Summary
In maturing retroviral virions CA protein assembles to form a capsid shell that is essential for infectivity. The structure of two folded domains (NTD and CTD) of CA is highly conserved among various retroviruses, and the capsid assembly pathway, although poorly understood, is thought to be conserved as well. In vitro assembly reactions with purified CA proteins of the Rous sarcoma virus (RSV) were used to define factors that influence the kinetics of capsid assembly and provide insights into the underlying mechanisms. CA multimerization was triggered by multivalent anions providing evidence that in vitro assembly is an electrostatically-controlled process. In the case of RSV, in vitro assembly was a well behaved nucleation-driven process that led to formation of structures with morphologies similar to those found in virions. Isolated RSV dimers when mixed with monomeric protein acted as efficient seeds for assembly, eliminating the lag phase that is characteristic of the monomer-only reaction. This demonstrates for the first time the purification of an intermediate on the assembly pathway. Differences in the intrinsic tryptophan fluorescence of monomeric protein and the assembly-competent dimer fraction suggest the involvement of the NTD in formation of the functional dimer. Furthermore, in vitro analysis of well characterized CTD mutants provides evidence for assembly dependence upon the second domain and suggests that the establishment of an NTD-CTD interface is a critical step in capsid assembly initiation. Overall the data provide clear support for a model whereby capsid assembly within the maturing virion is dependent upon formation of a specific nucleating complex that involves a CA dimer and is directed by additional virion constituents.
Introduction
A functional retroviral core structure forms during a complex multi-staged process in which assembly and egress of an immature particle is first coordinated by the Gag polyprotein. The action of the virally-encoded protease results in the reorganization of the internal virion constituents into a mature core structure that consists of RNA bound by viral proteins within a capsid shell.1 Characterization of mature retroviral capsids by cryo-electron tomography has led to the prediction that capsid formation is a de novo assembly process.2–4 Specifically, the occurrence of multiple core-like structures or multi-layered protein structures in some virus particles,2–4 as well as a fraction of CA protein that is not assembled into detectable structures,5–9 are difficult to reconcile with a model of capsid formation by a concerted condensation of the pre-existing CA-CA contacts that organized the Gag molecules in the immature particle.6,10–16 Instead, capsid formation may be initiated by the establishment of a unique, small complex of CA, perhaps in contact with other viral constituent(s), with further growth of the shell proceeding by the addition of monomers or small oligomers of CA that are liberated during proteolysis of Gag. In vitro CA assembly studies utilizing the proteins of the human immunodeficiency virus (HIV, a lentivirus) and the avian Rous sarcoma virus (RSV, an alpharetrovirus) have provided ancillary support for the de novo assembly mechanism.17–19 However, no structures that represent intermediates along the assembly pathway have been identified.
Capsids of both HIV and RSV exhibit a dramatic degree of polymorphism, i.e., particle-to-particle variation in morphology, as documented by electron tomographic analyses.2–4 This inherent variability has prevented the determination of a high resolution structure of any authentic retroviral capsid by conventional cryo-electron microscopy methods that involve averaging images of many presumably identical structures. Instead, the analysis of more regular, in vitro-assembled CA arrays has yielded a consensus model of protein organization in the core.20,21 The capsids are best described as protein lattices organized on the same geometric principles as fullerene carbon structures, with subunits arranged in hexamers interspersed with pentamers.22,23 A lack of regular spacing between pentamers results in a range of capsid morphologies and distinguishes retroviral cores from the icosahedral capsids found in numerous other viruses.23–25 The shape of an individual retroviral capsid is governed by the arrangement of the 12 pentamers that punctuate the hexameric lattice.21,23,24 An asymmetric but bipolar distribution (5 + 7) of pentamers can explain the cone-shaped cores that predominate in HIV, a more random distribution the irregular polyhedra of RSV.
Hexameric lattices of the HIV, RSV and murine leukemia virus CA proteins have been characterized extensively21,26–30 but until lately the pentameric capsomer has been only a mathematical conjecture without supporting physical evidence. However, the recent cryo-electron microscopy study of in vitro assembled icosahedra of the RSV CA has documented its ability to form both pentameric assemblages and pentamer-hexamer interactions that are consistent with the evolving models of retroviral capsid structures.31
Among retroviral CA proteins, the structure of each of the two α-helical domains (the N-terminal NTD and C-terminal CTD), as well as the intersubunit contacts that form the protein shell, are highly conserved.7,20,21,29,32–42 The first three NTD helices establish the intra-hexameric and intra-pentameric interfaces.20,30,31 Neighboring capsomers are joined via dimeric contacts between CTDs mediated by the second helix of the interacting domains.20,21,43 An NTD-CTD interdomain contact was originally inferred by a genetic study of RSV44 and has since been supported by biochemical, biophysical and structural studies with HIV and RSV.7,8,20,31 The biological importance of the NTD-CTD interface is confirmed by two HIV inhibitors that bind to CA in the region involved in the interdomain contact.41,45–47
The assembly pathway that produces these interactions in the maturing virion has not been defined. In vitro conditions under which CA can assemble into structures related to capsids have been developed with the HIV and RSV CA proteins.7,18,21,48 In each case, sigmoidal kinetics consistent with a rate-limited, nucleation-driven process were observed.17,18,49 Typically such nucleation-driven events are distinguished mechanistically from linear (isodesmic) polymerization, in which the association rate for all subunits is equal, by three criteria – the existence of a lag period preceding detectable complex formation, the requirement for a critical protein concentration, and the involvement of a nucleating oligomer that seeds further assembly.50 In the case of CA assembly, the lag period has been described17,18,49 but direct evidence of a nucleating complex is lacking.
A detailed examination of the initiation of RSV CA assembly in vitro is presented below. Under experimental conditions in which protein assembly is triggered by multivalent anions such as phosphate, the three criteria of nucleation-driven polymerization are met. Small CA oligomers ranging in size from dimer to tetramer were purified and shown to stimulate the assembly of monomeric CA, documenting that an oligomer as small as a dimer participates in nucleation. This work provides the strongest evidence to date that capsid assembly in situ is likewise dependent upon formation of a unique nucleating species and describes in vitro conditions that may be used to define the intermediate steps of the assembly pathway.
Results
Effects of salts on CA assembly in vitro
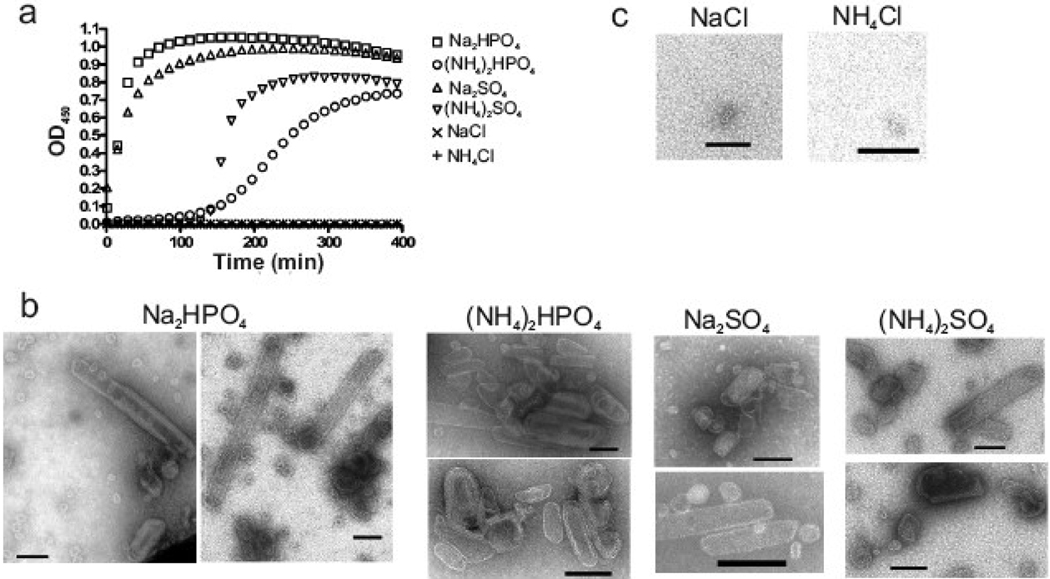
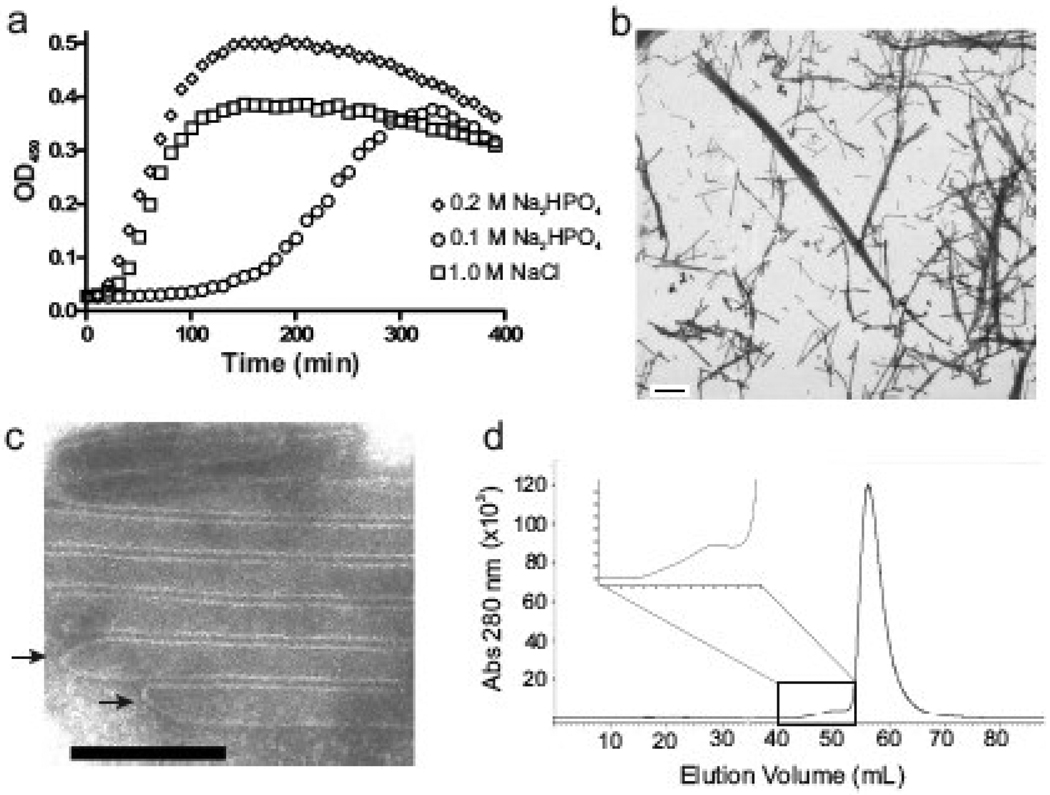
The RSV CA protein was purified from E. coli using a previously described protocol.18,51 Large planar arrays and small spheroidal complexes of CA protein were observed following ammonium sulfate [(NH4)2SO4] precipitation during purification (data not shown). Reasoning that protein assembly may have been triggered by ammonium sulfate, a turbidimetric assay was employed to test the influence of salts, pH and protein concentration on CA multimerization. With protein at 160 µM (4 mg/ml), the development of turbidity could be triggered by the addition of certain salts at 500 mM final concentration (Fig. 1a). In particular, sodium phosphate, ammonium phosphate, sodium sulfate, and ammonium sulfate, each of which contained a multivalent anionic group at the pH of 8.0 used in this experiment, caused an increase in optical density at 450 nm (OD450) (Fig. 1a). The phosphate salts were much more effective than the sulfate salts, in contrast to the recognized ability of these ions to cause salting-out of proteins,52 suggesting that phosphate has a specific influence on CA beyond altering protein solubility. Whereas sodium chloride and ammonium chloride at 500 mM failed to trigger any increase in turbidity (Fig. 1a) even after an extended incubation of 6 hours (data not shown).
Fig. 1.
Dibasic anion-induced multimerization of RSV CA. (a) CA protein was mixed with various salts at pH 8.0, and the resulting turbidity of the protein solution was followed by measurement of light scattering at 450 nm. The final concentrations were 160 µM protein and 500 mM salt. (b) EM analysis of the products formed in the turbid reactions containing phosphate and sulfate salts. (c) EM analysis of non-turbid samples containing chloride salts. All scale bars represent 100 nm.
To determine the nature of the light scattering material formed in the presence of divalent anion, precipitates were examined by negative staining and electron microscopy (EM). Each turbid sample contained organized structures that included angular capsid-like shells (Fig. 1b) similar to the irregular polyhedral capsids of native RSV virions4,53 as well as small spheres and tubular structures. We have previously shown that the same array of structures formed with CA assembled under slightly different conditions of pH 7.5 and lower protein concentration (Fig 3b–n of ref 18). The large angular, small spheroidal, and tubular shapes have each been observed with some frequency in authentic RSV particles4 and are consistent with formation of protein arrays in which the CA subunits are packed according to fullerene geometry.21,23,24 No clear bias for one type of structure or another was observed with any of the phosphate or sulfate salts. Consistent with the turbidity results, incubation of protein in ammonium chloride failed to induce assembly of any organized CA structures (Fig. 1c); similarly, examination of several grids from each of two independent reactions with sodium chloride identified only rare small spheroidal structures (Fig. 1c).
Fig. 3.
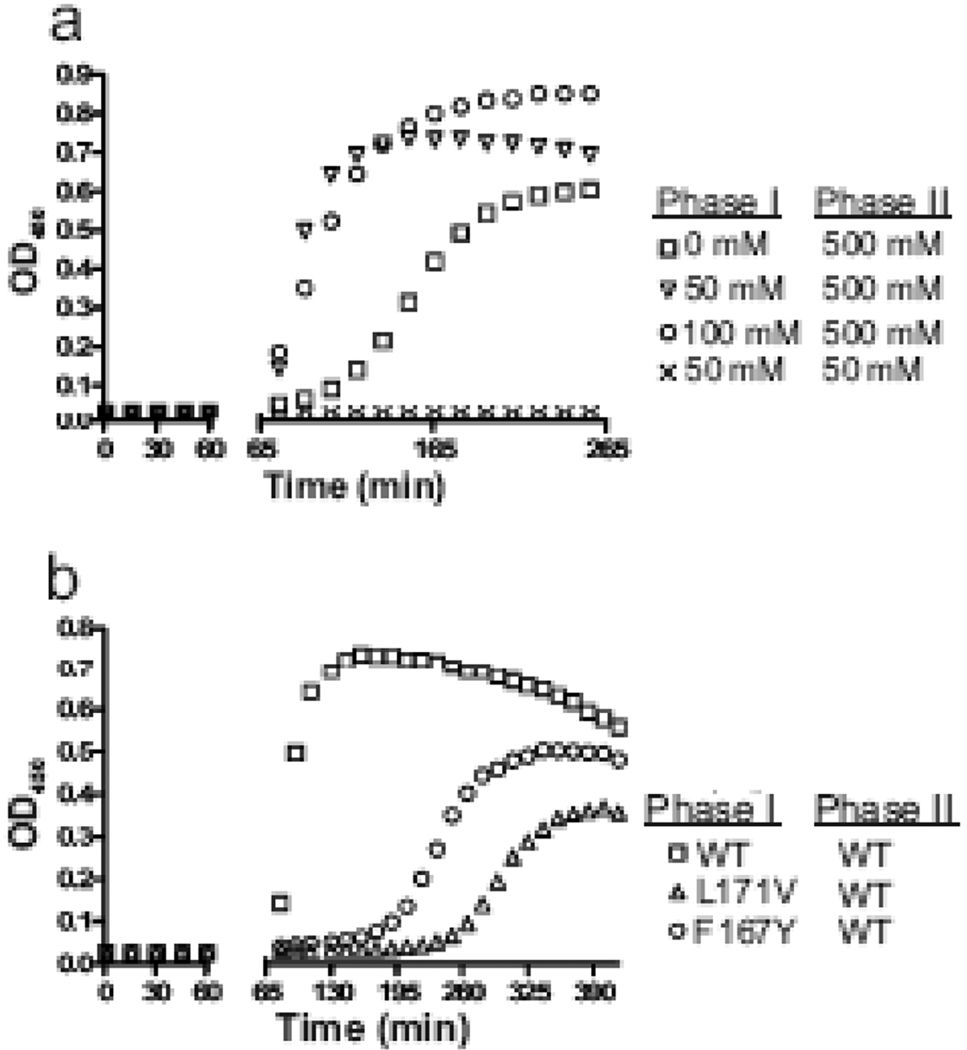
Low levels of sodium phosphate induce the formation of assembly seeds. (a) Monomeric RSV CA was incubated with 0, 50 or 100 mM sodium phosphate, pH 8.0, for 60 min (Phase I), after which the final sodium phosphate concentration was either raised to 500 mM or kept at 50 mM (Phase II). Additional protein was also added to maintain its concentration at 80 µM. (b) Two CTD CA mutations (F167Y and L171V) were tested for their influence on assembly initiation by incubating the mutant protein at 50 mM sodium phosphate pH 8.0 and then at 60 min spiking the reaction with additional salt and WT protein, as in part a.
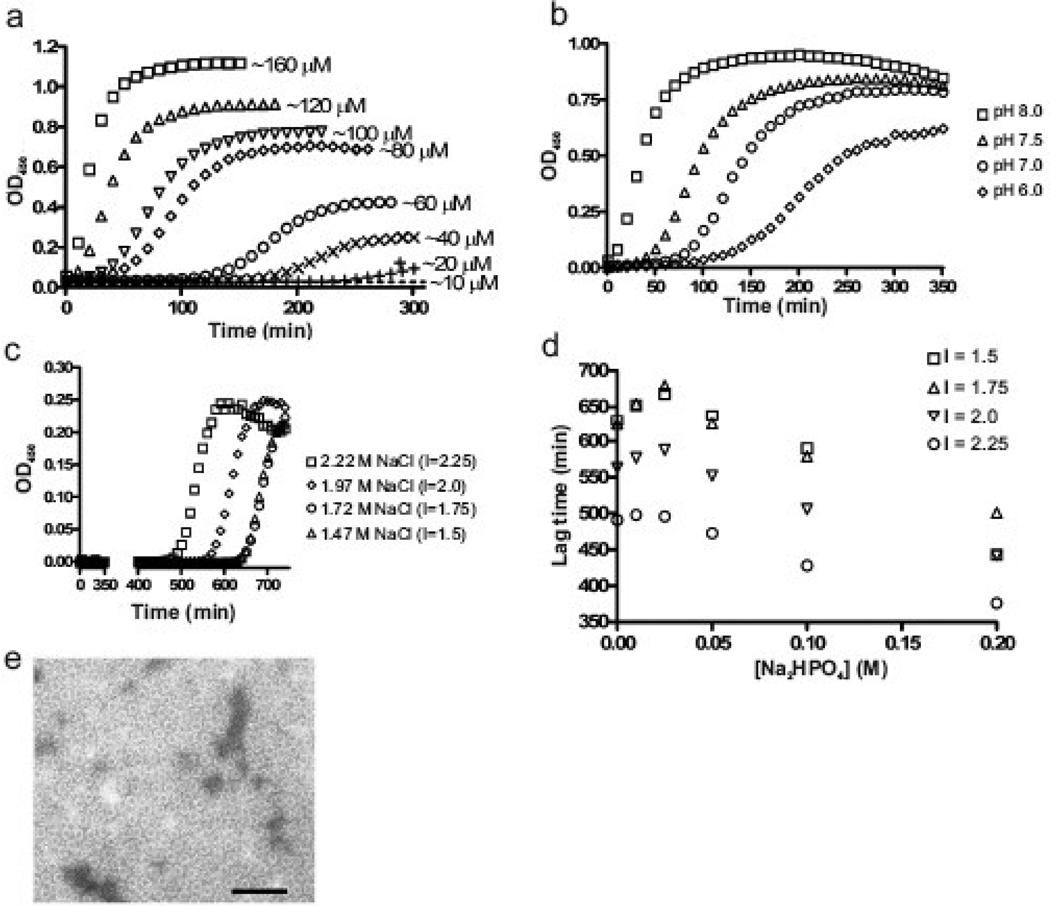
The extremely rapid assembly observed with sodium phosphate (Fig. 1a) was slowed when the protein concentration was reduced, and sigmoidal kinetics, with a clear lag followed by a rapid rise in turbidity, became evident (Fig. 2a). With 500 mM sodium phosphate to trigger assembly, the lag period increased and both the rate of assembly and the maximum turbidity decreased as the protein concentration was reduced over a range of 120 µM to 10 µM. Turbidity was undetectable in 300 min at the lowest concentration. We had previously shown that a small pool of CA protein remains soluble after microcentrifugation of turbid assembly reactions.18 Together these observations imply that assembly requires some critical concentration to proceed. To estimate this, samples containing 10–120 µM protein were initiated to assemble by addition of 500 mM sodium phosphate; 3–5 hrs after the maximal OD was reached the reactions were subjected to ultra-centrifugation. The pool of free CA that was resistant to pelleting was determined by absorbance at A280 to be ~ 15 µM (data not shown). This clear dependence of assembly upon protein concentration is consistent with a nucleation-driven process.54,55
Fig. 2.
Effects of protein concentration, pH, and ionic strength on CA protein assembly. (a) The turbidimetric profile of RSV CA protein ranging from 10 to 160 µM, induced to assemble by addition of 500 mM sodium phosphate at pH 8.0. (b) Influence of pH of the 500 mM sodium phosphate solution on assembly kinetics. The protein concentration was 120 µM. (c) Influence of ionic strength on assembly kinetics in 100 mM sodium phosphate, pH 8.0. The total ionic strength (I, as shown) was adjusted by the addition of sodium chloride. (d) Dependence of lag time on sodium phosphate concentration at various ionic strengths (I); for each curve, the sodium chloride level was decreased as phosphate increased to maintain constant ionic strength. The protein concentration in part c and d was 80 µM. (e) CA structures formed at low concentration of sodium phosphate. Monomeric CA at 400 µM was dialyzed against 50 mM salt at pH 8 and 4 °C until final protein concentration of 280 µM was reach and then examined by EM. Scale bar represents 100 nm.
Synergy of phosphate concentration and total ionic strength
Phosphate-driven assembly reactions were also sensitive to pH. The overall reaction kinetics increased as the pH of the sodium phosphate solution was increased from 6.0 to 8.0 (Fig. 2b). This is consistent with the observation that assembly is dependent upon the presence of multivalent ions since raising the pH of sodium phosphate drives the phosphate group towards a divalent state and further supports that in vitro CA multimerization is electrostatically-controlled. The effectiveness of 500 mM sodium phosphate in triggering assembly may reflect the contributions of both the dibasic anion as well as the overall ionic strength of the reactions solution.
To dissect the relative importance of these two variables, both the phosphate concentration and the total ionic strength (controlled by addition of sodium chloride) were varied systematically. In a typical experiment where sodium phosphate was held constant at 100 mM, pH 8, increasing the sodium chloride level caused a progressive shortening of the lag (Fig. 2c). The combined effects of the two variables are displayed in Fig. 2d where each curve illustrates the effect upon the lag of increasing phosphate while keeping the total ionic strength constant. Regardless of the total ionic strength, the lower limit at which the assembly begins (i.e., the calculated lag period begins to decrease) is ~ 25–50 mM phosphate. Without sodium phosphate a detectable turbidity occurred, but only after an extremely prolonged incubation (500–600 min) at very high concentrations of sodium chloride (1.5–2.25 M, Fig 2d).
To assess the formation of organized structures at low levels of both phosphate and ionic strength, monomeric CA was dialyzed against 50 mM sodium phosphate pH 8.0 at 4 °C for approximately 4 hours. EM examination showed the presence of a homogenous population of small spheroidal structures with a diameter of less than 20 nm (Fig. 2e). No larger tubular or polyhedral structures similar to those shown in Fig. 1b were observed under these conditions. We conclude that 50 mM sodium phosphate can trigger oligomerization of monomeric CA but the phosphate anion and total ionic strength are synergistic in supporting the formation of larger CA assemblies, including the capsid-like structures described above.
The hypothesis that 50 mM sodium phosphate may favor the formation of CA oligomers that are assembly-competent was tested directly by incubating 80 µM monomeric CA in the presence or absence of sodium phosphate at 50 mM and pH 8.0 for one hour before the sodium phosphate concentration was raised to 500 mM (Fig. 3a). Reactions that contained 50 or 100 mM sodium phosphate during the first incubation showed immediate assembly without an observable lag in the second phase. In contrast reactions that were not exposed to phosphate during pre-incubation developed turbidity only after a clear lag. When the sodium phosphate level was kept constant at 50 mM during the second phase, no increase in OD450 occurred. This result confirms that low levels of sodium phosphate can trigger the formation of nuclei that act without delay to seed the assembly of the monomer in phase 2.
Characterization of CA oligomers
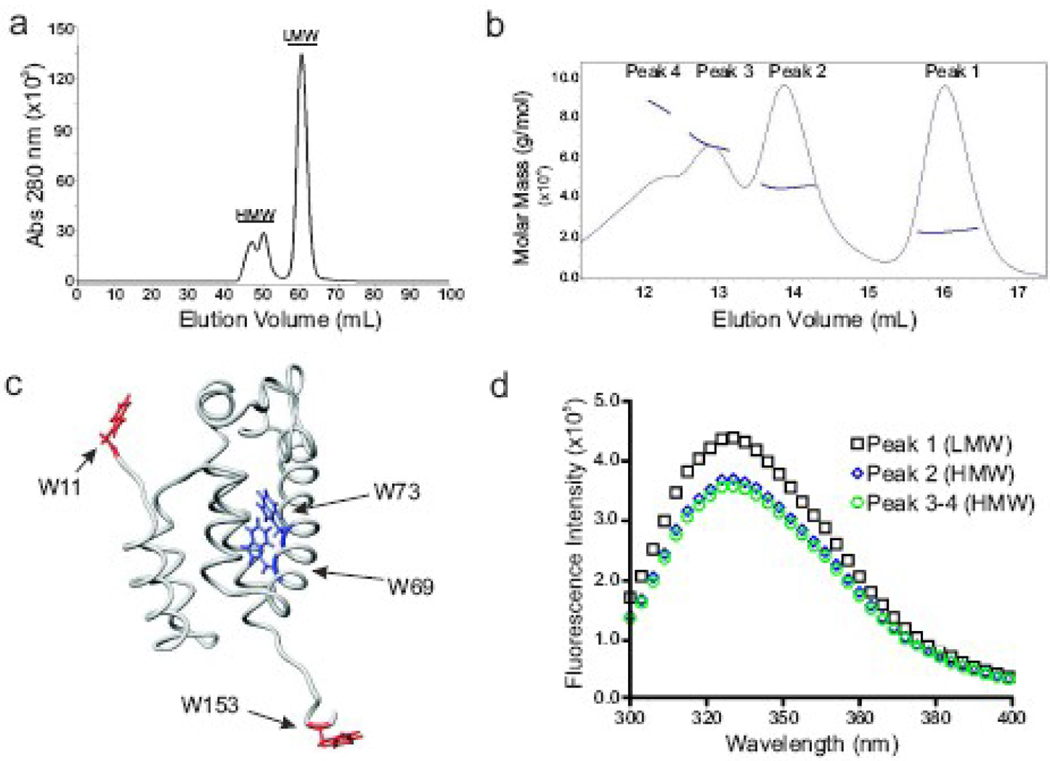
During purification two distinct populations of CA protein were observed by size exclusion chromatography (SEC) on Superdex S75 following separation by ion exchange chromatography (Fig. 4a). The majority of the protein eluted in a single peak with an apparent mass similar to the 25.5 kD expected of a CA monomer (Fig. 4a, labeled LMW). The remaining protein eluted as a cluster of peaks of larger mass (Fig. 4a, labeled HMW). Mass spectrometry revealed that both the LMW and HMW fractions contained a protein of 25.5 kD (specifically 25522 Daltons for LMW and 25508 Daltons for HMW), confirming that the protein was in fact authentic CA and that the vector-derived initiating methionine was removed. Proteins in the LMW and HMW fractions were indistinguishable by circular dichroism, exhibiting the expected high content of alpha helix and arguing against the possibility that the HMW fraction consisted of grossly misfolded aggregates (data not shown). Previous studies of the RSV CA protein detected no tendency of the protein to dimerize in vitro.38,56 The features of the purification protocol used here that allowed oligomerization are not clear but may potentially relate to the use of a ZYP-5052 autoinduction growth medium57 for E. coli to generate a high protein yield.
Fig. 4.
Identification and characterization of CA oligomers. (a) Size exclusion chromatography (SEC) on a preparative grade Superdex 75 column shows two distinct populations of protein, high molecular weight (HMW) and low molecular weight (LMW). (b) Analytical SEC and multi-angled light scattering (MALS) analysis of the HMW fraction. The molar mass of each peak was determined: peak 1 (monomer), peak 2 (dimer), peak 3 (trimer), and peak 4 (tetramer plus larger complexes). (c) The four tryptophan residues within in CA are shown on the structure of the NTD and linker region of RSV CA (PDB: 1d1d). Note that the protein in this model does not contain the β-hairpin structure at the N-terminus. The residues found in a hydrophilic region are red, and residues found within the hydrophobic environment are blue. (d) Intrinsic tryptophan fluorescence of the protein in the LMW and HMW fractions.
SEC coupled with dynamic multi-angle light scattering (MALS) was used to independently measure the oligomeric state of the proteins found within the S75 HMW fraction.58 The HMW fractions from multiple independent protein purifications were pooled, concentrated and analyzed by analytical SEC (Superdex 200) with on-line MALS (Fig. 4b). The protein eluted in four peaks with apparent masses of 22.5 kD (Peak 1 / monomer), 44.6 kD (Peak 2 / dimer), 66.1 kD (Peak 3 / trimer), and 84.8 kD (Peak 4 / tetramer). The pronounced leading edge of peak 4 suggests the presence of an oligomeric form larger than a tetramer. These numbers confirm the existence of discrete CA oligomers in the HMW material. Furthermore, the appearance of monomeric CA in the S200/MALS profile indicates that the HMW oligomers have a tendency to dissociate. The difference in the mass of the monomer measured by MALS and by MS is likely due to inaccuracies in the determination of protein concentration in each peak, a variable that often limits the accuracy of MALS59,60 especially at the low protein levels that occurs in these samples.
Intrinsic tryptophan fluorescence was utilized to probe for conformational differences between the monomer and oligomers. In the monomer two tryptophan residues, W69 and W73, are located within a hydrophobic pocket near the NTD-CTD interdomain interface (Fig. 4c). Two others lie in the β-hairpin and in the flexible linker connecting the NTD and CTD.56 The peak fluorescence signal of monomeric CA occurred at 325 nm (Fig. 4d), suggesting that residues within a hydrophobic environment, i.e., the W69 and W73, are responsible for the majority of the signal.61 The HMW oligomers, in comparison to the monomer, exhibited a ~30% decrease in overall intensity with no apparent shift in peak wavelength, indicating a change in the environment of the NTD hydrophobic pocket. Whether due to a change in conformation of each monomer or the effects of inter-subunit docking, this observation provides suggestive evidence that the NTD is involved in oligomerization.
Assembly activity of the CA dimer
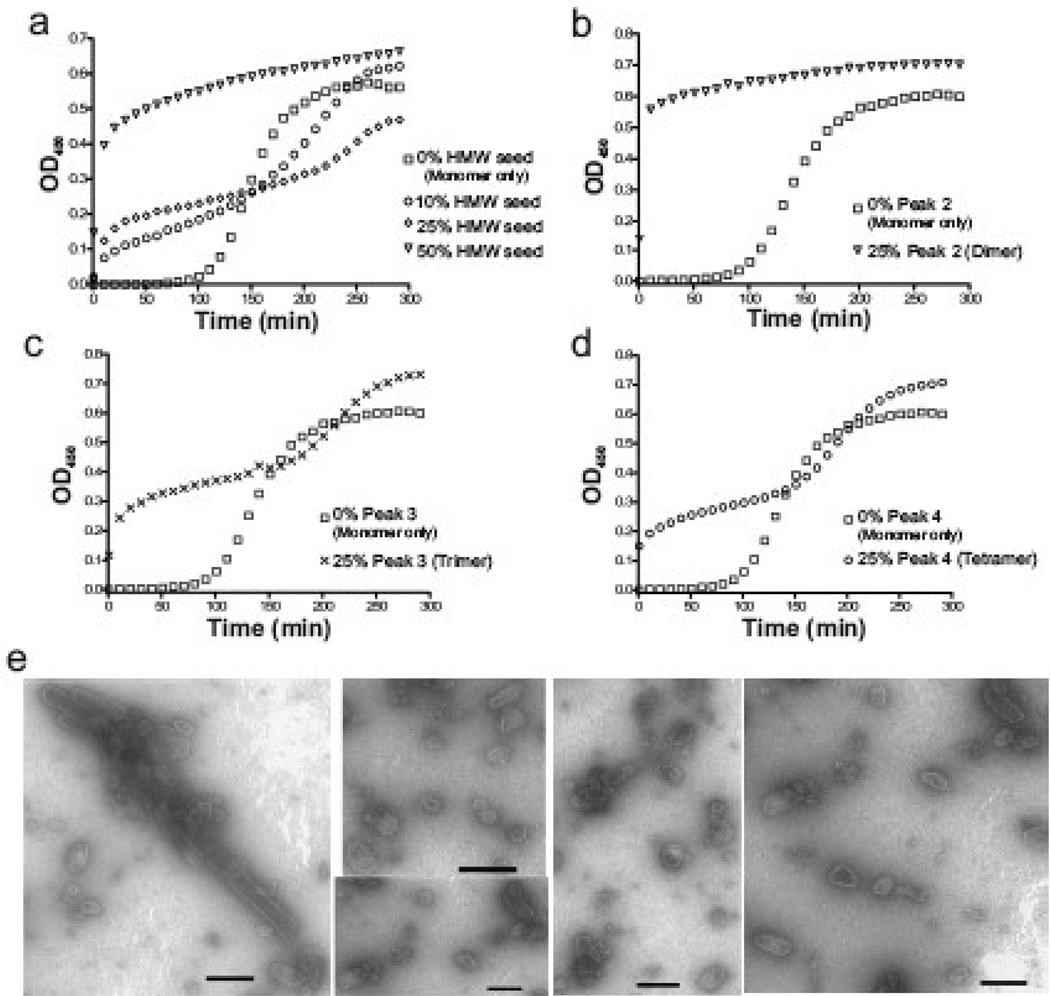
The hypothesis that the HMW oligomeric proteins represent on-pathway assembly intermediates was tested by adding the HMW oligomers to monomeric protein prior to the addition of sodium phosphate. When 50% (40 µM) of the total protein was added as HMW protein, the assembly proceeded very rapidly; the lag characteristic of monomer assembly disappeared (Fig. 5a). At lower ratios of HMW oligomers (10 or 25% of the total protein), the assembly occurred in three phases: 1) a rapid phase with an immediate increase in turbidity that is limited by depletion of the oligomers that seeded the initial burst of activity, 2) a slow growth phase and 3) a final growth phase with kinetics more like that of a monomer-only reaction. These results confirm that assembly-competent oligomers that can seed the formation of large CA structures are present in the HMW fraction.
Fig. 5.
Assembly seeding by the addition of the oligomeric CA. (a) Assembly kinetics of reactions containing monomeric CA mixed with protein from the HMW fraction. The total protein concentration was held constant at 80 µM and HMW protein was added at 8, 20, and 40 µM. (b–d) Each individual peak identified in Fig 4b was used to seed capsid assembly. Again, the total protein concentration was held at 80 µM. (e) EM micrographs of products of the peak 2 (dimer) seeded reaction (part b) with scale bars representing 100 nm.
To determine which HMW oligomer was responsible for nucleating assembly, reactions were seeded with protein from peak 2, 3 or 4. When a quarter of the total protein was contributed by peak 2 CA (dimer), assembly proceeded rapidly with no detectable lag, similar to that of the 50% HMW reaction (Fig. 5b). Reactions seeded with peak 3 or 4 were multiphasic, similar to those set up with small amounts of the mixed oligomers (Fig. 5c–d). The structures that formed by seeding with protein from either peak 2, 3 or 4 showed the same types of structures observed under standard monomer-only assembly conditions although with a somewhat reduced abundance of tubes (Fig. 5e). The results clearly show that the CA oligomers can function to nucleate in vitro capsid assembly with the dimer being most efficient. The multiphasic behavior observed with peak 3 and peak 4 protein likely reflects the addition of some active oligomers that immediately initiate assembly; sustained progress, however, requires de novo formation of additional oligomers from the monomers, hence the intermediate lag.
CTD involvement in early capsid assembly
The importance of the CTD for nucleation was tested using two well characterized CA mutants, F167Y and L171V, that have been previously proposed to cause a defect at initiating capsid assembly.18,44,62–64 Both substitutions alter conserved hydrophobic residues, result in non-infectious viruses with defects in core integrity,62,63 and prevent phosphate-triggered assembly in vitro.18 The opposing effects on in vitro assembly kinetics of the lethal mutations (i.e. an extremely prolonged lag) and second-site compensatory substitutions (i.e. rapid assembly kinetics) provides further evidence that the crippling effect of F167Y and L171V may be due to prevention of the nucleation step.18 When mixed with wild type (WT) protein the mutant CA proteins were co-assembled without altering the lag time, suggesting that the mutant were able to participate in the growth phase but not nucleation.18 Action of the lethal mutations on the nucleation step was tested directly in the two-phase assembly reaction described above (Fig. 3b). The mutant proteins were pre-incubated in low phosphate prior to the addition of monomeric WT protein and sodium phosphate at 500 mM. Both mutant proteins failed to cause the same immediate stimulation of assembly that was seen with the WT protein. Instead, the assembly detected after a considerable lag is likely nucleated by the WT protein added during the second phase. This confirms earlier interpretations that the integrity of the CTD is critical for the formation of the initiating oligomer.18 The deficit caused by the F167Y and L171V mutations may be due to influences on either the intradomain (CTD-CTD) and/or the interdomain (NTD-CTD) interface. However, the fact that both can be compensated by a second-site change in the NTD leads us to favor the interdomain interface.
HIV CA assembly in phosphate
HIV CA protein, unlike RSV CA protein, efficiently assembles under conditions of high sodium chloride concentration (1 – 2.5 M)21,48,49,65 but shows a marked preference for tube formation. We examined the assembly of monomeric HIV CA (Fig. 6a) under the conditions used here for the RSV protein. Assembly of the HIV protein at 80 µM occurred efficiently in 100 mM sodium phosphate, and the reaction exhibited sigmoidal kinetics with a discernable lag phase similar to that observed with the RSV protein in slightly higher phosphate. HIV CA showed a critical concentration of ~10 µM under 300 mM sodium phosphate assembly conditions (data not shown). Unlike RSV CA, however, the HIV protein formed almost exclusively long tubular structures which had an average diameter of ~45 nm (Fig. 6b–c) and resembled structures reported by others formed by high concentrations of sodium chloride.21,22,49,65 Some of the tubular polymers appeared to be capped (Fig. 6c) with structures that resembled the narrow ends of HIV cones. Finally, during SEC purification of the HIV protein, a small amount of protein eluted more rapidly than the monomeric CA (Fig. 6d). This is expected to represent dimers or higher oligomers, analogous to the RSV HMW fraction. However, these oligomers have not been obtained in sufficient quantity to test their ability to seed a monomer reaction.
Fig. 6.
Assembly of HIV CA. (a) Turbidimetric profile of monomeric HIV CA at 80 µM induced to assemble by 100 and 200 mM sodium phosphate or by 1.0 M NaCl, pH 8.0. (b) Assembled protein (at 200 mM sodium phosphate pH 8.0) examined at low magnification (2750x). The scale bar represents 1000 nm. (c) Higher magnification (35500x) of protein assembled at 250 mM sodium phosphate pH 8.0. The scale bar represents 100 nm. Arrows mark the closed ends of tubular structures. (d) SEC using a S75 preparative column shows the presence of HMW oligomers (inset) and LMW monomeric HIV CA protein.
Discussion
This study has defined the parameters that affect the initiation of in vitro assembly of capsid-like structures by the CA protein and provides important new insights into the potential mechanisms governing capsid formation in the maturing virion. Assembly of the RSV CA can be triggered by the multivalent anions sulfate and phosphate, with the latter causing a much more rapid assembly. The resulting structures are diverse in shape and include capsid-like polyhedral structures as well as tubes, small spheres and multilamellar structures. This range of morphologies suggests that phosphate-mediated assembly conditions support the formation of both pentameric and hexameric associations of CA monomers. Structural determination of two classes of small CA spheres by cryo-EM has confirmed this interpretation and provided the first view of a retroviral CA protein in pentameric symmetry and of pentamer-hexamer interactions.31
Phosphate-driven in vitro assembly of the RSV CA protein, followed turbidimetrically, provides a highly reproducible system and allows the quantitative assessment of reaction kinetics and the requirements for the initiation of assembly. The kinetic data we have obtained demonstrate that in vitro assembly of the RSV protein is a well-behaved nucleation-driven process. In particular, the phosphate-driven reactions exhibit a pronounced lag period which indicates the existence of an essential rate-limiting step, a critical protein concentration below which assembly fails to progress, and the ability of pre-formed oligomers to seed the assembly process effectively overcoming the rate-limited step. In this way retroviral capsid assembly shares some similarities with the processes that produce the icosahedral capsids of certain other viruses, such as bacteriophage P22 and polyoma virus.66–68
The in vitro behavior of mutant CA proteins described previously18 provides a strong argument that capsid assembly in the maturing virion is also nucleation-driven. Substitutions that alter highly conserved residues in the CTD and cripple capsid integrity in virions also interfere with nucleation of assembly in vitro (Fig. 3b). Second-site compensatory mutations in CA that restore capsid formation [Butan and Steven, unpublished] and correct the replication defect in virions44,64 also overcome the block to CA nucleation in vitro.18 These opposing effects of lethal and compensatory mutations on in vitro nucleation-driven assembly provide strong biochemical evidence in support of the proposal that capsid assembly begins in situ, as it does in vitro, with a specific nucleation event and proceeds through stepwise growth.2–4
Electrostatic control of capsid assembly
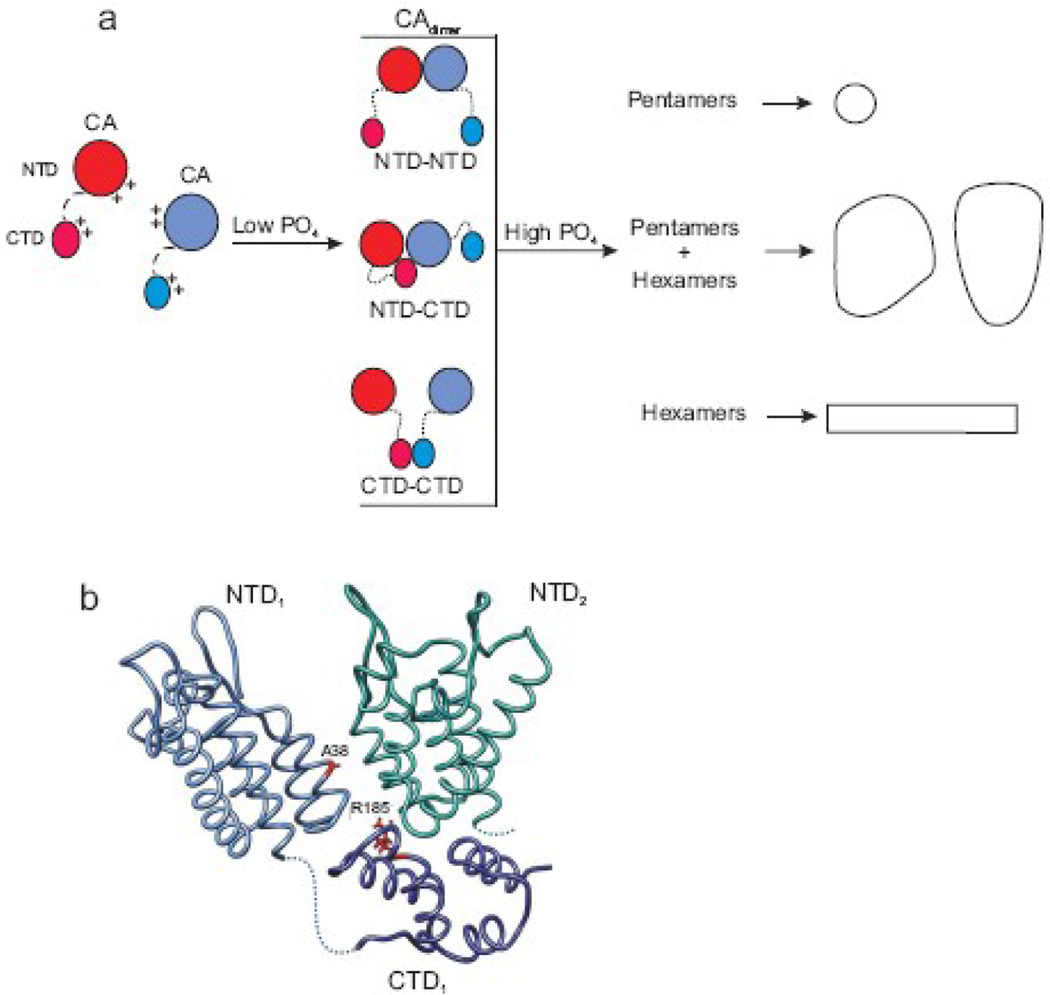
The strong requirement for a multivalent anion such as phosphate to stimulate efficient in vitro assembly suggests that shielding of basic residues in CA is needed in order to form assembly-competent oligomers that are presumably associated via some combination of the known NTD-NTD, CTD-CTD and NTD-CTD interfaces. Calculations using standard parameters in GRASP69 of RSV CA reveals a large net positive charge covering most of the surface of the NTD, whereas the CTD contains only small localized patches of both positive and negative charges (Maria Bewley, pers. comm.). Therefore, we suggest that phosphate ions at relatively low concentrations neutralize charge repulsion at one or more key points on the NTD to trigger assembly initiation (Fig. 7a). Continued growth by the formation of the full range of contacts needed to build a mature capsid requires conditions with a higher ionic strength. This is consistent with altered in vitro assembly rates of HIV mutants within a highly charged region of the NTD which independently led to the proposal that this domain is involved in a nucleation step.19 This interpretation receives support from the cryo-EM analysis of icosahedrally-organized RSV CA which identified a clustering of basic residues at the NTD-NTD and NTD-CTD interfaces that would require shielding in order to allow intersubunit contacts to form.31 The ability of the R185W charge-neutralization substitution in the interfacial region to reduce phosphate dependence and enhance nucleation of assembly is likewise consistent with electrostatic regulation of capsid assembly.18,31
Fig. 7.
Electrostatic control of CA assembly. (a) A schematic model of three possible dimeric (CAdimer) initiating complexes formed under low sodium phosphate levels. Multivalent anions favor the dimerization of CA monomers via NTD-NTD, NTD-CTD or CTD-CTD interactions by shielding specific positively charged residues. Additional salt is required to overcome electrostatic repulsion, allowing the full range of interactions and further multimerization into large structures. (b) The salt dependence of assembly, tryptophan fluorescence of dimeric CA, and modulation of nucleation by mutations suggest a model for the NTD-CTD interaction that initiates CA assembly, based upon Cardone et al.31 The NTD and CTD domains of one monomer are in blue with dotted line representing the interdomain region. The neighboring NTD is in green; its CTD is omitted for clarity. The locations of the compensatory substitutions A38V and R185W, mentioned in the text, are illustrated
The inorganic phosphate salts used to trigger assembly in vitro presumably act as a surrogate for other polyanionic compound(s) in the maturing virion. The candidates inside the particle include viral genomic RNA and cellular RNAs, acidic phospholipids, other envelope constituents and possibly nucleotides or other small molecules. The organization of the maturing virion with the membrane on the outside and RNA inside the nascent capsid shell may provide an ideal three-dimensional charge-stratification that serves to facilitate proper assembly. Three tomographic studies of RSV and HIV particles have each argued for a role of virion constituents in addition to the CA protein itself in the initiation of capsid formation during maturation.2–4 In particular, the model for HIV cone assembly described by Briggs et al.3 – specifically, assembly beginning with creation of the narrow end at the viral membrane, continuing with formation of the hexagonal lattice of the sides, and finishing with closure of the shell when the growing cone approaches the distant side of the membrane – is appealing because it predicts a role for the membrane in directing pentamer insertion at the two poles. Tomographic analysis of RSV virions led to the prediction that the membrane or the Env glycoprotein plays a role in controlling capsid morphology.4 The absence of such regulating features of virion structure may explain the wider distribution of structures produced in vitro compared to in situ RSV capsids; polyhedral shapes are less common and spheres and rods more abundant in vitro than in virions.4,18
The assembly of HIV CA is also strongly dependent upon pH and electrostatic influences.19,49,70–73 The marked preference for the formation of tubular protein arrays rather than the cones that predominate in virions suggests that the HIV protein favors the formation of hexamers over pentamers under the conditions studied. We have shown here that assembly of the HIV CA can be triggered by sodium phosphate at relatively low concentrations, but unlike the RSV protein, the resulting structures are almost entirely tubes similar to those that assemble in high concentrations of sodium chloride.21,48,49 Thus, the sodium phosphate assembly conditions remain an insufficient substitute for some feature of the virion that is needed to facilitate the formation of CA pentamers and the conical HIV capsid.
Potential role of a CA dimer in nucleating assembly
The data presented here show that the RSV CA protein is capable of existing as a dimer and suggest that its tendency to do so in vitro is favored by the presence of divalent anions. Furthermore, the dimer immediately stimulates assembly when mixed with monomeric protein in the presence of phosphate ions, arguing that the dimer is involved in the rate-limited assembly step detected by these kinetic experiments. In contrast, the HIV CA protein readily dimerizes in vitro,34,74 but no assembly-active oligomers have been isolated as yet nor has the structure of the dimer of full-length CA been determined. Whether in the maturing retrovirion a dimeric form of CA is released from Gag by proteolysis or whether CA exists transiently as a monomer that must undergo refolding events in its NTD and/or CTD to activate its dimerization activity is unknown at present. Either scenario is consistent with the data presented here that indicate a key role of the dimer in initiating capsid assembly. Dimerization itself or the addition of the next subunit to the dimer may be the actual rate-limiting event detected in vitro.
The precise intermolecular interfaces through which the assembly-competent dimer forms are undefined. However, two observations suggest that formation of the NTD-CTD interface is a critical step that occurs early on the pathway of capsid assembly. The difference in the intrinsic tryptophan fluorescence of monomers and oligomers suggests that the neighborhood around W69 and W73 (on helix 4 in the hydrophobic core of the NTD) undergoes a slight change of environment upon formation of the assembly-active dimers. Such a shift would not be expected if the sole point of contact in formation of the dimer is via the CTDs. Furthermore, the two lethal substitutions in the CTD that cause a failure of CA assembly nucleation (Fig. 3b) are corrected by a compensating change (A38V) that lies at the NTD-NTD interface.18,31,64 Therefore, we suggest that the NTD-CTD interdomain interface described in RSV pentamers, in which the CTD docks beneath the intersection of two NTDs (Fig. 7b) is a likely model for the contact site in the assembly active dimer. This tripartite interface is a slight variation of the interdomain contacts in HIV CA hexamers but invokes interaction of the CTD with its own NTD as well as with the NTD of the adjacent subunit in the pentamer.
Genetic analyses and inhibitor-binding studies with HIV have documented the essential nature of the NTD-CTD contacts to capsid functionality.20,41,45–47,75 However this study is the first to propose an indispensable role for the NTD-CTD interface specifically in the initiation of capsid assembly. Ultimately, understanding the entire pathway of capsid morphogenesis in the maturing virion will be needed for the further development of maturation inhibitors and for understanding the mechanisms of resistance to such drugs. This work demonstrates that the capsid assembly pathway may be defined via in vitro approaches. In particular, the determination of high-resolution structures of oligomeric intermediates in capsid assembly and further definition of the roles of other virion constituents in the process will be critical for the development of a complete model for maturation.
Materials and Methods
Protein expression and purification
RSV CA protein was expressed in Escherichia coli (BL21 DE3) using a pET-24(+) plasmid and purified by ammonium sulfate precipitation, cation exchange (DEAE) and size exclusion chromatography (SEC) as previously described.18 For most experiments protein from 3 or more preparations were used. The CA oligomers were isolated by pooling and concentrating the HMW fraction from ~20 independent protein preps. HIV CA protein (p24) was similarly purified using the pWISP98-85 plasmid.76 The following extinction coefficient values were used for the RSV CA: 24980 M−1cm−1 for WT, L171V, and 26470 M−1cm−1 for F167Y.77 An extinction coefficient value of 33585 M−1cm−1 was used for HIV CA. The structural images of CA shown in Fig 4c and Fig 7b were made using Chimera.78
In vitro capsid assembly
Capsid assembly was monitored by optical density at 450 nm (OD450) using a spectrophotometer as previously described.18 UV-Transparent 96-well microplates (BD Falcon and Corning) were used for 100 µl reactions and UV-Transparent half area 96-well microplates (Corning) were used for 50 µl reactions. To circumvent capsids from settling at the bottom of the well the reactions were mixed for 3 sec prior to each reading. Typical protein concentrations used for assembly were 80 µM (2 mg/ml) to 160 µM (4 mg/ml). CA protein was induced to assemble by the addition of salts. For most reactions sodium phosphate pH 8.0 was utilized. When other salts were used the pH was controlled by dissolving the salt in 20 mM Tris-HCl pH 8.0. Determination of the lag time was done using the modified Gompertz equation.79 Unless otherwise noted all assembly reactions were performed at room temperature. For clarity, only one in twenty data points are shown on most of the graphs and only a representative graph is shown from multiple repeated experiments.
Electron microscopy (EM)
Capsids were assembled as described above except the reactions were mixed for 3 sec every 2.5 min. Upon reaching the maximum OD450 the products of the in vitro assembly reactions were removed from the spectrophotometer and allowed to settle overnight at 4 °C prior to being analyzed by EM. The samples were applied to a formvar-coated grid, washed with water or 0.1 M KCl, stained with 2% uranyl acetate and visualized using a 120 kV TEM (Phillips) operating at 60 kV. Phosphotungstic acid was used in place of uranyl acetate for samples containing sodium sulfate.
Critical concentration determination
The assembly reactions were carried out as described above. The final sodium phosphate concentration for RSV CA was 500 mM and 300 mM for HIV CA protein. The protein concentration varied from 10 to 160 µM. The reactions were allowed to proceed for an additional 3 to 5 hours after reaching the maximum OD450. To decrease evaporation during the long incubations the plates were covered with sealing tape. The samples were centrifuged in an ultracentrifuge at 66,000 × g for 30 min at 20 °C. The protein concentration remaining in the supernatant was determined.
Multi-angle light scattering (MALS)
Determination of protein mass was performed by SEC coupled MALS at room temperature. Oligomeric protein (HMW fractions) from multiple protein preps were pooled and concentrated. The protein was applied to a Superdex 200 HR 10/300 GL column (Amersham Pharmacia) equilibrated in 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1 mM EDTA, and 0.1% sodium azide. The scattered intensity of the complexes eluted off the column was recorded at 18 angles using a Dawn Heleos II multiangle laser light scattering detector (Wyatt Technology). The protein concentration in each complex was monitored by changes in the refractive index using an Optilab rEX refractive index detector (Wyatt Technology). The molecular weight of each complex in the various chromatographic peaks was determined using the ASTRA software (Wyatt Technology).
Circular dichroism spectroscopy and intrinsic tryptophan fluorescence
Protein was diluted in 50 mM sodium phosphate pH 7.5, 50 mM NaCl, and 0.1 mM EDTA to a final concentration of 0.1 mg/ml. A Jasco J-710 spectropolarimeter operating in the far-UV region of the spectrum was used for circular dichroism. Intrinsic tryptophan fluorescence was performed using a 290 nm excitation wavelength on a PTI QuantaMaster luminescence spectrometer.
Acknowledgements
Our grateful appreciation is extended to Roland Meyers of the Penn State College of Medicine core facilities for assistance with EM analyses, to J. Bernard Heymann, Giovanni Cardone and Alasdair Steven for insights into the intersubunit interactions, to Eric Barklis for supplying plasmid, and to Maria Bewley for helpful discussions and Grasp modeling. Funding for this research was provided by the NIH grant CA100322 (R.C.C.), the Pennsylvania Department of Health (R.C.C., J.M.F., and I.J.R.) and the Penn State Cancer Institute (R.C.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus H, editors. Retroviruses. Woodbury, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 2.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J. Mol. Biol. 2005;346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs JA, Grunewald K, Glass B, Forster F, Krausslich HG, Fuller SD. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Butan C, Winkler DC, Heymann JB, Craven RC, Steven AC. RSV Capsid Polymorphism Correlates with Polymerization Efficiency and Envelope Glycoprotein Content: Implications that Nucleation Controls Morphogenesis. J. Mol. Biol. 2008;376:1168–1181. doi: 10.1016/j.jmb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 7.Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE. Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J. Mol. Biol. 2003;325:759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- 8.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE., Jr Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 2004;11:676–677. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- 9.Lanman J, Prevelige PE., Jr Kinetic and mass spectrometry-based investigation of human immunodeficiency virus type 1 assembly and maturation. Adv. Virus Res. 2005;64:285–309. doi: 10.1016/S0065-3527(05)64009-2. [DOI] [PubMed] [Google Scholar]
- 10.Ako-Adjei D, Johnson MC, Vogt VM. The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles. J. Virol. 2005;79:13463–13472. doi: 10.1128/JVI.79.21.13463-13472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briggs JAG, Johnson MC, Simon MN, Fuller SD, Vogt VM. Cryo-electron Microscopy Reveals Conserved and Divergent Features of Gag Packing in Immature Particles of Rous Sarcoma Virus and Human Immunodeficiency Virus. J. Mol. Biol. 2006;355:157–168. doi: 10.1016/j.jmb.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov D, Tsodikov OV, Kasanov J, Ellenberger T, Wagner G, Collins T. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc. Natl. Acad. Sci USA. 2007;104:4353–4358. doi: 10.1073/pnas.0609477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nandhagopal N, Simpson AA, Johnson MC, Francisco AB, Schatz GW, Rossmann MG, Vogt VM. Dimeric Rous Sarcoma Virus Capsid Protein Structure Relevant to Immature Gag Assembly. J. Mol. Biol. 2004;335:275–282. doi: 10.1016/j.jmb.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 14.Phillips JM, Murray PS, Murray D, Vogt VM. A molecular switch required for retrovirus assembly participates in the hexagonal immature lattice. EMBO J. 2008;27:1411–1420. doi: 10.1038/emboj.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilk T, Gross I, Gowen BE, Rutten T, de Haas F, Welker R, Krausslich HG, Boulanger P, Fuller SD. Organization of immature human immunodeficiency virus type 1. J. Virol. 2001;75:759–771. doi: 10.1128/JVI.75.2.759-771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrera FN, del Alamo M, Mateu MG, Neira JL. Envelope lipids regulate the in vitro assembly of the HIV-1 capsid. Biophys. J. 2008;94:L8–L10. doi: 10.1529/biophysj.107.118083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purdy JG, Flanagan JM, Ropson IJ, Rennoll-Bankert KE, Craven RC. Critical role of conserved hydrophobic residues within the major homology region in mature retroviral capsid assembly. J. Virol. 2008;82:5951–5961. doi: 10.1128/JVI.00214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douglas CC, Thomas D, Lanman J, Prevelige PE. Investigation of N-terminal domain charged residues on the assembly and stability of HIV-1CA. Biochemistry. 2004;43:10435–10441. doi: 10.1021/bi049359g. [DOI] [PubMed] [Google Scholar]
- 20.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of Full-Length HIV-1 CA: A Model for the Mature Capsid Lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 22.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly Properties of the Human Immunodeficiency Virus Type 1 CA Protein. J. Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 24.Heymann JB, Butan C, Winkler DC, Craven RC, Steven AC. Irregular and semi-regular polyhedral models for Rous sarcoma virus cores. Comput. Math. Meth. Medicine. 2008;9:197–210. doi: 10.1080/17486700802168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen TT, Bruinsma RF, Gelbart WM. Continuum Theory of Retroviral Capsids. Phys. Rev. Lett. 2006;96:078102(1)–078102(4). doi: 10.1103/PhysRevLett.96.078102. [DOI] [PubMed] [Google Scholar]
- 26.Ganser BK, Cheng A, Sundquist WI, Yeager M. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J. 2003;22:2886–2892. doi: 10.1093/emboj/cdg276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayo K, McDermott J, Barklis E. Hexagonal organization of Moloney murine leukemia virus capsid proteins. Virology. 2002;298:30–38. doi: 10.1006/viro.2002.1452. [DOI] [PubMed] [Google Scholar]
- 28.Mayo K, Huseby D, McDermott J, Arvidson B, Finlay L, Barklis E. Retrovirus capsid protein assembly arrangements. J. Mol. Biol. 2003;325:225–237. doi: 10.1016/s0022-2836(02)01176-2. [DOI] [PubMed] [Google Scholar]
- 29.Mortuza GB, Haire LF, Stevens A, Smerdon SJ, Stoye JP, Taylor IA. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature. 2004;431:481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
- 30.Mortuza GB, Dodding MP, Goldstone DC, Haire LF, Stoye JP, Taylor IA. Structure of B-MLV capsid amino-terminal domain reveals key features of viral tropism, gag assembly and core formation. J. Mol. Biol. 2008;376:1493–1508. doi: 10.1016/j.jmb.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 31.Cardone G, Purdy JG, Cheng N, Craven RC, Steven AC. Visualization of a missing link in retrovirus capsid assembly. Nature. 2009;457:694–698. doi: 10.1038/nature07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alcaraz LA, del Alamo M, Barrera FN, Mateu MG, Neira JL. Flexibility in HIV-1 assembly subunits: solution structure of the monomeric C-terminal domain of the capsid protein. Biophys. J. 2007;93:1264–1276. doi: 10.1529/biophysj.106.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berthet-Colominas C, Monaco S, Novelli A, Sibai G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 35.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 36.Jin Z, Jin L, Peterson DL, Lawson CL. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J. Mol. Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- 37.Khorasanizadeh S, Campos-Olivas R, Summers MF. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 38.Kingston RL, Fitzon-Ostendorp T, Eisenmesser EZ, Schatz GW, Vogt VM, Post CB, Rossmann MG. Structure and self-association of the Rous sarcoma virus capsid protein. Structure. 2000;8:617–628. doi: 10.1016/s0969-2126(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 39.Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, Lee BM, Gorbalenya AE, Tong L, McClure J, Ehrlich LS, Summers MF, Carter C, Rossmann MG. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 40.Mortuza GB, Goldstone DC, Pashley C, Haire LF, Palmarini M, Taylor WR, Stoye JP, Taylor IA. Structure of the Capsid Amino-Terminal Domain from the Betaretrovirus, Jaagsiekte Sheep Retrovirus. J. Mol. Biol. 2008;386:1179–1192. doi: 10.1016/j.jmb.2008.10.066. [DOI] [PubMed] [Google Scholar]
- 41.Ternois F, Sticht J, Duquerroy S, Krausslich HG, Rey FA. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat. Struct. Mol. Biol. 2005;12:678–682. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 42.Wong HC, Shin R, Krishna NR. Solution Structure of a Double Mutant of the Carboxy-Terminal Dimerization Domain of the HIV-1 Capsid Protein. Biochemistry. 2008;47:2289–2297. doi: 10.1021/bi7022128. [DOI] [PubMed] [Google Scholar]
- 43.Worthylake DK, Wang H, Yoo S, Sundquist WI, Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 44.Bowzard JB, Wills JW, Craven RC. Second-site suppressors of Rous sarcoma virus CA mutations: Evidence for interdomain interactions. J. Virol. 2001;75:6850–6856. doi: 10.1128/JVI.75.15.6850-6856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, Hill CP. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J. Mol. Biol. 2007;373:355–366. doi: 10.1016/j.jmb.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sticht J, Humbert M, Findlow S, Bodem J, Muller B, Dietrich U, Werner J, Krausslich HG. A peptide inhibitor of HIV-1 assembly in vitro. Nat. Struct. Mol. Biol. 2005;12:671–677. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- 47.Tang C, Loeliger E, Kinde I, Kyere S, Mayo K, Barklis E, Sun Y, Huang M, Summers MF. Antiviral inhibition of the HIV-1 capsid protein. J. Mol. Biol. 2003;327:1013–1020. doi: 10.1016/s0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- 48.Gross I, Hohenberg H, Krausslich HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 49.Lanman J, Sexton J, Sakalian M, Prevelige PE. Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 2002;76:6900–6908. doi: 10.1128/JVI.76.14.6900-6908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frieden C. Protein aggregation processes: In search of the mechanism. Protein Sci. 2007;16:2334–2344. doi: 10.1110/ps.073164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovari LC, Momany CA, Miyagi F, Lee S, Campbell S, Vuong B, Vogt VM, Rossmann MG. Crystals of Rous sarcoma virus capsid protein show a helical arrangement of protein subunits. Virology. 1997;238:79–84. doi: 10.1006/viro.1997.8807. [DOI] [PubMed] [Google Scholar]
- 52.Baldwin RL. How Hofmeister ion interactions affect protein stability. Biophys. J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kingston RL, Olson NH, Vogt VM. The organization of mature Rous sarcoma virus as studied by cryoelectron microscopy. J. Struct. Biol. 2001;136:67–80. doi: 10.1006/jsbi.2001.4423. [DOI] [PubMed] [Google Scholar]
- 54.Oosawa F, Kasai M. A theory of linear and helical aggregations of macromolecules. J. Mol. Biol. 1962;4:10–21. doi: 10.1016/s0022-2836(62)80112-0. [DOI] [PubMed] [Google Scholar]
- 55.Prevelige PE, Jr, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campos-Olivas R, Newman JL, Summers MF. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 57.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 58.Wyatt PJ. Light scattering and the absolute characterization of macromolecules. Analytica Chimica Acta. 1993;272:1–40. [Google Scholar]
- 59.Utepbergenov DI, Fanning AS, Anderson JM. Dimerization of the scaffolding protein ZO-1 through the second PDZ domain. J. Biol. Chem. 2006;281:24671–24677. doi: 10.1074/jbc.M512820200. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi T, Adachi K. Hemoglobin equilibrium analysis by the multiangle laser light-scattering method. Biochem. Biophys. Res. Commun. 2002;290:1382–1387. doi: 10.1006/bbrc.2002.6362. [DOI] [PubMed] [Google Scholar]
- 61.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cairns TM, Craven RC. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. J. Virol. 2001;75:242–250. doi: 10.1128/JVI.75.1.242-250.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Craven RC, Leuredupree AE, Weldon RA, Wills JW. Genetic-Analysis of the Major Homology Region of the Rous-Sarcoma Virus Gag Protein. J. Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lokhandwala PM, Nguyen TL, Bowzard JB, Craven RC. Cooperative role of the MHR and the CA dimerization helix in the maturation of the functional retrovirus capsid. Virology. 2008;376:191–198. doi: 10.1016/j.virol.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kainov DE, Butcher SJ, Bamford DH, Tuma R. Conserved intermediates on the assembly pathway of double-stranded RNA bacteriophages. J. Mol. Biol. 2003;328:791–804. doi: 10.1016/s0022-2836(03)00322-x. [DOI] [PubMed] [Google Scholar]
- 67.Salunke DM, Caspar DL, Garcea RL. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys. J. 1989;56:887–900. doi: 10.1016/S0006-3495(89)82735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 69.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 70.del Alamo M, Neira JL, Mateu MG. Thermodynamic dissection of a low affinity protein-protein interface involved in human immunodeficiency virus assembly. J. Biol. Chem. 2003;278:27923–27929. doi: 10.1074/jbc.M304466200. [DOI] [PubMed] [Google Scholar]
- 71.del Alamo M, Mateu MG. Electrostatic repulsion, compensatory mutations, and long-range non-additive effects at the dimerization interface of the HIV capsid protein. J. Mol. Biol. 2005;345:893–906. doi: 10.1016/j.jmb.2004.10.086. [DOI] [PubMed] [Google Scholar]
- 72.Ehrlich LS, Agresta BE, Carter CA. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ehrlich LS, Liu T, Scarlata S, Chu B, Carter CA. HIV-1 Capsid Protein Forms Spherical (Immature-Like) and Tubular (Mature-Like) Particles in Vitro: Structure Switching by pH-induced Conformational Changes. Biophys. J. 2001;81:586–594. doi: 10.1016/S0006-3495(01)75725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rose S, Hensley P, O'Shannessy DJ, Culp J, Debouck C, Chaiken I. Characterization of HIV-1 p24 self-association using analytical affinity chromatography. Proteins. 1992;13:112–119. doi: 10.1002/prot.340130204. [DOI] [PubMed] [Google Scholar]
- 75.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoo S, Myszka DG, Yeh C, McMurray M, Hill CP, Sundquist WI. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 77.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucl. Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 79.Zwietering MH, Jongenburger I, Rombouts FM, van 't Riet K. Modeling of the Bacterial Growth Curve. Appl. Environ. Microbiol. 1990;56:1875–1881. doi: 10.1128/aem.56.6.1875-1881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]