scholar]
- 29.Devine MO, Duke SO, Fedtke C, editors. Physiology of Herbicide Action. Englewood Cliffs, NJ: Prentice Hall; 1993. [Google Scholar]
- 30.Jasieniuk M, Brule-Babel AL, Morrison IN. The evolution and genetics of herbicide resistance in weeds. Weed Sci. 1996;44:176–193. [Google Scholar]
- 31.Van Eerd LL, Stephenson GR, Kwiatkowski J, Grossmann K, Hall JC. Physiological and biochemical characterization of quinclorac resistance in a false cleavers (Galium spurium L.) biotype. J Agric Food Chem. 2005;53:1144–1151. doi: 10.1021/jf048627e. [DOI] [PubMed] [Google Scholar]
- 32.Fukumori F, Hausinger RP. Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J Biol Chem. 1993;268:24311–24317. [PubMed] [Google Scholar]
- 33.Luo L, et al. An assay for Fe(II)/2-oxoglutarate-dependent dioxygenases by enzyme-coupled detection of succinate formation. Anal Biochem. 2006;353:69–74. doi: 10.1016/j.ab.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 34.Weigel D, Glazebrook J. Arabidopsis: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 35.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 36.Zeng P, Vadnais DA, Zhang Z, Polacco JC. Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill] Plant Cell Rep. 2004;22:478–482. doi: 10.1007/s00299-003-0712-8. [DOI] [PubMed] [Google Scholar]
- 37.Memelink J, Swords K, Harry J, Hoge C. Southern, Northern, and Western Blot Analysis. Plant Molecular Biology Manual. 1994;F1:1–23. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.